Abstract
Site-specific nucleases (SSNs) have drawn much attention in plant biotechnology due to their ability to drive precision mutagenesis, gene targeting or allele replacement. However, when devoid of its nuclease activity, the underlying DNA-binding activity of SSNs can be used to bring other protein functional domains close to specific genomic sites, thus expanding further the range of applications of the technology. In particular, the addition of functional domains encoding epigenetic effectors and chromatin modifiers to the CRISPR/Cas ribonucleoprotein complex opens the possibility to introduce targeted epigenomic modifications in plants in an easily programmable manner. Here we examine some of the most important agronomic traits known to be controlled epigenetically and review the best studied epigenetic catalytic effectors in plants, such as DNA methylases/demethylases or histone acetylases/deacetylases and their associated marks. We also review the most efficient strategies developed to date to functionalize Cas proteins with both catalytic and non-catalytic epigenetic effectors, and the ability of these domains to influence the expression of endogenous genes in a regulatable manner. Based on these new technical developments, we discuss the possibilities offered by epigenetic editing tools in plant biotechnology and their implications in crop breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Significance Statement
As the role of epigenetics in the control of gene expression is being progressively unveiled, it is becoming apparent that epigenetic modifiers are underexploited breeding tools. This article reviews the epigenetic modifications in plants and their key molecular players, and evaluates various technologies for programmable epigenomic editing, including recent applications for specific and tunable regulation of gene expression. The review provides an overview of the latest developments in plant epigenomic editing and discusses the potential of these tools for their application in crop breeding.
Amit Dhingra, Department of Horticulture, Washington State University, Pullman, WA USA
Introduction
The phenotypic traits found in eukaryotic organisms are not only due to the polymorphisms present in their genomes. Traditionally, it was thought that only DNA sequence variants could be inherited in subsequent generations, and therefore the phenotypic variation was only due to such transmissible DNA polymorphisms. However, this paradigm was broken with the arrival of epigenetics. The modification of the chromatin structure plays a fundamental role in the functionality of the genome and the regulation of gene expression. These chromatin modifications are carried out by covalent epigenetic marks on DNA or associated histones, which are also responsible for the maintaining of the shape and structure of a nucleosome. It has also been shown in different studies that these epigenetic marks can be heritable (Wolffe and Matzke 1999; Henderson and Jacobsen 2007) or they can vary at different stages of development or through of the interaction of the organism with the environment. The study of epigenetics not only offers a vision of how modifying the structure of DNA can affect plant gene expression, but also opens the door to new gene editing technologies to generate and select those phenotypes of interest in an efficient and specific way. In this review, we will present how new editing technologies, in particular CRISPR systems, can be combined with epigenetic effectors and transcription regulatory domains to offer programmable systems for epigenetic editing and transcriptional regulation in plant species of agronomic interest.
The agronomic value of the study of the epigenetics in plants
The epigenetic regulatory mechanisms occur during development and also as an adaptation to the environment. The molecular basis of these adaptive systems can be especially attractive from an agronomic point of view, since finding the epigenetic mechanisms that influence important agronomic traits such as yield, biomass, flowering time, resistance to abiotic stresses or fruit ripening will allow the improvement of crops in an specific and efficient way.
Heterosis or “hybrid vigour” is the improved or increased function in a biological quality occurring in a hybrid offspring. Heterosis brings a lot of attention due to its obvious agronomic value. It has been classically explained by different mechanisms such as dominance, over-dominance, and epistasis (Shull 1908; Bruce 1910; Jones 1917). However, recent studies suggest the involvement of epigenetic components that result in differential expression of genes between parentals and their progeny (Groszmann et al. 2011; Barber et al. 2012; Chodavarapu et al. 2012; Zhang et al. 2016). Some authors suggest a direct relationship between the epigenetic and the phenotypic variation found in several plant species (Jin et al. 2008; Shen et al. 2012). An example of heterosis associated with dynamic epigenetic marks was described by Ni et al. (2009). They showed that the circadian clock genes CCA1 (CIRCADIAN CLOCK ASSOCIATED 1) and LHY (LATE ELONGATED HYPOCOTYL) present transcriptional repression due to histone modifications. The presence of these marks in Arabidopsis hybrids was associated with an increase in biomass. These studies were also applied to rice (He et al. 2010), where the parental contribution of epigenetic states in the progeny was analysed and shown to affect to differential gene expression in hybrids. These findings can give clues on how to improve crop yields by engineering epigenetic marks.
Flowering, one of the best studied developmental events in plant physiology, is also partially controlled by epigenetics. Overlapping environmental stimuli are regulating the timing of flowering such as temperature or photoperiod. An example is vernalization, which causes early flowering through exposure to low temperatures (Luo and He 2020). This process has allowed plant species to adapt their reproductive cycles to the most favourable annual season and allows also the control of flowering in certain species through heat treatment. The two of the main genes involved in the vernalization in Arabidopsis, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) (Whittaker and Dean 2017; Li et al. 2018c; Zhang and Jiménez-Gómez 2020), are strong flowering repressors. FLC encodes a MADS-Box domain protein that represses flowering and, likewise, FRI is required for FLC transcriptional activation. It has been shown that low temperature regulates the expression of the flowering repression genes through a complex epigenetic mechanism of histone and DNA methylation that silences the FLC locus (Sung et al. 2006; Wood et al. 2006; Shafiq et al. 2014). This mechanism is mediated by the action of Polycomb proteins (Amasino 2010; Kim and Sung 2013). The understanding of the vernalization pathway has allowed the transfer of this process to the field, obtaining early flowering in wheat through the treatment with the cytosine methyltransferase inhibitor 5-azacytidine (Brock and Davidson 1994; Horváth et al. 2003). Also, one of the first methyltransferase mutants studied in Arabidopsis showed early flowering, and this phenotype was inherited to the progeny (Finnegan et al. 1998). However, these approaches may not be suitable for field application since they lack specificity and generate pleiotropic problems due to the alteration of global methylation patterns (Ronemus et al. 1996). Furthermore, the general derepression of transposable elements might be armful for the stability of the plant genome (Underwood et al. 2017). Thus, a specific approach can be performed when only the histone epigenetic marks related with FLC and FT gene are selectively edited (Jeong et al. 2015; Han et al. 2019).
We are starting to understand how epigenetics mediates both immediate and long-term stress responses. Plants can “memorize” a stress situation and transmit the active defensive state to the progeny (Wibowo et al. 2016; Furci et al. 2019; Cong et al. 2019). For instance, it has been found that increased salt concentrations induce differential patterns of DNA methylation in rice (Wang et al. 2011; Karan et al. 2012), and that the expression of stress responsive genes involves phosphorylation of histone H3 and acetylation of histone H4 in Arabidopsis and tobacco (Sokol et al. 2007). Cold stress is known to induce DNA demethylation in different plant species (Shan et al. 2013; Song et al. 2015a), and also upregulates histone deacetylases leading to H3 and H4 histone deacetylation and the subsequent activation of responsive genes such as COR (cold-regulated) (Zhu et al. 2008; Hu et al. 2011).
Finally, fruit ripening has been also shown to be partially controlled by dynamic epigenetic mechanisms. In tomato, the DNA Methyltransferase Domains Rearranged Methyltransferase 2 (SlDML2) regulates genes involved in fruit ripening (Lang et al. 2017). Spontaneous mutations that induce hypermethylation in the promoter region of the Colorless Non-Ripening (CNR) gene result in a non-ripening phenotype (Manning et al. 2006). In apple, DNA methylation at the MdMYB10 gene modulates its expression, influencing the pigmentation of the fruit (El-Sharkawy et al. 2015). Moreover, it has been found that the global DNA methylation levels in citrus increase during ripening (Huang et al. 2019). Understanding the mechanisms that regulate fruit ripening in different plant species of agricultural interest has made possible to obtain late-maturing plant varieties (Liu et al. 2016a) or with greater fruit quality (Liu et al. 2015; Osorio et al. 2020).
The influence of epigenetic marks on gene expression
The status of chromatin influences the accessibility of the transcriptional machinery and therefore, gene expression. The chromatin status is strongly regulated by epigenetic marks that respond to environmental stimuli or specific physiological conditions. Epigenetic marks include DNA methylation and histone modification and numerous studies have identified their link with transcriptional activation or repression of genes (Fig. 1).
Schematic diagram of epigenetic regulation and the epigenetic marks associated to gene expression and gene repression. The epigenetic effectors and marks that modulate the “relaxed” or “condensed” state of chromatin are represented. The condensed representation of the chromatin include DNA methylation, generated by the methyltransferase MET, histone dimethylation and monomethylation H3K9, carried out by histone methyltransferases (HMT) and histone deacetylation carried out by histone deacetylases (HDACs). The transition of condensed state to more relaxed is carried out by JMJ histone demethylases, that removes the trimethylation of H3K27 and ROS1, a DNA demethylase. The relaxed stated of the chromatin, include histone acetylation carried out by histone acetyltranferase (HAT) and histone trimethylation of H3K4. A representation of gene transcription is included in the relaxed section of the chromatin. Created with BioRender.com
DNA methylation
DNA methylation refers, classically, to the addition of a methyl group to the 5th carbon of the cytosines to form 5-methylcytosine (5mC), although it has been recently found in mammals that there are some additional DNA modifications based on methylation such as 3-methylcytosine (Sadakierska-Chudy et al. 2015). In plants, 5mC DNA methylation is found mainly in symmetrical CG and CHG sequences, but also to some extent in non-symmetrical CHH sequence contexts (Kumar et al. 2018), where H is A, C or T. DNA methylation plays a prominent role in gene silencing and it is mainly found in heterochromatic regions, such as pericentromeric and telomeric regions (Zhang et al. 2018b). Moreover, DNA methylation found around gene regulatory regions, usually comprising promoters and terminators, is linked to gene repression. On the other hand, demethylation of promoter regions is related to active gene expression (Zilberman et al. 2007). Although this is the general rule, it has been recently found that DNA methylation can promote gene expression through the SU(VAR)3‐9 DNA methyl-readers SUVH1 and SUVH3 (Harris et al. 2018). DNA methylation status is controlled by DNA methylases and demethylases, which can also interact with other modulators in response to different stimuli to control gene expression.
Depending on the cytosine context (CG, CHG or CHH) methylation is mediated by a different type of methyltransferases. CG methylation, the most abundant in plant genomes, is carried out mostly by Methyltransferase1 (MET1), an orthologue of the mammalian DNA (cytosine-5)-methyltransferase 1 (DNMT1) (Cokus et al. 2008). MET1 maintains the methylation status acting over hemi-methylated CG dinucleotides during DNA replication (Aufsatz et al. 2004). CHG methylation is maintained by CHROMOMETHYLASE 3 (CMT3) (Bartels et al. 2018), with a marked preference for CWG (W = A or T) cytosines (Gouil and Baulcombe 2016). Finally, the CHH methylation is maintained by DNA-methyltransferase 2 (DRM2) via RNA-directed DNA methylation pathway (RdDM) (Matzke and Mosher 2014) and CHROMOMETHYLASE 2 (CMT2), which is mostly active in pericentromeric heterochromatin and long transposable elements (Bewick et al. 2017; Harris and Zemach 2020).
Methylation is a reversible process and can be reversed actively by specific demethylases. In plants, the active demethylation is mediated mostly by DNA glycosylases through a base-excision-repair pathway, although another mechanism such as passive demethylation or blocking de novo methylation pathway has been reported in early stages of development and imprinting (Penterman et al. 2007). In Arabidopsis, a group of glycosylases have been found to be involved in active demethylation, namely DEMETER (DME) and REPRESSOR OF SILENCING 1 (ROS1) (Choi et al. 2002; Morales-Ruiz et al. 2006). Also, DEMETER-LIKE 2 (DML2) and DEMETER-LIKE 3 (DML3) proteins show an active role in demethylation processes (Ortega-Galisteo et al. 2008).
Histone modifications
Covalent modifications in histones can alter chromatin structure and change the accessibility of the transcriptional machinery, causing changes in gene expression. The structure of the histones allow a greater variety of modifications than DNA, and therefore creates a “histone code” that is usually linked to active or repressive states of chromatin. Modifications are usually located at the N-terminal tails of histones and, in some cases, at nucleosome core regions (Zhao et al. 2019b). The most studied histone modifications involve methylation, acetylation, ubiquitination, and phosphorylation. For the purpose of this review we will focus in the description of methylation and acetylation, which are the epigenetic marks whose edition that have been attempted so far.
The process of histone methylation is one of the most frequently observed in epigenetic regulation and the proteins involved in methylation and demethylation have been identified also in plants. Histone methylation takes place at lysine and arginine residues. Lysines can present mono-, di- or trimethylation, usually located in lysines 4, 9, 27 and 36 of histone H3 (Zhang and Reinberg 2001; Cheng et al. 2020). Arginine residues present only mono and di-methylation at N-terminal tails of arginine 2, 8, 7, 17 and 26 of Histone H3 and arginine 3 of Histone 4 (Bedford and Richard 2005). It has been shown that lysine and arginine methylation are involved in transcriptional regulation, RNA processing, nuclear transport, DNA-damage repair and signal transduction (Liu et al. 2010). Also, the number of methyl groups have different biological significances. For example, trimethylations of H3K4 and H3K36 are present in transcriptionally active genes, whereas trimethylation of H3K27 and mono- and dimethylation in H3K9 are associated with transcriptional repression (Tariq et al. 2003; Wang et al. 2016).
In Arabidopsis, a wide variety of proteins are involved in controlling the histone methylation status. Histone methyltransferases (HMTs) involved in lysine methylation contain the conserved so-called SET domains that catalyse the transfer of one or multiple methyl groups to the ε-nitrogen of specific lysine residues (Xiao et al. 2016; Zhou et al. 2020). The H3K4 methylation is mediated by various proteins of the ARABIDOPSIS TRITHORAX (ATX) family (Alvarez-Venegas et al. 2003; Saleh et al. 2008; Chen et al. 2017), with homologous found in rice and maize (Springer et al. 2003; Choi et al. 2014). H3K9 is methylated by the methyltransferase SUVH family such as KRYPTONITE (KYP), also known as SUVH4, SUVH5, SUVH6 or SUVR4 (Ebbs et al. 2005; Li et al. 2018b). The proteins of Polycomb group that form Polycomb Repressive Complex 2 (PRC2) are responsible for the trimethylation of H3K27. Interesting, it has been found that ARABIDOPSIS TRITHORAX RELATED 5 and 6, (ATXR5 and ATXR6) can also target H3K27 by monomethylation, producing the opposite biological effect despite targeting the same histone residue (Jacob et al. 2009; Wiles and Selker 2017; Zhou et al. 2020). Finally, the epigenetic mark H3K36 is mostly regulated by EARLY FLOWERING IN SHORT DAYS (EFS)/SET DOMAIN GROUP 8 (SDG8) methyltransferases, but other members of the TRITHORAX family, such as SDG26 can catalyse the reaction at specific loci (Xu et al. 2008; Berr et al. 2015; Liu et al. 2019).
Histone demethylation is a dynamic reaction that shows the reversibility of the histone methylation process. Histone demethylases can remove the methyl group from tri- to di- methylations, from di- to mono methylations and mono to non-methylated lysine (Xiao et al. 2016). They are classified based on the number of methylations and the target lysine. The firstly identified demethylase was the human Lysine Specific Demethylase 1 (LSD1) (Shi et al. 2004). This cluster of demethylases performs a FAD-dependent oxidation reaction to remove the methyl group from lysine residues. In Arabidopsis, four LSD1 homologs have been identified; LSD1-like 1 (LDL1), LSD1-like 2 (LDL2), LSD1-like 3 (LDL3) and FLOWERING LOCUS D (FLD) (Jiang et al. 2007). As with the human LSD1, the Arabidopsis homologs are only able to remove groups from mono- and di-methylated H3K4.
The second group of demethylases belong to the JmjC domain-containing protein family (JMJ), which are subdivided, in Arabidopsis, in five subfamilies. These demethylases can recognize and remove the methyl group from mono-, di- and trimethylated lysine histones residues through the action of Fe (II) and α-ketoglutarate (αKG) cofactors (Duan et al. 2018). For example, JMJ14, a flowering repressor, and JMJ15 and JMJ18, flowering activators, were a identified as H3K4demethylases in Arabidopsis (Lu et al. 2010; Yang et al. 2012a, b), and JMJ703 in rice (Cui et al. 2013). The JMJ proteins involved in H3K9 demethylation comprises JMJ25 (or IBM1) and JMJ27 in Arabidopsis (Saze and Kakutani 2007; Dutta et al. 2017). and JMJ706 in rice (Sun and Zhou 2008). The Histone H3K27 can be demethylated by several proteins such as JMJ11 (ELF6), JMJ12 (REF6) and JMJ13, which are involved in flowering (Lu et al. 2010; Gan et al. 2015; Zheng et al. 2019) in Arabidopsis and JMJ705 in rice (Li et al. 2013). The demethylation over histone H3K36 is not so well elucidated, but some studies relate the JMJ12 (REF6) protein with reduced H3K36me2 in FLC locus (Gan et al. 2015) and, more recently, the protein JMJ30 as a possible demethylation effector over H3K36 (Cheng et al. 2020).
Histone methylation takes also place in arginine residues. The arginine methyltransferases (PRMTs) are a group of evolutionarily conserved methyltransferases that can replace the 5-hydrogen of the guanidine nitrogen atoms present in the arginine by methyl moieties to form different methylated arginine isomers. Depending on the isomer and symmetrical or asymmetrical methylation that is formed, the PRMTs are divided into four subgroups (Type I, II, III and IV) (Ahmad and Cao 2012; Hartley and Lu 2020). In Arabidopsis, the H3R2 and H3R17 can be asymmetrically di-methylated by AtPRMT4a/b and H4R3 can be symmetrically di-methylated by AtPRMT5, AtPRMT10 and AtPRMT1a/b. Both epigenetic marks are involved in flowering time, but performing different roles (Liu et al. 2010). Unfortunately, the mechanisms of the arginine demethylation are less understood, and this activity is linked to certain JmjC domain-containing proteins (Cho et al. 2012; Li et al. 2018a).
Histone acetylation takes place at lysine residues through the addition of an acetyl group, which is provided by acetyl-CoA. The acetylation neutralizes the positive charge of the amino residues and increases the size of the lysine tail, which causes relaxation of chromatin and creates a landing pad for the transcriptional machinery. In addition to increase the accessibility, the acetylation located in the histones also can recruit reader proteins, as chromatin remodelers (Marmorstein and Zhou 2014). For this reason, the epigenetic mark of acetylation is linked to transcriptionally active genes (Liu et al. 2016b). In plants, the lysine residues that are frequently acetylated are K9, K14, K18, K23 and K27 of histone H3 and residues K5, K8, K12, K16 and K20 of histone H4. Histones can present up to four acetylations. Acetylations are often linked to specific responses in plants; for example H3K9/14ac or H3K27ac correlate with activation of developmental and stress-induced genes (Weiste and Dröge-Laser 2014; Kim et al. 2017b). Similarly, histones can also undergo active deacetylation that represses transcription (Chen and Tian 2007), this linked to a great variety of processes such as photomorphogenesis (Benhamed et al. 2006; Jang et al. 2011) or flowering (Yu et al. 2011).
Histones acetyltransferases (HATs) identified in plants can be divided into four families, HAC, HAF, HAG and HAM. The HAC group contains HATs of the p300/CREB binding protein family. The HAF group is formed by HATs from the TATA-binding protein-associated factor (TAFII250) family. The HAG group contains HATs of the general control non-repressible 5-related N-terminal acetyltransferase (GNAT) family. Finally, the HAM group is formed by the MYST family (Liu et al. 2016b; Jiang et al. 2020). The acetyl group can be removed by other proteins named histone deacetylases (HDACs), which are linked with transcriptional repression. HDACs are well conserved proteins and they can be divided into four subclasses. In Arabidopsis, the class I contains 6 protein members, HDA6, HDA7, HDA9, HDA10, HDA17, and HDA19. Class II contains five protein members, HDA5, HDA8, HDA14, HDA15, and HDA18. The class III, which comprises sirtuin-like members, present fewer homologs in Arabidopsis than in humans, with only two members SRT1 and SRT2. Finally, the HDA2 protein is the unique component of class IV of HDACs in plants (Alinsug et al. 2009; Yu et al. 2017; Zhang et al. 2018a; Zhao et al. 2019a; Chen et al. 2020). Finaly, a group of four plant-specific HDACs, HD2A, HD2B, HD2C, and HD2D (Bourque et al. 2016) are involved in biotic and abiotic stress resistance (Luo et al. 2012; Ding et al. 2012; Han et al. 2016; Park et al. 2018).
Non-catalytic domains that regulate the gene expression
Whereas catalytic domains in epigenetic effector proteins directly modify the chromatin structure to modulate gene expression, other non-catalytic regulatory domains are capable of recruiting epigenetic effectors, ultimately regulating the transcriptional activity of the target gene. Other regulators do not recruit effectors but interact with the transcriptional machinery in other forms, also influencing transcription.
In general, non-catalytic regulatory domains have been isolated from transcription factors (TFs), which usually comprise, next to DNA-recognition and binding domains, an effector domain that recruit chromatin modifiers and transcriptional machinery (Ma 2011; Yamasaki et al. 2013; Song et al. 2015b). One of the first described transcriptional regulation domains came from the yeast GAL4 TF, whose trans-activation domain (TAD) was identified and isolated. Likewise, the DNA-binding domain of this TF was also isolated and synthetically fused to other potential TADs, which allowed not only the identification of more activation and repression domains, but also demonstrated the orthogonality of the TADs and, therefore, its potential to operate in various species and various genomic contexts (Keegan et al. 1986; Hope and Struhl 1986; Hebbes et al. 1988). The modular nature on many transcriptional regulation domains led to the identification of powerful viral TADs, such as the VP16 domain of the herpes simplex virus (Campbell et al. 1984; Carey et al. 1990), which proved to be a powerfull activator also in plants (Moore et al. 1998; Schwechheimer et al. 1998). In addition, this domain offered the possibility of increasing its transcriptional activation potential through the fusion of several repetitions in tandem, originating the synthetic activation domains VP64, VP128, etc. These synthetic domains offered a greater activation range in target genes (Ordiz et al. 2002; Li et al. 2017) and also the ability to maintain this activation in the following generations (Utley et al. 1998; Guan et al. 2002).
On the other hand, endogenous regulatory domains have also been identified in plants that can offer either activation or repression. Two examples of plant TADs are the ERF2 (m) and the EDLL domains, both from the Ethylene Response Factor family (ERF) (Tiwari et al. 2012; Li et al. 2013). However, the ERF family has also proteins with identified transcriptional repression domains (TRDs, EAR motifs), with which it has been possible to obtain efficient transcriptional repression. A remarkable example is ERF3, (Ohta et al. 2001; Tiwari et al. 2004) from which widely-used TRDs SRDX and BRD are derived (Utley et al. 1998; Hiratsu et al. 2003).
Programmable epigenetic editors for regulation the gene expression
Epigenetic tools has emerged as important assets in crop breeding. The first attempts to take advantage of epigenetic control of gene expression involved the mutation or deletion of specific epigenetic effectors, thus generating new epigenetic mutants with potential agronomic interest. For example, ddm1 and met1 hypomethylated mutants in Arabidopsis cause transposon mobilization. These mutant backgrounds show multiple phenotypes in plant structure such as the short and compact inflorescence found in bns (BONSAI) plants (Saze and Kakutani 2007). However, the great variety of genes and transposons targeted by these effectors makes this approach very unspecific and unpredictable, generating in most cases aberrant phenotypes and developmental abnormalities (Kakutani et al. 1996; Tsukahara et al. 2009).
Target-specific epigenetic interventions require programming the DNA binding specifity of the effectors. The traditional approach for this involves the ectopic expression of heterologous TFs under the control of inducible promoters, therefore connecting promoter-specified inputs to a cascade of TF-targeted activated/repressed genes as output (Li et al. 2013; Petolino and Davies 2013). A limitation of this approach is that it does not allow free selection of the output response, as the collection of target genes is restricted by the DNA binding specificities of the TFs employed. An alternative is the engineering of specific binding stites in the promoters of the target genes (Kumar et al. 2015; Mohan et al. 2017; Dong et al. 2019; Bai et al. 2020). However, the complexity of the design and the effort required to generate a specific synthetic promoters driving each downstream gene is a factor to consider.
It was not until the discovery of artificial zinc fingers (ZFs) and transcription activator-like effectors (TALEs) that the possibility of creating custom-programmable epigenetic factors arose, opening the field to bioengineering and synthetic biology approaches. The artificial ZFs are custom specific DNA binding domains that typically recognize 3–6 nucleotide triplets. The initial engineering approach based on producing double-strand DNA breaks (DBS) with the fusion of Folk1 nucleases (Durai et al. 2005), rapidly evolve to artificial TFs containing transcriptional regulator domains or epigenetic effectors (Shrestha et al. 2018). Likewise, the engineered TALEs share many similarities in operation and structure with ZFs but offer a higher degree of specificity. They are proteins derived from Xanthomonas bacteria to aid the infection of plant species and promote the expression of host genes. TALEs consist of a specific and customizable DNA binding domain comprising tandem repeats arrays of amino acids, which can recognize a specific DNA target sequence (Boch et al. 2009; Moore et al. 2014). However, the nature of TALEs requires of a new design of the protein for each target sequence, what makes them efficient but labor-intesive tools.
In plants, several works describe ZF and TALEs being employed as artificial TFs, thus enabling programable gene regulation. The first ZFs examples targeted the APETAL3 gene in Arabidopsis. The VP64 TAD and the mSin3 interaction domain (SID) TRD were fused to an APETAL3 ZF, yielding the expected transcription changes and generating alternated floral patterns (Guan et al. 2002). In parallel, engineered TALEs were also proved as efficient programable regulators in plants. Interesting examples are the regulation of EGL3 and KNAT endogenous genes in Arabidopsis (Morbitzer et al. 2010), or the regulation of the AtPAP1 transgene in tobacco (Liu et al. 2014). There are few examples in plants of ZF or TALE programmable regulators using catalytic epigenetic effectors. A remarkable exception is the specific and programmable demethylation of the FWA gene in Arabidopsis obtained with ZF technology using the catalytic domain of human TEN-ELEVEN TRANSLOCATION1 (TET1) (Gallego-Bartolomé et al. 2018), a dioxygenase involved in the demethylation of DNA (Chen et al. 2014). In a related example, a ZF fused to DNAJ1 methylation reader, which form complexes with SUVH1 and SUVH3 proteins, increased the expression of adjacent genes through the recognition of DNA methylations (Harris et al. 2018).
The potential of CRISPR in epigenomic editing in plants
In the last decade, the CRISPR/Cas systems emerged as new versatile programmable effectors. They offer a wide range of applications with high efficiency and specificity, avoiding the main problem that limited previous tools, since CRISPR/Cas eliminates the need to make a new protein for each target (Waryah et al. 2018; Arya et al. 2020). The simplicity and versatility of this system based on a small RNA guide (sgRNA) and an effector Cas protein was proved initially in human cells (Cong et al. 2013; Mali et al. 2013; Cho et al. 2013) and later extended to all types of organisms (Sanders et al. 2018; Kim et al. 2019; Shi et al. 2019; Schuster and Kahmann 2019) including plant crops, oferring new possibilities to plant breeding (Veillet et al. 2019; Gao 2019; Shao et al. 2020; Zaynab et al. 2020). Moreover, CRISPR systems can work as programmable epigenetic editors and efficient transcriptional regulators. Following a similar strategy as that described for ZFs and TALEs, endonuclease-inactivated versions of Cas9 protein, known as “dead Cas9” or dCas9, obtained by mutagenesis of specific aminoacids in RuvC1 and HNH nuclease domains (Qi et al. 2013), can be transformed in easily programmable activation (CRISPRa) or repression factors (CRISPRi), depending on the effector domain that is attached to the Cas9 protein. This approach has been tested extensively, using both catalytic and non-catalytic domains, for the regulation gene expression with remarkable results in mammalian cells and other organisms, such bacteria and fungi (Shakirova et al. 2020; Zhang et al. 2020), opening new perspectives to its application in plant systems.
Transcriptional regulation strategies for dCas9
The initial strategies to generate programmable transcriptional effectors in plants based on CRISPR-dCas9 made use of direct fusions to the C-terminal of the Cas9 protein (Fig. 2a). TAL, VP64 and EDLL were the first non-catalytic domains attached to dCas9 for transcriptional activation in Arabidopsis and N. benthamiana. Following the same strategy, plant-derived BRD and SDRX were employed for transcriptional repression (Piatek et al. 2015; Vazquez-Vilar et al. 2016). In all cases, the activation/repression efficiency obtained was only moderate. However, a remarkable improvement in gene regulation was achieved when several domains were fused in tandem (Fig. 2b). The activation TV (6xTAL–VP128) domain, and the activation VPR (VP64-P65-RTA) domain (Chavez et al. 2015; Li et al. 2017, 2019), were the most successful examples of CRISPRa tandem fusions used in Arabidopsis and rice, achieving up to 190 fold activation in AtCDG1 gene when employing dCas9-TV (Li et al. 2017). At the same time, in the CRISPRi approach the tandem fusion of the SRDX repressor domain was employed, obtaining repressions of up to 80% in Arabidopsis miRNA target genes (Lowder et al. 2015). Regarding catalytic epigenetic domains, so far most examples have been tested only in mammalian cells, such as the histone demethylase LSD1, the human DNA demethylase TET1, the methyltransferase DNMT3A or the plant DNA demethylase ROS1 (Kearns et al. 2015; Vojta et al. 2016; Liu et al. 2016c; Devesa-Guerra et al. 2020). These examples prove that a direct fusion of these elements to dCas9 can change the epigenetic status in the target sequence. This makes its application to plants very promising, considering the orthogonality of the catalytic domains (Ji et al. 2018). A pionerering example in plants is the successful epigenetic regulation of the AREB1/ABF2 gene in Arabidopsis (Roca Paixão et al. 2019). The AREB1/ABF2 gene is involved in the ABA signaling pathway and its loss of function presents a phenotype of sensitivity to drought stress. In this work, aimed to specifically increase the expression of AREB1 and to obtain drought resistant plants, a programmable epigenetic editor was developed based on the direct fusion to dCas9 of the P300 catalytic domain derived from the Arabidopsis histone acetyltransferase 1 (AtHAC1). As distinctive feature in this approach, P300 was fused to the N-terminal end of Cas9 instead of the C-terminal end as it is usually described. Although the transcriptional activation of the gene was modest, reaching only twofold, a substantial increase in the survival rate was obtained when plants were exposed to drought stress.
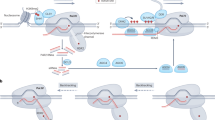
Representation of Cas9 strategies for gene regulation. a Cas9 strategy based on the direct fusion of the effector domain in C- terminal. b Cas9 strategy based on the direct fusion in tandem of the effector domains in C- terminal. c SAM aptamer-based Cas9 strategy. A direct fusion of effector domains could be added in C-terminal. d scRNA aptamer-based Cas9 strategy. A direct fusion of effector domains could be added in C-terminal. e SunTag strategy with ten tandem repeats of GCN4 peptide in C-terminal that are recognized by ScFv fused to a effector domain. Created with BioRender.com
In a further elaboration of the technique, two new strategies were designed to recruit an even greater number of regulatory domains or combinations of them in a single dCas9 molecule, thus generating a new battery of CRISPR/dCas9-based regulation tools. The first type of strategies, represented by the so-called SAM (Synergistic Activation Mediator) and scRNA (scaffolding RNAs) approaches, are based on the permissiveness of the gRNA scaffold to accept RNA aptamers in its structure, which bind with high specificity other proteins such as the coat protein of the MS2 virus (Zhang et al. 2015; Konermann et al. 2015; Zalatan et al. 2015). This provides new anchoring sites in the dCas9 complex, increasing its ability to recruit regulatory domains. Also, SAM (Fig. 2c) and scRNA (Fig. 2d) are compatible with the direct fusion strategy discussed above, so that it allows different combinations of regulatory domains to be recruited simultaneously. The use of both SAM and scRNA approaches improved dramatically the activation rates in mammalian cells and in plants (Lowder et al. 2015, 2018; Park et al. 2017; Lee et al. 2019), and opened the way to further combinatorial optimizations of the system, as shown with the combination of EDLL and VPR domains in the recently described dCasEV2.1 activation system (Selma et al. 2019). In addition, the presence of an RNA aptamer also allows the incorporation of epigenetic catalytic effectors. In Arabidopsis, a scRNA strategy involving the histone acetyltransferase catalytic domain P300 produced a H3K27ac enrichment in the FT target gene. In the same work, a similar strategy using the histone methyltransferase KRYPTONITE was shown to produce a H3K9me2 enrichment in the same gene. However, although the epigenetic marks were evident, the effects on flowering produced by these marks were mild (Lee et al. 2019).
A second type of strategies uses multi-epitope tags to attach several regulatory domains to a single dCas9 molecule. The so-called SunTag strategy (Tanenbaum et al. 2014) fuses dCas9 to tandem GCN4 peptide repeats, which are recognized by a single chain antibody (ScFv) fused to a regulatory domain (Fig. 2e). Although the first versions of the Suntag strategy offered good results in mammalian cells for gene activation (Gilbert et al. 2014), the same effects were not achieved in plants, offering lower activation rates than the previously mentioned strategies (Selma et al. 2019). However, a new version of this strategy appeared shortly after. In this optimization, the size of the linker sequence separating tandem GCN4 epitopes changed from seven to twenty-two amino acids (Suntag22a), avoiding possible steric impediments (Morita et al. 2016). The new Suntag22a version offered very high activation when used with the non-catalityc VP64 TAD (Papikian et al. 2019). Furthermore, in another remarkable achievement, this platform was successfully used in the catalytic modification of epigenetic marks, first in mammal cells (Morita et al. 2016; Huang et al. 2017) and later in plants, by attaching the catalytic domains of TET1, involved in demethylation, and the NtDRM2 methylase (Gallego-Bartolomé et al. 2018; Papikian et al. 2019) respectively. In both studies, the FWA gene, involved in flowering time, was chosen as the target for epigenetic regulation. The study carried out by Gallego-Bartolomé et al. (2018) employing the dCas9-Suntag-TET1 strategy, achieved specific demethylation of the FWA promoter, and therefore, late flowering phenotypes in Arabidopsis. Likewise, the strategy carried out by Papikian et al. (2019), was based in the same molecular mechanism, but in this case dCas9-Suntag-DRM2 produced target demethylation on the FWA promoter, generating early flowering phenotypes. The epigenetic mutations obtained with these systems were transfered to the next generation of plants, which showed the same flowering phenotype as their parents. The programmable transcriptional and epigenetic effectors based on CRISPR described in plants are summarize in Table 1.
Transcriptional regulation using dCas12a and dCas12b
After the success of Cas9, new nucleases belonging to other types of the CRISPR/Cas family have emerged. In plants, Cas12a has been presented as a good alternative to Cas9 as a genetic editor (Kim et al. 2017a; Bernabé-Orts et al. 2019), showing a remarkable efficiency for targeted mutations. Following the same strategy used with dCas9, dCas12a versions have been also generated (Tang et al. 2017) (Fig. 3a) and used as programmable epigenetic editors in plants (Table 1). However, the sgRNA architecture of dCas12a does not allow the same modifications as dCas9 sgRNA and, for this reason, the aptamer-based strategies for gene regulation cannot be applied. On the other hand, transcriptional activation and repression has been achieved using a direct fusion strategy (Tang et al. 2017; Tak et al. 2017), and although catalytic epigenetic effectors are not reported in plants using the Cas12a SunTag strategy, there are very likely to work, as it was earlier shown in mammalian cells (Kim et al. 2017a; Zhang et al. 2018c).
More recently, a new Cas12b protein from Alicyclobacillus acidiphilus has emerged as a novel programable effector in plants (Fig. 3b). This new Cas version can generate targeted mutations with moderate efficiency, and transcriptional regulators have been engineered using its “dead” version fused to the TV domain (Table 1). The dCas12b tool shows more flexibility than the dCas12a in its sgRNA structure, allowing the addition of RNA aptamers for MS2 protein recognition, and showing acceptables rates of gene activation in rice (Ming et al. 2020).
Perspectives of epigenomic editing applied to crop breeding
As reviewed above, the rapidly evolving CRISPR technologies offer new crop breeding tools in the form of programmable epigenetic editors. As mentioned, examples of successful epigenetic edition are already in place in model organisms, as for the regulation of flowering time in Arabidopsis through the manipulation of DNA methylation of the FWA gene (Gallego-Bartolomé et al. 2018; Papikian et al. 2019). As can be easily inferred, fine regulation of flowering time could be applied to crops to obtain early or late-flowering varieties, or even varieties that do not flower as a strategy to increase biomass. Importantly, it has been demonstrated that, at least for targeted DNA methylation at the FWA locus, modifications are inherited in subsequent generations (Papikian et al. 2019), this being an important prerequisite in crop breeding where trait stability needs to be ensured. Other promising breeding trait for epigenetic editing, especially relevant in the context of climate change, is drought stress resistance. Very recently, Arabidopsis lines have been generated with improved tolerance to drought by activating the AREB1/ABF2 genes using a dCas9 fused to a histone acetyltransferase (Roca Paixão et al. 2019). Being able to apply this type of technology to edible crops in areas with poor irrigation systems or dry climates could help to alleviate the effects of climate change in crop production.
Although most examples of directed epimutations described to date were developed in model species, it is not difficult to envision epigenetic versions of well-established alleles conferring favourable traits in crop species. Extensive research was carried out on the activation and repression of rice genes though CRISPR-based effectors (Li et al. 2017; Lowder et al. 2018; Ming et al. 2020). Potential candidates for epigenetic down-regulation in rice affecting grain size are GW2, GW5 or TGW6 genes. Likewise, the nutritional properties of the grain could be increased with the accumulation of essential microelements or health-related metabolites through the epigenetic activation of e.g., genes of the nicotianamine synthase (NAS) family, or the carotenoid synthesis pathway in the endosperm (Zheng et al. 2010; Xu et al. 2008; Fiaz et al. 2019). As new epigenetic mechanisms regulating traits of interest are being discovered, the number of potential targets of the new programmable editors also grows. For example, possible interesting targets for breeding are those mentioned previously in this review, such as the ripening genes RIN and and CNR in tomato (Liu et al. 2016a; Lang et al. 2017; Li et al. 2020), the clock genes CCA1 and LHY (Ni et al. 2009), or the FLC and COR genes involved in cold resistance (Yang et al. 2014; Park et al. 2018) (Fig. 4).
Representation of target traits for crop improvement using programmable epigenetic editors. Epigenetic regulation of genes involved in phenotypic traits of interest as possible targets of programmable epigenetic editors. The RIN and CNR genes are selected as targets of fruit ripening. Early ripening included the demethylation of H3K27me3 associated to RIN gen and DNA demethylation of RIN and CNR promoters. Late ripening is represented by the enrichment of H3K27me3 marks associated to RIN and DNA methylation in RIN and CNR promoters (Liu et al. 2016a; Lang et al. 2017; Li et al. 2020). The clock genes CCA1 and LHY are negative regulators of growth and biomass improvement (Ni et al. 2009) and can be repressed by the reduction of the associated active epigenetic marks H3K9Ac and H3K4Me2. Drought resistance is linked to the expression of AREB1, which expression can be increased thought the enrichment of histone acetylation (Roca Paixão et al. 2019). FWA gene regulates the flowering time. FWA activation though DNA demethylation promotes late flowering and FWA repression though DNA methylation promotes early flowering (Yang et al. 2014; Park et al. 2018). Created with BioRender.com
Epigenetic editions could bring new sources of genetic variation to crop breeding, enabling trait associated features that are not readily accessible to classical and/or directed mutagenesis as e.g. tissue/organ specificity or environmental plasticity. Interestingly, next to functional and agronomical considerations, legal and regulatory aspects need to be considered when discussing the future of epigenetic breeding. It is uncertain what will be the legal status of epigenetically modified crops, which, according to its definition, cannot be considered strictly speaking as genetically modified. Taking as reference the EU directive 2001/18/EC on the deliberate release of Genetically Modified Organism, where GMOs are defined as those organisms whose “genetic material has been altered”, it is reasonable to argue that histones can not be considered as genetic material and, consequently, modifications in histone-based epigenetic marks should not fall within the scope of this directive. Similar uncertain regulatory scenarios are likely to occur regarding DNA epigenetic marks. Although transgenics is currently the most common method to deliver CRISPR/Cas and gRNAs to the plant cells, new delivery methods based on nanoparticles, biolistics and viral vectors are being developed that could circumvent the need for transgenic intermediates (Uranga et al. 2020; Ariga et al. 2020). In this scenario, epimutations obtained through non-transgenic editing tools could circumvent current GMO regulations and this would undoubtedly be a strong incentive for the development of new tools and techniques.
References
Ahmad A, Cao X (2012) Plant PRMTs broaden the scope of arginine methylation. J Genet Genomics 39:195–208. https://doi.org/10.1016/j.jgg.2012.04.001
Alinsug MV, Yu C-W, Wu K (2009) Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol 9:37. https://doi.org/10.1186/1471-2229-9-37
Alvarez-Venegas R, Pien S, Sadder M et al (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13:627–637. https://doi.org/10.1016/s0960-9822(03)00243-4
Amasino R (2010) Seasonal and developmental timing of flowering. Plant J 61:1001–1013. https://doi.org/10.1111/j.1365-313X.2010.04148.x
Ariga H, Toki S, Ishibashi K (2020) Potato virus X vector-mediated DNA-free genome editing in plants. Plant Cell Physiol 61:1946–1953. https://doi.org/10.1093/pcp/pcaa123
Arya SS, Mahto BK, Ramkumar TR, Lenka SK (2020) Sharpening gene editing toolbox in Arabidopsis for plants. J Plant Biochem Biotechnol 29:769–784. https://doi.org/10.1007/s13562-020-00606-4
Aufsatz W, Mette M, Matzke A, Matzke M (2004) The role of MET1 in RNA-directed de novoand maintenance methylation of CG dinucleotides. Plant Mol Biol 54:793–804. https://doi.org/10.1007/s11103-004-0179-1
Bai J, Wang X, Wu H et al (2020) Comprehensive construction strategy of bidirectional green tissue-specific synthetic promoters. Plant Biotechnol J 18:668–678. https://doi.org/10.1111/pbi.13231
Barber WT, Zhang W, Win H et al (2012) Repeat associated small RNAs vary among parents and following hybridization in maize. Proc Natl Acad Sci USA 109:10444–10449. https://doi.org/10.1073/pnas.1202073109
Bartels A, Han Q, Nair P et al (2018) Dynamic DNA methylation in plant growth and development. Int J Mol Sci 19:2144. https://doi.org/10.3390/ijms19072144
Bedford MT, Richard S (2005) Arginine methylation: an emerging regulatorof protein function. Mol Cell 18:263–272. https://doi.org/10.1016/j.molcel.2005.04.003
Benhamed M, Bertrand C, Servet C, Zhou D-X (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18:2893–2903. https://doi.org/10.1105/tpc.106.043489
Bernabé-Orts JM, Casas-Rodrigo I, Minguet EG et al (2019) Assessment of Cas12a-mediated gene editing efficiency in plants. Plant Biotechnol J 17:1971–1984. https://doi.org/10.1111/pbi.13113
Berr A, Shafiq S, Pinon V et al (2015) The trxG family histone methyltransferase SET DOMAIN GROUP 26 promotes flowering via a distinctive genetic pathway. Plant J 81:316–328. https://doi.org/10.1111/tpj.12729
Bewick AJ, Niederhuth CE, Ji L et al (2017) The evolution of chromomethylases and gene body DNA methylation in plants. Genome Biol 18:65. https://doi.org/10.1186/s13059-017-1195-1
Boch J, Scholze H, Schornack S et al (2009) Breaking the code of DNA binding specificity of TAL-Type III effectors. Science 326:1509–1512. https://doi.org/10.1126/science.1178811
Bourque S, Jeandroz S, Grandperret V et al (2016) The evolution of HD2 proteins in green plants. Trends Plant Sci 21:1008–1016. https://doi.org/10.1016/j.tplants.2016.10.001
Brock RD, Davidson JLS (1994) 5-azacytidine and gamma rays partially substitute for cold treatment in vernalizing winter wheat
Bruce AB (1910) The mendelian theory of heredity and the augmentation of vigor. Science 32:627628. https://doi.org/10.1126/science.32.827.627-a
Campbell ME, Palfreyman JW, Preston CM (1984) Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol 180:1–19. https://doi.org/10.1016/0022-2836(84)90427-3
Carey M, Leatherwood J, Ptashne M (1990) A potent GAL4 derivative activates transcription at a distance in vitro. Science 247:710–712. https://doi.org/10.1126/science.2405489
Chavez A, Scheiman J, Vora S et al (2015) Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12:326–328. https://doi.org/10.1038/nmeth.3312
Chen H, Kazemier HG, de Groote ML et al (2014) Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res 42:1563–1574. https://doi.org/10.1093/nar/gkt1019
Chen L-Q, Luo J-H, Cui Z-H et al (2017) ATX3, ATX4, and ATX5 encode putative H3K4 methyltransferases and are critical for plant development. Plant Physiol 174:1795–1806. https://doi.org/10.1104/pp.16.01944
Chen X, Ding AB, Zhong X (2020) Functions and mechanisms of plant histone deacetylases. Sci China Life Sci 63:206–216. https://doi.org/10.1007/s11427-019-1587-x
Chen ZJ, Tian L (2007) Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta 1769:295–307. https://doi.org/10.1016/j.bbaexp.2007.04.007
Cheng K, Xu Y, Yang C et al (2020) Histone tales: lysine methylation, a protagonist in Arabidopsis development. J Exp Bot 71:793–807. https://doi.org/10.1093/jxb/erz435
Cho J-N, Ryu J-Y, Jeong Y-M et al (2012) Control of seed germination by light-induced histone arginine demethylation activity. Dev Cell 22:736–748. https://doi.org/10.1016/j.devcel.2012.01.024
Cho SW, Kim S, Kim JM, Kim J-S (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232. https://doi.org/10.1038/nbt.2507
Chodavarapu RK, Feng S, Ding B et al (2012) Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci USA 109:12040–12045. https://doi.org/10.1073/pnas.1209297109
Choi SC, Lee S, Kim S-R et al (2014) Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol 164:1326–1337. https://doi.org/10.1104/pp.113.228049
Choi Y, Gehring M, Johnson L et al (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110:33–42. https://doi.org/10.1016/S0092-8674(02)00807-3
Cokus SJ, Feng S, Zhang X et al (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219. https://doi.org/10.1038/nature06745
Cong L, Ran FA, Cox D et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi.org/10.1126/science.1231143
Cong W, Miao Y, Xu L et al (2019) Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol 19:282. https://doi.org/10.1186/s12870-019-1887-7
Cui X, Jin P, Cui X et al (2013) Control of transposon activity by a histone H3K4 demethylase in rice. Proc Natl Acad Sci U S A 110:1953–1958. https://doi.org/10.1073/pnas.1217020110
Devesa-Guerra I, Morales-Ruiz T, Pérez-Roldán J et al (2020) DNA methylation editing by CRISPR-guided excision of 5-methylcytosine. J Mol Biol 432:2204–2216. https://doi.org/10.1016/j.jmb.2020.02.007
Ding B, del Bellizzi M, R, Ning Y, et al (2012) HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24:3783–3794. https://doi.org/10.1105/tpc.112.101972
Dong H, Liu L, Fan X et al (2019) The artificial promoter rMdAG2I confers flower-specific activity in malus. Int J Mol Sci. https://doi.org/10.3390/ijms20184551
Duan C-G, Zhu J-K, Cao X (2018) Retrospective and perspective of plant epigenetics in China. J Genet Genomics 45:621–638. https://doi.org/10.1016/j.jgg.2018.09.004
Durai S, Mani M, Kandavelou K et al (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33:5978–5990. https://doi.org/10.1093/nar/gki912
Dutta A, Choudhary P, Caruana J, Raina R (2017) JMJ27, an Arabidopsis H3K9 histone demethylase, modulates defense against Pseudomonas syringae and flowering time. Plant J 91:1015–1028. https://doi.org/10.1111/tpj.13623
Ebbs ML, Bartee L, Bender J (2005) H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol 25:10507–10515. https://doi.org/10.1128/MCB.25.23.10507-10515.2005
El-Sharkawy I, Liang D, Xu K (2015) Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J Exp Bot 66:7359–7376. https://doi.org/10.1093/jxb/erv433
Fiaz S, Ahmad S, Noor MA et al (2019) Applications of the CRISPR/Cas9 system for rice grain quality improvement: perspectives and opportunities. Int J Mol Sci 20. https://doi.org/10.3390/ijms20040888
Finnegan EJ, Genger RK, Kovac K et al (1998) DNA methylation and the promotion of flowering by vernalization. Proc Natl Acad Sci USA 95:5824–5829. https://doi.org/10.1073/pnas.95.10.5824
Furci L, Jain R, Stassen J et al (2019) Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. Elife 8:e40655. https://doi.org/10.7554/eLife.40655
Gallego-Bartolomé J, Gardiner J, Liu W et al (2018) Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc Natl Acad Sci U S A 115:E2125–E2134. https://doi.org/10.1073/pnas.1716945115
Gan E-S, Xu Y, Ito T (2015) Dynamics of H3K27me3 methylation and demethylation in plant development. Plant Signal Behav 10:e1027851. https://doi.org/10.1080/15592324.2015.1027851
Gao C (2019) Precision plant breeding using genome editing technologies. Transgenic Res 28:53–55. https://doi.org/10.1007/s11248-019-00132-7
Gilbert LA, Horlbeck MA, Adamson B et al (2014) Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159:647–661. https://doi.org/10.1016/j.cell.2014.09.029
Gouil Q, Baulcombe DC (2016) DNA methylation signatures of the plant chromomethyltransferases. PLOS Genet 12:e1006526
Groszmann M, Greaves IK, Albert N et al (2011) Epigenetics in plants-vernalisation and hybrid vigour. Biochim Biophys Acta 1809:427–437. https://doi.org/10.1016/j.bbagrm.2011.03.006
Guan X, Stege J, Kim M et al (2002) Heritable endogenous gene regulation in plants with designed polydactyl zinc finger transcription factors. Proc Natl Acad Sci U S A 99:13296–13301. https://doi.org/10.1073/pnas.192412899
Han Q, Bartels A, Cheng X et al (2019) Epigenetics regulates reproductive development in plants. Plants (Basel, Switzerland). https://doi.org/10.3390/plants8120564
Han Z, Yu H, Zhao Z et al (2016) AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front Plant Sci 7:310. https://doi.org/10.3389/fpls.2016.00310
Harris CJ, Scheibe M, Wongpalee SP et al (2018) A DNA methylation reader complex that enhances gene transcription. Science 362:1182–1186. https://doi.org/10.1126/science.aar7854
Harris KD, Zemach A (2020) Contiguous and stochastic CHH methylation patterns of plant DRM2 and CMT2 revealed by single-read methylome analysis. Genome Biol 21:194. https://doi.org/10.1186/s13059-020-02099-9
Hartley A-V, Lu T (2020) Modulating the modulators: regulation of protein arginine methyltransferases by post-translational modifications. Drug Discov Today. https://doi.org/10.1016/j.drudis.2020.06.031
He G, Zhu X, Elling AA et al (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22:17–33. https://doi.org/10.1105/tpc.109.072041
Hebbes TR, Thorne AW, Crane-Robinson C (1988) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J 7:1395–1402
Henderson IR, Jacobsen SE (2007) Epigenetic inheritance in plants. Nature 447:418–424. https://doi.org/10.1038/nature05917
Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34:733–739. https://doi.org/10.1046/j.1365-313x.2003.01759.x
Hope IA, Struhl K (1986) Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46:885–894. https://doi.org/10.1016/0092-8674(86)90070-x
Horváth E, Szalai G, Janda T et al (2003) Effect of vernalisation and 5-azacytidine on the methylation level of DNA in wheat (Triticum aestivum L., cv. Martonvásár 15). Plant Sci 165:689–692. https://doi.org/10.1016/S0168-9452(03)00221-8
Hu Y, Zhang L, Zhao L et al (2011) Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS ONE 6:e22132–e22132. https://doi.org/10.1371/journal.pone.0022132
Huang H, Liu R, Niu Q et al (2019) Global increase in DNA methylation during orange fruit development and ripening. Proc Natl Acad Sci 116:1430–1436. https://doi.org/10.1073/pnas.1815441116
Huang Y-H, Su J, Lei Y et al (2017) DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol 18:176. https://doi.org/10.1186/s13059-017-1306-z
Jacob Y, Feng S, LeBlanc CA et al (2009) ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16:763–768. https://doi.org/10.1038/nsmb.1611
Jang I-C, Chung PJ, Hemmes H et al (2011) Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell 23:459–470. https://doi.org/10.1105/tpc.110.080481
Jeong HJ, Yang J, Yi J, An G (2015) Controlling flowering time by histone methylation and acetylation in Arabidopsis and rice. J Plant Biol 58:203–210. https://doi.org/10.1007/s12374-015-0219-1
Ji L, Jordan WT, Shi X et al (2018) TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Nat Commun 9:895. https://doi.org/10.1038/s41467-018-03289-7
Jiang D, Yang W, He Y, Amasino RM (2007) Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19:2975–2987. https://doi.org/10.1105/tpc.107.052373
Jiang J, Ding AB, Liu F, Zhong X (2020) Linking signaling pathways to histone acetylation dynamics in plants. J Exp Bot 71:5179–5190. https://doi.org/10.1093/jxb/eraa202
Jin H, Hu W, Wei Z et al (2008) Alterations in cytosine methylation and species-specific transcription induced by interspecific hybridization between Oryza sativa and O. officinalis. Theor Appl Genet 117:1271–1279. https://doi.org/10.1007/s00122-008-0861-9
Jones DF (1917) Dominance of linked factors as a means of accounting for heterosis. Proc Natl Acad Sci USA 3:310–312. https://doi.org/10.1073/pnas.3.4.310
Kakutani T, Jeddeloh JA, Flowers SK et al (1996) Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci U S A 93:12406–12411. https://doi.org/10.1073/pnas.93.22.12406
Karan R, DeLeon T, Biradar H, Subudhi PK (2012) Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE 7:e40203–e40203. https://doi.org/10.1371/journal.pone.0040203
Kearns NA, Pham H, Tabak B et al (2015) Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12:401–403. https://doi.org/10.1038/nmeth.3325
Keegan L, Gill G, Ptashne M (1986) Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231:699–704. https://doi.org/10.1126/science.3080805
Kim D-H, Sung S (2013) Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25:454–469. https://doi.org/10.1105/tpc.112.104760
Kim D-H, Yu J, Park JC et al (2019) Targeted cytochrome P450 3045C1 (CYP3045C1) gene mutation via CRISPR-Cas9 ribonucleoproteins in the marine rotifer Brachionus koreanus. Hydrobiologia 844:117–128. https://doi.org/10.1007/s10750-018-3854-y
Kim H, Kim S-T, Ryu J et al (2017a) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8:14406. https://doi.org/10.1038/ncomms14406
Kim J-M, To TK, Matsui A et al (2017b) Acetate-mediated novel survival strategy against drought in plants. Nat plants 3:17097. https://doi.org/10.1038/nplants.2017.97
Konermann S, Brigham MD, Trevino AE et al (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588. https://doi.org/10.1038/nature14136
Kumar S, AlAbed D, Whitteck JT et al (2015) A combinatorial bidirectional and bicistronic approach for coordinated multi-gene expression in corn. Plant Mol Biol 87:341–353. https://doi.org/10.1007/s11103-015-0281-6
Kumar S, Chinnusamy V, Mohapatra T (2018) Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front Genet 9:640
Lang Z, Wang Y, Tang K et al (2017) Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci 114:E4511–E4519. https://doi.org/10.1073/pnas.1705233114
Lee JE, Neumann M, Duro DI, Schmid M (2019) CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants. PLoS ONE 14:e0222778. https://doi.org/10.1371/journal.pone.0222778
Li J, Blue R, Zeitler B et al (2013) Activation domains for controlling plant gene expression using designed transcription factors. Plant Biotechnol J 11:671–680. https://doi.org/10.1111/pbi.12057
Li S, Ali S, Duan X et al (2018a) JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep 23:389–403. https://doi.org/10.1016/j.celrep.2018.03.051
Li X, Harris CJ, Zhong Z et al (2018b) Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation. Proc Natl Acad Sci U S A 115:E8793–E8802. https://doi.org/10.1073/pnas.1809841115
Li Z, Jiang D, He Y (2018c) FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat Plants 4:836–846. https://doi.org/10.1038/s41477-018-0250-6
Li Z, Jiang G, Liu X et al (2020) Histone demethylase SlJMJ6 promotes fruit ripening by removing H3K27 methylation of ripening-related genes in tomato. New Phytol 227:1138–1156. https://doi.org/10.1111/nph.16590
Li Z, Wang F, Li J-F (2019) Targeted transcriptional activation in plants using a potent dead Cas9-derived synthetic gene activator. Curr Protoc Mol Biol 127:e89. https://doi.org/10.1002/cpmb.89
Li Z, Zhang D, Xiong X et al (2017) A potent Cas9-derived gene activator for plant and mammalian cells. Nat Plants 3:930–936. https://doi.org/10.1038/s41477-017-0046-0
Liu B, Liu Y, Wang B et al (2019) The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment. Nat Commun 10:2999. https://doi.org/10.1038/s41467-019-10850-5
Liu C, Lu F, Cui X, Cao X (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61:395–420. https://doi.org/10.1146/annurev.arplant.043008.091939
Liu D-D, Zhou L-J, Fang M-J et al (2016a) Polycomb-group protein SlMSI1 represses the expression of fruit-ripening genes to prolong shelf life in tomato. Sci Rep 6:31806. https://doi.org/10.1038/srep31806
Liu R, How-Kit A, Stammitti L et al (2015) A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci 112:10804–10809. https://doi.org/10.1073/pnas.1503362112
Liu W, Rudis MR, Peng Y et al (2014) Synthetic TAL effectors for targeted enhancement of transgene expression in plants. Plant Biotechnol J 12:436–446. https://doi.org/10.1111/pbi.12150
Liu X, Yang S, Yu C-W et al (2016b) Histone acetylation and plant development. Enzyme 40:173–199. https://doi.org/10.1016/bs.enz.2016.08.001
Liu XS, Wu H, Ji X et al (2016c) Editing DNA methylation in the mammalian genome. Cell 167:233-247.e17. https://doi.org/10.1016/j.cell.2016.08.056
Lowder LG, Zhang D, Baltes NJ et al (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169:971–985. https://doi.org/10.1104/pp.15.00636
Lowder LG, Zhou J, Zhang Y et al (2018) Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol Plant 11:245–256. https://doi.org/10.1016/j.molp.2017.11.010
Lu F, Cui X, Zhang S et al (2010) JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 20:387–390
Luo M, Wang Y-Y, Liu X et al (2012) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63:3297–3306. https://doi.org/10.1093/jxb/ers059
Luo X, He Y (2020) Experiencing winter for spring flowering: a molecular epigenetic perspective on vernalization. J Integr Plant Biol 62:104–117. https://doi.org/10.1111/jipb.12896
Ma J (2011) Transcriptional activators and activation mechanisms. Protein Cell 2:879–888. https://doi.org/10.1007/s13238-011-1101-7
Mali P, Yang L, Esvelt KM et al (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826. https://doi.org/10.1126/science.1232033
Manning K, Tör M, Poole M et al (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952. https://doi.org/10.1038/ng1841
Marmorstein R, Zhou M-M (2014) Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6:a018762. https://doi.org/10.1101/cshperspect.a018762
Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15:394–408. https://doi.org/10.1038/nrg3683
Ming M, Ren Q, Pan C et al (2020) CRISPR-Cas12b enables efficient plant genome engineering. Nat plants 6:202–208. https://doi.org/10.1038/s41477-020-0614-6
Mohan C, Jayanarayanan AN, Narayanan S (2017) Construction of a novel synthetic root-specific promoter and its characterization in transgenic tobacco plants. 3 Biotech 7:234
Moore I, Gälweiler L, Grosskopf D et al (1998) A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci U S A 95:376–381. https://doi.org/10.1073/pnas.95.1.376
Moore R, Chandrahas A, Bleris L (2014) Transcription activator-like effectors: a toolkit for synthetic biology. ACS Synth Biol 3:708–716. https://doi.org/10.1021/sb400137b
Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI et al (2006) DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci 103:6853–6858. https://doi.org/10.1073/pnas.0601109103
Morbitzer R, Römer P, Boch J, Lahaye T (2010) Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A 107:21617–21622. https://doi.org/10.1073/pnas.1013133107
Morita S, Noguchi H, Horii T et al (2016) Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol 34:1060–1065. https://doi.org/10.1038/nbt.3658
Ni Z, Kim E-D, Ha M et al (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457:327–331. https://doi.org/10.1038/nature07523
Ohta M, Matsui K, Hiratsu K et al (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13:1959–1968. https://doi.org/10.1105/tpc.010127
Ordiz MI, Barbas CF 3rd, Beachy RN (2002) Regulation of transgene expression in plants with polydactyl zinc finger transcription factors. Proc Natl Acad Sci U S A 99:13290–13295. https://doi.org/10.1073/pnas.202471899
Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldán-Arjona T (2008) Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol 67:671–681. https://doi.org/10.1007/s11103-008-9346-0
Osorio S, Carneiro RT, Lytovchenko A et al (2020) Genetic and metabolic effects of ripening mutations and vine detachment on tomato fruit quality. Plant Biotechnol J 18:106–118. https://doi.org/10.1111/pbi.13176
Papikian A, Liu W, Gallego-Bartolomé J, Jacobsen SE (2019) Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun 10:729. https://doi.org/10.1038/s41467-019-08736-7
Park J-J, Dempewolf E, Zhang W, Wang Z-Y (2017) RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS ONE 12:e0179410. https://doi.org/10.1371/journal.pone.0179410
Park J, Lim CJ, Shen M et al (2018) Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc Natl Acad Sci U S A 115:E5400–E5409. https://doi.org/10.1073/pnas.1721241115
Penterman J, Uzawa R, Fischer RL (2007) Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol 145:1549–1557. https://doi.org/10.1104/pp.107.107730
Petolino JF, Davies JP (2013) Designed transcriptional regulators for trait development. Plant Sci 201–202:128–136. https://doi.org/10.1016/j.plantsci.2012.12.006
Piatek A, Ali Z, Baazim H et al (2015) RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13:578–589. https://doi.org/10.1111/pbi.12284
Qi LS, Larson MH, Gilbert LA et al (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. https://doi.org/10.1016/j.cell.2013.02.022
Roca Paixão JF, Gillet F-X, Ribeiro TP et al (2019) Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci Rep 9:8080. https://doi.org/10.1038/s41598-019-44571-y
Ronemus MJ, Galbiati M, Ticknor C et al (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273:654–657. https://doi.org/10.1126/science.273.5275.654
Sadakierska-Chudy A, Kostrzewa RM, Filip M (2015) A comprehensive view of the epigenetic landscape part I: DNA methylation, passive and active DNA demethylation pathways and histone variants. Neurotox Res 27:84–97. https://doi.org/10.1007/s12640-014-9497-5
Saleh A, Alvarez-Venegas R, Yilmaz M et al (2008) The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20:568–579. https://doi.org/10.1105/tpc.107.056614
Sanders SM, Ma Z, Hughes JM et al (2018) CRISPR/Cas9-mediated gene knockin in the hydroid Hydractinia symbiolongicarpus. BMC Genomics 19:649. https://doi.org/10.1186/s12864-018-5032-z
Saze H, Kakutani T (2007) Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J 26:3641–3652. https://doi.org/10.1038/sj.emboj.7601788
Schuster M, Kahmann R (2019) CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet Biol 130:43–53. https://doi.org/10.1016/j.fgb.2019.04.016
Schwechheimer C, Smith C, Bevan MW (1998) The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: design of modular transcription factors for high-level expression. Plant Mol Biol 36:195–204. https://doi.org/10.1023/a:1005990321918
Selma S, Bernabé-Orts JM, Vazquez-Vilar M et al (2019) Strong gene activation in plants with genome-wide specificity using a new orthogonal CRISPR/Cas9-based programmable transcriptional activator. Plant Biotechnol J 17:1703–1705
Shafiq S, Berr A, Shen W-H (2014) Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol 201:312–322. https://doi.org/10.1111/nph.12493
Shakirova KM, Ovchinnikova VY, Dashinimaev EB (2020) Cell reprogramming with CRISPR/Cas9 based transcriptional regulation systems. Front Bioeng Biotechnol 8:882
Shan X, Wang X, Yang G et al (2013) Analysis of the DNA methylation of maize (Zea mays L.) in response to cold stress based on methylation-sensitive amplified polymorphisms. J Plant Biol 56:32–38. https://doi.org/10.1007/s12374-012-0251-3
Shao X, Wu S, Dou T et al (2020) Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol J 18:17–19. https://doi.org/10.1111/pbi.13216
Shen H, He H, Li J et al (2012) Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24:875–892. https://doi.org/10.1105/tpc.111.094870
Shi T-Q, Gao J, Wang W-J et al (2019) CRISPR/Cas9-based genome editing in the filamentous fungus fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth Biol 8:445–454. https://doi.org/10.1021/acssynbio.8b00478
Shi Y, Lan F, Matson C et al (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953. https://doi.org/10.1016/j.cell.2004.12.012
Shrestha A, Khan A, Dey N (2018) Cis–trans engineering: advances and perspectives on customized transcriptional regulation in plants. Mol Plant 11:886–898. https://doi.org/10.1016/j.molp.2018.05.008
Shull GH (1908) The composition of a field of maize. J Hered 4:296–301. https://doi.org/10.1093/jhered/os-4.1.296
Sokol A, Kwiatkowska A, Jerzmanowski A, Prymakowska-Bosak M (2007) Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227:245–254. https://doi.org/10.1007/s00425-007-0612-1
Song Y, Liu L, Feng Y et al (2015a) Chilling- and freezing-induced alterations in cytosine methylation and its association with the cold tolerance of an alpine subnival plant, Chorispora bungeana. PLoS ONE 10:e0135485–e0135485. https://doi.org/10.1371/journal.pone.0135485
Song Z-T, Sun L, Lu S-J et al (2015b) Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc Natl Acad Sci U S A 112:2900–2905. https://doi.org/10.1073/pnas.1419703112
Springer NM, Napoli CA, Selinger DA et al (2003) Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol 132:907–925. https://doi.org/10.1104/pp.102.013722
Sun Q, Zhou D-X (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci U S A 105:13679–13684. https://doi.org/10.1073/pnas.0805901105
Sung S, He Y, Eshoo TW et al (2006) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires like heterochromatin protein 1. Nat Genet 38:706–710. https://doi.org/10.1038/ng1795
Tak YE, Kleinstiver BP, Nuñez JK et al (2017) Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat Methods 14:1163–1166. https://doi.org/10.1038/nmeth.4483
Tanenbaum ME, Gilbert LA, Qi LS et al (2014) A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159:635–646. https://doi.org/10.1016/j.cell.2014.09.039
Tang X, Lowder LG, Zhang T et al (2017) A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17018. https://doi.org/10.1038/nplants.2017.18
Tariq M, Saze H, Probst AV et al (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci 100:8823–8827. https://doi.org/10.1073/pnas.1432939100
Tiwari SB, Belachew A, Ma SF et al (2012) The EDLL motif: A potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J 70:855–865. https://doi.org/10.1111/j.1365-313X.2012.04935.x
Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16:533–543. https://doi.org/10.1105/tpc.017384
Tsukahara S, Kobayashi A, Kawabe A et al (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature 461:423–426. https://doi.org/10.1038/nature08351
Underwood CJ, Henderson IR, Martienssen RA (2017) Genetic and epigenetic variation of transposable elements in Arabidopsis. Curr Opin Plant Biol 36:135–141. https://doi.org/10.1016/j.pbi.2017.03.002
Uranga M, Aragonés V, Selma S, et al (2020) Efficient Cas9 multiplex editing using unspaced gRNA arrays engineering in a Potato virus vector. bioRxiv 2020.06.25.170977. https://doi.org/10.1101/2020.06.25.170977
Utley RT, Ikeda K, Grant PA et al (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498–502. https://doi.org/10.1038/28886
Vazquez-Vilar M, Bernabé-Orts JM, Fernandez-del-Carmen A et al (2016) A modular toolbox for gRNA-Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods 12:1–12. https://doi.org/10.1186/s13007-016-0101-2
Veillet F, Perrot L, Chauvin L et al (2019) Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci. https://doi.org/10.3390/ijms20020402
Vojta A, Dobrinić P, Tadić V et al (2016) Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 44:5615–5628. https://doi.org/10.1093/nar/gkw159
Wang J, Meng X, Yuan C et al (2016) The roles of cross-talk epigenetic patterns in Arabidopsis thaliana. Brief Funct Genomics 15:278–287. https://doi.org/10.1093/bfgp/elv025
Wang W, Zhao X, Pan Y et al (2011) DNA methylation changes detected by methylation-sensitive amplified polymorphism in two contrasting rice genotypes under salt stress. J Genet Genomics 38:419–424. https://doi.org/10.1016/j.jgg.2011.07.006
Waryah CB, Moses C, Arooj M, Blancafort P (2018) Zinc fingers, TALEs, and CRISPR systems: a comparison of tools for epigenome editing. Methods Mol Biol 1767:19–63. https://doi.org/10.1007/978-1-4939-7774-1_2
Weiste C, Dröge-Laser W (2014) The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat Commun 5:3883. https://doi.org/10.1038/ncomms4883
Whittaker C, Dean C (2017) The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu Rev Cell Dev Biol 33:555–575. https://doi.org/10.1146/annurev-cellbio-100616-060546
Wibowo A, Becker C, Marconi G et al (2016) Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife 5:e13546. https://doi.org/10.7554/eLife.13546
Wiles ET, Selker EU (2017) H3K27 methylation: a promiscuous repressive chromatin mark. Curr Opin Genet Dev 43:31–37. https://doi.org/10.1016/j.gde.2016.11.001
Wolffe AP, Matzke MA (1999) Epigenetics: regulation through repression. Science 286:481–486. https://doi.org/10.1126/science.286.5439.481
Wood CC, Robertson M, Tanner G et al (2006) The Arabidopsisthaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A 103:14631–14636. https://doi.org/10.1073/pnas.0606385103
Xiao J, Lee U-S, Wagner D (2016) Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr Opin Plant Biol 34:41–53. https://doi.org/10.1016/j.pbi.2016.08.002
Xu L, Zhao Z, Dong A et al (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28:1348–1360. https://doi.org/10.1128/MCB.01607-07
Yamasaki K, Kigawa T, Seki M et al (2013) DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci 18:267–276. https://doi.org/10.1016/j.tplants.2012.09.001
Yang H, Han Z, Cao Y et al (2012a) A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet 8:e1002664. https://doi.org/10.1371/journal.pgen.1002664
Yang H, Howard M, Dean C (2014) Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24:1793–1797. https://doi.org/10.1016/j.cub.2014.06.047
Yang H, Mo H, Fan D et al (2012b) Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep 31:1297–1308. https://doi.org/10.1007/s00299-012-1249-5
Yu C-W, Liu X, Luo M et al (2011) HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol 156:173–184. https://doi.org/10.1104/pp.111.174417
Yu C-W, Tai R, Wang S-C et al (2017) HISTONE DEACETYLASE6 acts in concert with histone methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell 29:1970–1983. https://doi.org/10.1105/tpc.16.00570
Zalatan JG, Lee ME, Almeida R et al (2015) Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160:339–350. https://doi.org/10.1016/j.cell.2014.11.052
Zaynab M, Sharif Y, Fatima M et al (2020) CRISPR/Cas9 to generate plant immunity against pathogen. Microb Pathog 141:103996. https://doi.org/10.1016/j.micpath.2020.103996
Zhang F, Wang L, Ko EE et al (2018a) Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30:153–166. https://doi.org/10.1105/tpc.17.00671
Zhang H, Lang Z, Zhu J-K (2018b) Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19:489–506. https://doi.org/10.1038/s41580-018-0016-z
Zhang L, Jiménez-Gómez JM (2020) Functional analysis of FRIGIDA using naturally occurring variation in Arabidopsis thaliana. Plant J 103:154–165. https://doi.org/10.1111/tpj.14716
Zhang Q, Wang D, Lang Z et al (2016) Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci 113:E4248–E4256. https://doi.org/10.1073/pnas.1607851113
Zhang S, Guo F, Yan W et al (2020) Recent advances of CRISPR/Cas9-based genetic engineering and transcriptional regulation in industrial biology. Front Bioeng Biotechnol 7:459
Zhang X, Wang W, Shan L et al (2018c) Gene activation in human cells using CRISPR/Cpf1-p300 and CRISPR/Cpf1-SunTag systems. Protein Cell 9:380–383
Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15:2343–2360. https://doi.org/10.1101/gad.927301
Zhang Y, Yin C, Zhang T et al (2015) CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep 5:1–14. https://doi.org/10.1038/srep16277
Zhao L, Peng T, Chen C-Y et al (2019a) HY5 interacts with the histone deacetylase HDA15 to Repress Hypocotyl Cell Elongation In Photomorphogenesis. Plant Physiol 180:1450–1466. https://doi.org/10.1104/pp.19.00055
Zhao T, Zhan Z, Jiang D (2019) Histone modifications and their regulatory roles in plant development and environmental memory. J Genet Genomics 46:467–476. https://doi.org/10.1016/j.jgg.2019.09.005
Zheng L, Cheng Z, Ai C et al (2010) Nicotianamine, a novel enhancer of rice iron bioavailability to humans. PLoS One 5:e10190. https://doi.org/10.1371/journal.pone.0010190
Zheng S, Hu H, Ren H et al (2019) The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat Commun 10:1303. https://doi.org/10.1038/s41467-019-09310-x
Zhou H, Liu Y, Liang Y et al (2020) The function of histone lysine methylation related SET domain group proteins in plants. Protein Sci 29:1120–1137. https://doi.org/10.1002/pro.3849
Zhu J, Jeong J, Zhu Y et al (2008) Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci U S A 105:4945–4950. https://doi.org/10.1073/pnas.0801029105
Zilberman D, Gehring M, Tran RK et al (2007) Genome-wide analysis of Arabidopsisthaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39:61–69. https://doi.org/10.1038/ng1929
Acknowledgements
This work has been funded by the PID2019-108203RB-100 Plan Nacional I+D project from the Spanish Ministry of Science and Innovation. Sara Selma is recipient of FPI fellowships associated with BIO2016-78601-R Grant from the same funding source.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Selma, S., Orzáez, D. Perspectives for epigenetic editing in crops. Transgenic Res 30, 381–400 (2021). https://doi.org/10.1007/s11248-021-00252-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-021-00252-z