Abstract
Hrip1 is a novel hypersensitive response-inducing protein secreted by Alternaria tenuissima that activates defense responses and systemic acquired resistance in tobacco. This study investigates the role that Hrip1 plays in responses to abiotic and biotic stress using transgenic Arabidopsis thaliana expressing the Hrip1 gene under the control of the stress-inducible rd29A promoter or constitutive cauliflower mosaic virus 35S promoter. Bioassays showed that inducible Hrip1 expression in rd29A∷Hrip1 transgenic lines had a significantly higher effect on plant height, silique length, plant dry weight, seed germination and root length under salt and drought stress compared to expression in 35S∷Hrip1 lines and wild type plants. The level of enhancement of resistance to Botrytis cinerea by the 35S∷Hrip1 lines was higher than in the rd29A∷Hrip1 lines. Moreover, stress-related gene expression in the transgenic Arabidopsis lines was significantly increased by 200 mM NaCl and 200 mM mannitol treatments, and defense genes in the jasmonic acid and ethylene signaling pathway were significantly up-regulated after Botrytis inoculation in the Hrip1 transgenic plants. Furthermore, the activity of some antioxidant enzymes, such as peroxidase and catalase increased after salt and drought stress and Botrytis infection. These results suggested that the Hrip1 protein contributes to abiotic and biotic resistance in transgenic Arabidopsis and may be used as a useful gene for resistance breeding in crops. Although the constitutive expression of Hrip1 is suitable for biotic resistance, inducible Hrip1 expression is more responsive for abiotic resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are continuously being challenged with biotic and abiotic stresses that have adverse effects on growth and cause severe losses in crop production. Indeed, it has been estimated that approximately 70 % of yield reduction is the result of abiotic and biotic stresses. Thus, there is an urgent need for improving the tolerance of crops to stress. The transgenic approach is one of the many tools available for modern plant improvement programs, and transgenic plant have been well recognized as the most promising and effective strategy for food security against environmental stress.
Microbial elicitors and effectors produced from plant pathogens during plant-pathogen interactions modulate plant defense systems or trigger plant defense responses (Hogenhout et al. 2009). A number of microbially derived protein elicitors capable of inducing immune responses, including harpin protein, flagellin, elicitin, activator, and glycoprotein elicitor, have been isolated from plant pathogenic bacteria, viruses, oomycete, and fungi (Ebel 1998; Beissmann and Beisener 1990). Protein elicitors are known to be involved in both biotic and abiotic stress responses and function as activators of systemic acquired resistance (SAR) in plants as a result of adversity and pathogen infection (Nürnberger 1999; Nürnberger and Scheel 2001). A 14-kDa hypersensitive response-inducing protein (MoHrip1) secreted by Magnaporthe oryzae activates the early events of the tobacco defense response and also notably elicits the expression of pathogenesis-related (PR) genes and signal transduction-related genes, improving systemic resistance to M. oryzae in rice seedlings (Chen et al. 2012). A protein elicitor, PebC1, from B. cinerea was found to promote tomato resistance to drought and pathogen stress after exogenous application on tomato leaves (Zhang et al. 2010). The PemG1 protein was purified from Magnaporthe grisea, and pemG1-overexpresing transgenic rice showed enhanced resistance against rice blast after inoculation with M. grisea spores (Qiu et al. 2009).
Protein elicitors have resulted in improved pathogen resistance in transgenic plants. The gene encoding harpin was shown to activate pathogen resistance when expressed in transgenic tobacco and rice (Peng et al. 2004; Shao et al. 2008), and flagellin expression also conferred disease resistance in transgenic rice (Takakura et al. 2008). Transgenic tobacco plants expressing the cryptogein elicitor enhanced resistance against fungal pathogens such as Thielaviopsis basicola, Erysiphe cichoracearum, and Botrytis cinerea (Keller et al. 1999). Thus, broad-spectrum disease resistance in a plant can be achieved through the expression of genes encoding elicitors from microbes.
Hrip1 is a novel hypersensitive response-inducing protein from the necrotrophic fungus Alternaria tenuissima, and, as a type of exogenous elicitor, induces calcium influx, medium alkalinization, and the expression of several defense-related genes after infiltration in tobacco leaves (Kulye et al. 2012). In the present study, we investigated the functions of Hrip1 in transgenic Arabidopsis thaliana plants by expressing the protein under the control of the stress-inducible rd29A promoter and constitutive cauliflower mosaic virus (CaMV) 35S promoter. These experiments were performed under abiotic and biotic stress conditions, and attempts were made to determine potential of Hrip1 for engineering stress-tolerant transgenic plants.
Materials and methods
Plant growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used in this study. Seeds were surface-sterilized with 0.05 % NaClO for 15 min, rinsed with distilled water, and then stratified for 3 day on ½-strength MS medium at 4 °C prior to germination. The plants were grown under a 16:8-h light:dark cycle at 22 °C and 60 % humidity for 3–4 weeks prior to the treatments.
Construction of expression constructs and transformation of Arabidopsis
The plant expression constructs were based on the pCAMBIA1301 plasmid vector (CAMBIA, Canberra, Australia). The open reading frame of the Hrip1 gene was amplified by PCR using gene-specific primers. The expected PCR fragments were cloned into the PMD18-T vector and sequenced. These fragments were then cloned into the binary vector pCAMBIA1301-35S. Four recombinant plasmids, 35S∷Hrip1, rd29A∷Hrip1, 35S∷Hrip1∷GUS, and rd29A∷Hrip1∷GUS, were introduced into Agrobacterium tumefaciens strain GV3101. Agrobacterium-mediated transformation of Arabidopsis (Col-0) was performed using the floral dip method (Clough and Bent 1998).
Polymerase chain reaction (PCR) analysis, semi-quantitative RT-PCR analysis and quantitative real-time RT-PCR
Seeds from transgenic plants were selected on ½-strength MS medium containing 25 mg mL−1 hygromycin, and hygromycin-resistant T0 plants were confirmed by PCR using the gene-specific primers described previously (Kulye et al. 2012). Homozygous (T3 and T4) plants of 35S∷Hrip1 and rd29A∷Hrip1 independent transgenic lines were used for the functional analyses.
Total RNA was extracted from detached Arabidopsis leaves at the indicated times after treatment using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The DNase-treated RNA was reverse-transcribed using Prime Script™ RTase (TaKaRa, Ohtsu, Japan). Semi-quantitative RT-PCR was performed using the following program: 95 °C for 3 min, 27 cycles of 94 °C for 30 s, 59 °C for 30 s, and 72 °C for 40 s, followed by 72 °C for 10 min.
Real-time quantitative (q) RT-PCR was performed using SYBR green (TaKaRa, Ohtsu, Japan) and a CFX96 real-time PCR instrument (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions. All reactions were performed in triplicate. The thermal cycling consisted of a hold at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. After amplification, the samples were kept at 55 °C for 1 min, and the temperature was raised gradually by 0.5 °C every 10 s to perform the melting curve analysis. Arabidopsis ACT2 (accession no. U41998) was amplified as an internal control. The primers used for qRT-PCR are listed in Supplemental Table 1. Each relative expression level was analyzed with CFX96 software using the Normalized Expression method. Significant differences between the mock and treatment conditions were assessed using SPSS (Version 16.0).
Western blot assay
Total plant protein for western blotting was extracted from 4-week-old wild-type, 35S∷Hrip1, and rd29A∷Hrip1 Arabidopsis plants using precooled extraction buffer (0.025 mol L−1 K2HPO4, 0.025 mol L−1 KH2PO4, and 2 mmol L−1 EDTA, pH 8.0). After centrifugation at 12,000×g for 10 min at 4 °C, the supernatants were transferred into clean tubes, immediately frozen with liquid N2, and stored at −80 °C. For western blotting, total protein was separated by electrophoresis on 12 % sodium dodecyl sulfate (SDS)-polyacrylamide gels, and the proteins were transferred to PVDF membranes by semi-dry electroblotting. The membranes were incubated for 3 h with Hrip1 antibody (1:1,000 dilution); alkaline phosphatase-tagged goat anti-rabbit IgG [diluted 1:2,000 in Tris-buffered saline with Tween (TBST)] was used as the secondary antibody. The staining reaction was performed using a freshly prepared solution of BCIP/NBT (E116, Amresco, Solon, OH, USA) diluted in TBST. The reaction was stopped with dd-H2O after the signals were detected.
Histochemical GUS assay
GUS staining was essentially performed according to Jefferson (1987). Sample tissues were incubated in staining buffer [50 mM Na2HPO4–NaH2PO4, pH 7.3, 10 mM EDTA, 2 mM K4Fe (CN)6, 2 mM K3Fe(CN)6, and 0.1 % Triton X-100] with 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-b-d-glucuronide acid (X-Gluc) overnight at 37 °C. The seedlings were then incubated in 95 % ethanol solution to remove the chlorophyll from the plant tissues. The GUS staining patterns were recorded under a dissecting microscope (Nikon SMZ800).
Salt and drought stress tolerance assays of seedlings and adult plants
For the salt stress tests, T4 seeds from 35S∷Hrip1 and rd29A∷Hrip1 transgenic and WT plants were sown on ½-strength MS plates supplemented with different concentrations of NaCl and mannitol (0, 50, 75, 125, 150, 200, and 300 mM). The plates were placed in a growth chamber under a 16:8-h light:dark cycle at 22 °C. Germination was scored for five consecutive days and was considered to have occurred when the radicle penetrated the seed coat. The relative root length was calculated as follows: (root length on MS medium containing different NaCl or mannitol/the average of root length on MS medium) × 100 %.
For the salt stress evaluation, 3-week-old plants were irrigated at 2-day intervals with 250 mM NaCl for 20 days. For the drought tolerance evaluation, 3-week-old plants were not watered for 25 days prior to rewatering. The survival rates of the plants were recorded on day 5 after the stress treatment.
Inoculation of plants with Botrytis cinerea
Botrytis cinerea strain BC4221 was cultured on potato dextrose agar (PDA) at 22 °C for 10 days in the dark. The conidia were collected, and the spore suspension was adjusted to the appropriate concentration of 2 × 105 conidia mL−1. A detached leaf from a 3-week-old Arabidopsis plant was placed on wet filter, inoculated with two droplets of spore suspension, and incubated at 22 °C for 48 h. The lesion diameter was measured at 2 days after inoculation. The experiment was repeated three times with different plant batches, and a statistical analysis of the results was performed by a randomized block ANOVA.
For a whole-plant test, 3-week-old soil-grown plants were sprayed with a spore suspension and covered with transparent plastic film for 48 h to keep the humidity high; the plants were then incubated in a growth chamber under a 12:12-h light:dark dark cycle. The disease symptoms were photographed, and the surviving plants were assessed at the indicated time points. All pathogen infection experiments were repeated three times with different plants batches, and each repeat used three lines of different plant lines.
Measurement of leaf POD and CAT activities
Peroxidase (POD) and catalase (CAT) were measured using the guaiacol method and ultraviolet spectroscopy test (Zhang et al. 2006). Each treatment consisted of 10 individual Arabidopsis plants and was repeated three times. The middle leaves of young Arabidopsis plants at the same position were used to measure the peroxidase (POD) and catalase (CAT) activities at 24 h post-treatment.
Data analysis
The data for this study were analyzed by a one-way analysis of variance (ANOVA) using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA), and Duncan’s Multiple Range test was used to detect significant differences between the means. Significant differences were evaluated at the 5 % level.
Results
Generation of transgenic plants and molecular analysis of regenerants
The full-length Hrip1 sequence was cloned under the control of the CaMV35S and rd29A promoters; a total of 11 CaMV35S∷Hrip1 and 13 rd29A∷Hrip1 independent transgenic lines were generated. The T1 lines segregated 3:1 for hygromycin resistance, indicating a single T-DNA insertion in the transgenic lines. Three lines of 35s:Hrip1 and rd29A:Hrip1 were chosen for further analysis and were named 35S∷Hrip1-1, -2, and -3 and rd29A∷Hrip1-1, -2, and -3. The 35S∷Hrip1 and rd29A∷Hrip1 plants were detected at the mRNA and protein levels by RT-PCR and western blotting, respectively, and three independent T3 homozygous 35S∷Hrip1 and rd29A∷Hrip1 lines showed expression (Supplemental Figure 1a). We also analyzed the expression of the 35S∷Hrip1∷GUS and rd29A∷Hrip1∷GUS genes in the transgenic plants. Although the 35s∷Hrip1∷GUS line displayed no difference between the normal and salt stress conditions, the rd29A∷Hrip1∷GUS fusion protein was significantly induced under drought and salt stresses in the transgenic plant (Supplemental Figure 1b).
Hrip1 expression improves plant growth
To examine the possible phenotypes of the transgenic lines, T3 progeny of the Hrip1-expressing and wild-type (WT) plants were grown in the greenhouse under identical conditions. The plant height, dry weight, and silique length were increased by 1.35-, 1.50-, and 1.30-fold in rd29A∷Hrip1 and 1.12-, 1.22-, and 1.13-fold in 35S∷Hrip1 compared to WT (Fig. 1; Table 1), differences between WT and transgenic plants were significant, with the rd29A∷Hrip1 lines showing improved growth over the 35S∷Hrip1 lines.
Hrip1 expression increases tolerance to salt and drought stresses in plants
We compared seed germination and root growth of WT, 35S∷Hrip1, and rd29A∷Hrip1 plants on MS medium supplemented with NaCl or mannitol. The seed germination rate of the transgenic lines were significantly higher than that of WT on both MS medium and MS supplemented with 75 mM NaCl or 50 mM mannitol (Supplemental Figure 2a). Under normal growth conditions (MS medium), the WT seeds germinated at a rate 58.33 % lower than the Hrip1-exepressing lines after 1 day; however, germination of the wild-type and transgenic seeds reached 100 % after 2 days (Supplemental Figure 2b).
After 2 days on MS medium (75 mM NaCl), WT seed germination was only 45.56 %, whereas the rd29A∷Hrip1 and 35S∷ Hrip1 transgenic lines exhibited 97.22 and 70.56 % seed germination, respectively (Supplemental Figure 2b). On MS medium supplemented with 50 mM mannitol, the WT seed germination rate was 78.89 %, though the rd29A∷Hrip1 and 35S∷Hrip1 transgenic seed germination rates reached 100 % after 2 days (Supplemental Figure 2b).
The seedling root growth was measured under normal and stress conditions on MS medium. Under NaCl and mannitol stress conditions, WT root growth was limited to a greater degree than that of the transgenic lines (Supplemental Figure 3a). Seedlings grown on a medium supplemented with different concentrations of NaCl or mannitol exhibited decreased root growth compared to that on MS medium alone (Supplemental Figure 3b). The relative root length of the rd29A∷Hrip1 lines and 35S∷Hrip1 lines was 83.15 and 82.22 %, respectively, compared to 58 % for WT at day 7 under 150 mM NaCl stress (Supplemental Figure 3b); the relative root length of the rd29A∷Hrip1 lines and 35S∷Hrip1 lines was 91.72 and 91.37 %, respectively, under 200 mM mannitol stress and 85.50 and 77.43 % under 300 mM mannitol stress compared to 59 % for WT (Supplemental Figure 3b).
The plant survival rate was also measured in 21-day-old plants grown on soil under normal, 250 mM NaCl, or water-withheld conditions. The transgenic plants showed stronger survival than the WT plants (Fig. 2). The survival rates of the rd29A∷Hrip1 and 35S∷Hrip1 lines were 1.80 and 1.29 times, respectively, of that of WT after 250 mM NaCl for 20 days and 2.20 and 1.57 times of that of WT, respectively, after dehydration stress for 25 days (Supplemental Table 2). These results showed that the osmotic tolerance to salt and drought stress was increased in the Hrip1-expressing plants, with the rd29A∷Hrip1 plants showing increased tolerance to salt and drought in comparison to the 35S∷Hrip1 plants.
Hrip1 expression enhances tolerance to Botrytis stress
In a detached-leaf assay (thirty leaves for every repeat), the leaf lesion size on day 2 post-inoculation was used to investigate resistance to Botrytis. In the WT plants, necrotic lesions caused by B. cinerea were visible as early as 2 days post-inoculation (dpi), whereas the lesions at 2 dpi were limited in the 35S∷Hrip1 and rd29A∷Hrip1 lines. The average lesion diameter was 4.21 ± 1.32 mm in WT and 2.24 ± 0.40 and 0.24 ± 0.02 mm in the rd29A∷Hrip1 lines and 35S∷Hrip1 lines, respectively (Supplemental Table 3). Thus, the 35s∷Hrip1 and rd29A∷Hrip1-expressing plants exhibited resistance to Botrytis infection.
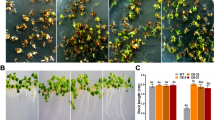
In a whole-plant assay, inoculum (2 × 105 spore mL−1) was used to challenge 3-week-old WT, 35S∷Hrip1, and rd29A∷Hrip1 plants, and resistance was determined by the percentage of surviving plants. Botrytis infection caused necrotic lesions in some leaf tissue at 2 days dpi, and the then lesions continuously spread, ultimately resulting in the death of some plants. In contrast, the 35S∷Hrip1 and rd29A∷Hrip1 plants showed small lesions and a reduction in disease incidence (Fig. 3a). At 4 dpi, the plant survival of WT, rd29A∷Hrip1, and 35S∷Hrip1 plants was 37.43, 53.22, and 63.47 %, respectively, with the rd29A∷Hrip1 and 35S∷Hrip1 rates 1.42 times and 1.67 times that of WT. After 13 days inoculation, survival rates of whole plants were significantly decreased. At 15 dpi, most of the wild type plants has died, only 4.79 % WT plants survived, whereas rd29A∷Hrip1 and 35S∷Hrip1 survival was 21.19 and 33.39 % (Fig. 3b). These results indicate that the potential of B. cinerea resistance in Arabidopsis was enhanced by expressing the Hrip1 gene, with the 35S∷Hrip1 plants exhibiting a greater resistance than the rd29A∷Hrip1 plants.
Growth of transgenic and wild-type Arabidopsis plants treated with Botrytis. a Disease symptoms of Arabidopsis plants at 0, 4, 11, and 13 days after inoculation with 2 × 105 Botrytis spores mL−1. b Plant survival rate at 4, 11, 13, and 15 days after inoculation. The error bars represent the SD from three biological replicates, and each replicate contained at least 30 plant leaves for each genotype. *, **, ***Significant differences at P < 0.05
Expression of the Hrip1 gene increases POD and CAT activities in Arabidopsis
Increased activities of antioxidant enzymes in plants are correlated with increased tolerance to salt, drought, and pathogen stresses (Liu and Zhu 1997; Shan et al. 2007; Huang et al. 2009; Zong et al. 2009; Ning et al. 2010; Zhu et al. 2013). Thus, POD and CAT activities were detected in Hrip1-transformed plants and WT plants for investigating responses to salt and drought stresses and Botrytis infection. The POD and CAT activities in the rd29A∷Hrip1 lines and 35S∷Hrip1 lines were higher than in WT, regardless of the treatments (Fig. 4a, b), indicating that Hrip1 expression resulted in enhanced resistance to biotic and abiotic stresses in Arabidopsis, partly due to increases in antioxidant enzyme activity.
POD and CAT activities in transgenic and wild-type Arabidopsis plants treated with NaCl, mannitol, and Botrytis. The enzyme activities were measured at 24 h after stress treatment. Each bar represents an average of three replicates, and the bars indicate the SD from three biological replicates. Different * indicate significant differences in comparison with WT at P < 0.05
Transcription of resistance-related genes in response to biotic and abiotic stresses
To elucidate the molecular mechanism by which Hrip1 expression enhances resistance to abiotic response in Arabidopsis, the expression of cold-responsive genes P5CS2 and DREB2A and drought-responsive genes NCED3 and LOS6 were monitored by real-time PCR. The tested marker genes exhibited increased expression levels in the Hrip1-expressing plants compared to WT under 200 mM NaCl or 200 mM mannitol treatment for 0, 1.5, and 10.5 h (Supplemental Figure 4a, b). The expression level of these genes in the rd29A∷Hrip1 lines was substantially higher than in the 35S∷Hrip1 lines, in agreement with the results of the salt and drought tolerance tests.
Plant defense in response to pathogen attack is regulated by a complex network of signaling pathways. The salicylic acid (SA)-dependent and jasmonic acid (JA)/ethylene (ET)-dependent pathways are two major pathogen defense signaling pathways; PR1 and BGL2 are marker genes in the SA pathway, and Thi2.1 and PR4 are marker genes in the JA/ET pathway. A real-time PCR analysis showed that PR1 and BGL2 were hardly induced in the wild-type and transgenic plants with Botrytis infection (Supplemental Figure 4c), whereas Thi2.1 and PR4 expression was up-regulated in the Hrip1-expressing plants compared to WT at 1.5 and 10.5 h after Botrytis inoculation (Supplemental Figure 4d). These results indicated that the JA/ET signaling pathways may be involved in the pathogen stress tolerance of Hrip1-transgenic plants. Furthermore, the expression level of these marker genes in the 35S∷Hrip1 lines were substantially higher than in the rd29A∷Hrip1 lines, consistent with the Botrytis stress resistance experiment outputs.
Discussion
Plants develop mechanisms to cope with and adapt to different types of abiotic and biotic stresses imposed by the frequently adverse environment. Plant responses to different stresses are highly complex and involve changes at the transcriptional, cellular, and physiological levels (Atkinson and Urwin 2012). In addition, microbial elicitors produced from plant pathogens during plant-pathogen interactions modulate plant defense systems and trigger plant defense responses (Hogenhout et al. 2009). In recent years, some of these protein elicitors have been purified from plant pathogens and have been shown to exhibit the ability to enhance plant resistance to pathogens or abiotic stress. In this study, the Hrip1 elicitor was successfully expressed as an exogenous gene in Arabidopsis under the control of the stress-inducible rd29A promoter and constitutive CaMV35S promoter. Our results suggest that the expression of Hrip1 in Arabidopsis both enhances plant disease resistance and also improves salt and drought tolerance. Our findings indicate that Hrip1 expression in Arabidopsis may be involved in inducing or reducing certain master regulators that connect both biotic and abiotic stress response pathways, potentially providing an opportunity for developing broad-spectrum stress-tolerant crop plants. Furthermore, the constitutively expressing plants showed strong resistance to biotic stress, whereas inducible expression of Hrip1 was more responsive to abiotic stress. This study offers an approach to generating stress-resistant crops through differential expression based on the specific requirements.
The overexpression of genes under a constitutive promoter partly prevents plant growth and reduces its production because of energy competition and decreased protein and RNA synthesis. Therefore, the use of a stress-inducible plant promoter is a trend for cultivating new varieties of crops with stress tolerance. The rd29A promoter is a drought and other stress-inducible promoter, the use of which improves stress tolerance and also eliminates the negative impact on growth. Many genes related to stress tolerance appear to be overproduced under the control of the CaMV35S promoter, even under control conditions; in contrast, such genes are rapidly overproduced only in response to stress treatment under the rd29A promoter (Kasuga et al. 1999). For example, the expression of Morus indica cv. K2 genes in tobacco using the stress inducible promoter rd29A produced transgenic plants that were able to tolerate salt and drought stresses more efficiently than plants with the constitutive CaMV35S promoter; however, in terms of Botrytis tolerance, transgenic plants with the Morus indica cv. K2 gene performed better than WT (Das et al. 2011). Our results are consistent with this previous report, though the expression of Hrip1 under the CaMV35S promoter did not result in a negative effect on plant growth. The Hrip1-transgenic Arabidopsis with the rd29A stress inducible promoter more efficiently tolerated salt and drought stresses than the transgenic plants with the CaMV35S constitutive promoter; in addition, the rd29A∷Hrip1 transgenic lines showed improved plant height, silique length, and biomass compared to the 35S∷Hrip1 lines. Nonetheless, the 35S∷Hrip1 transgenic plants performed better than the rd29A∷Hrip1 transgenic lines with Botrytis stress. We consider that the different biological effects may be due to the level of Hrip1 expression under the different promoters. We observed strong expression of the 35S∷Hrip1∷GUS fusion gene in the leaves and roots and weak expression of the rd29A∷Hrip1∷GUS fusion genes in the stamens, leaves, and siliques under normal growth conditions (data not shown). To address various attacks, plants have developed diverse adaptation strategies, including constitutive chemical mechanical barriers and inducible defense systems (Klarzynski et al. 2003; Thatcher et al. 2005). Abiotic stress leads to the accumulation of reactive oxygen species, which causes damage to cellular structures, particularly membranes. The overexpression of ZmMPK7 (Zea mays mitogen-activated protein kinase 7) in transgenic tobacco enhanced POD activity, which in turn protected against ROS-mediated injury during osmotic stress (Zong et al. 2009), and ZmMPK5 (Zea mays mitogen-activated protein kinase 5) enhanced the expression of the antioxidant genes CAT1, Capx, and GR1 and the total activities of the antioxidant enzymes CAT, ascorbate, POD, glutathione reductase, and superoxide dismutase (Lin et al. 2009; Zhang et al. 2006). In the present study, the POD and CAT activities were increased in the Hrip1-expressing plants compared to WT under both biotic and abiotic stress conditions. The POD and CAT activities were higher in rd29A∷Hrip1 than in 35S∷Hrip1 under abiotic stress, whereas the results were the opposite under biotic stress (Fig. 4). These results indicate the Hrip1-transgenic plants regulated tolerance to biotic and abiotic stresses with different adaptation strategies via different regulatory pathways.
Plant defense in response to pathogen attack apparently involves two major pathogen defense signaling pathways: an SA-dependent pathway and a JA/ET-dependent pathway (Kunkel and Brooks 2002). In this study, we observed the expression levels of four genes (PR1, BGL2, PR4, and Thi2.1) involved in SA- or JA/ET-mediated signal transduction in the Hrip1-expressing Arabidopsis (Supplemental Figure 4). Two marker genes (PR4 and Thi2.1) involved in the JA/ET signal pathway were markedly induced at 1.5 h and 10.5 h post-Botrytis treatment in the transgenic plants, whereas the relative expression BGL2 and PR1 involved in the SA signaling pathway was hardly increased at any time point. These results indicate that pathogen stress tolerance induced by Hrip1 may primarily involve in the JA/ET signaling pathway. However, the signaling pathway involved in Hrip1-mediated signal transduction remains unclear and needs to be further elucidated.
Abbreviations
- CaMV35S:
-
Cauliflower mosaic virus 35S promoter
- rd29A:
-
Promoter of desiccation-responsive rd29A gene in Arabidopsis thaliana
- POD:
-
Peroxidase
- CAT:
-
Catalase (Kong et al. 2011)
- dpi:
-
Days post inoculate
- hpi:
-
Hours post inoculate
- SA:
-
Salicylic acid
- JA/ET:
-
Jasmonates/ethylene
References
Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63:3523–3543
Beissmann B, Beisener HJ (1990) Isolation and purity determination of a glycoprotein elicitor from wheat stem rust by medium-pressure liquid chromatography. J Chromatogr A 521:187–197
Chen MJ, Zeng HM, Qiu DW, Guo LH, Yang XF, Shi HX, Zhou TT, Zhao J (2012) Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS One 7:e37654
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium: mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Das M, Chauhan H, Chhibbar A, Haq QMR, Khurana P (2011) High-efficiency transformation and selective tolerance against biotic and abiotic stress in mulberry, Morus indica cv. K2, by constitutive and inducible expression of tobacco osmotin. Transgenic Res 20:231–246
Ebel J (1998) Oligoglucoside elicitor-mediated activation of plant defense. BioEssays 20:569–576
Hogenhout SA, Van der Hoorn RAL, Terauchi R, Kamoun S (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22:115–122
Huang JG, Yang M, Liu P, Yang GD, Wu CA, Zheng CC (2009) GhDREB1 enhances abiotic stress tolerance, delays GA-mediated development and represses cytokinin signaling in transgenic Arabidopsis. Plant Cell Environ 32:1132–1145
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnol 17:287–291
Keller H, Pamboukdjian N, Ponchet M, Poupet A, Delon R, Verrier JL, Roby D, Ricci P (1999) Pathogen-induced elicitin production in transgenic tobacco generates a hypersensitive response and nonspecific disease resistance. Plant Cell 11:223–235
Klarzynski O, Descamps V, Plesse B, Yvin JC, Kloareg B, Fritig B (2003) Sulfated fucan oligosaccharides elicit defense responses in tobacco and local and systemic resistance against tobacco mosaic virus. Mol Plant Microbe Interact 16:115–122
Kong XP, Pan JW, Zhang MY, Xing X, Zhou Y, Liu Y, Li DP, Li DP (2011) ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant Cell Environ 34:1291–1303
Kulye M, Liu H, Zhang YL, Zeng HM, Yang XF, Qiu DW (2012) Hrip1, a novel protein elicitor from necrotrophic fungus, Alternaria tenuissima, elicits cell death, expression of defence-related genes and systemic acquired resistance in tobacco. Plant Cell Environ 35:2104–2120
Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5:325–331
Lin F, Ding HD, Wang JX, Zhang H, Zhang A, Zhang Y, Tan MP, Dong W, Jiang MY (2009) Positive feedback regulation of maize NADPH oxidase by mitogen-activated protein kinase cascade in abscisic acid signalling. J Exp Bot 60:3221–3238
Liu J, Zhu JK (1997) Proline accumulation and salt-stress induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol 114:591–596
Ning J, Li X, Hicks LM, Xiong L (2010) A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol 152:876–890
Nürnberger T (1999) Signal perception in plant pathogen defense. Cell Mol Life Sci 55:167–182
Nürnberger T, Scheel D (2001) Signal transmission in the plant immune response. Trends Plant Sci 6:372–379
Peng JL, Bao ZL, Ren HY, Wang JS, Dong HS (2004) Expression of harpinXoo in transgenic tobacco induces pathogen defense in the absence of hypersensitive cell death. Phytopathology 94:1048–1055
Qiu DW, Mao JJ, Yang XF, Zeng HM (2009) Expression of an elicitor-encoding gene from Magnaporthe grisea enhances resistance against blast disease in transgenic rice. Plant Cell Rep 28:925–933
Shan DP, Huang JG, Yang YT, Guo YH, Wu CA, Yang GD, Gao Z, Zheng CC (2007) Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol 176:70–81
Shao M, Wang J, Dean RA, Lin Y, Gao X, Hu S (2008) Expression of a harpin-encoding gene in rice confers durable nonspecific resistance to Magnaporthe grisea. Plant Biotechnol J 6:73–81
Takakura Y, Che FS, Ishida Y, Tsutsumi F, Kurotani K, Usami S, Isogai A, Imaseki H (2008) Expression of a bacterial flagellin gene triggers plant immune responses and confers disease resistance in transgenic rice plants. Mol Plant Pathol 9:525–529
Thatcher LF, Anderson JP, Singh KB (2005) Plant defence responses: what have we learnt from Arabidopsis? Funct Plant Biol 32:1–19
Zhang A, Jiang MY, Zhang JH, Tan MP, Hu XL (2006) Mitogen activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol 141:475–487
Zhang YH, Yang XF, Liu Q, Zhang YL, Zeng HM, Yuan JJ, Mao JJ (2010) Purification of novel protein elicitor from Botrytis cinerea that induces disease resistance and drought tolerance in plants. Microbiol Res 165:142–151
Zhu F, Zhang P, Meng YF, Xu F, Zhang DW, Cheng J, Lin HH, Xi DH (2013) Alpha-momorcharin, a RIP produced by bitter melon, enhances defense response in tobacco plants against diverse plant viruses and shows antifungal activity in vitro. Planta 237:77–88
Zong XJ, Li DP, Gu LK, Li DQ, Liu LX, Hu XL (2009) Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 229:485–495
Acknowledgments
We thank Dr. Jianping Yang (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China) for assistance with Arabidopsis planting. This work was supported by the National technology research and development program (863 program) project (Grant Nos. 2011AA10A205 and 2012AA101504), The State Key Laboratory for Biology of Plant Disease and Insect Pests, Institute of Plant protection, Chinese Academy of Agricultural science, No. 12 Zhong-guan-cun South Street, Beijing, China.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
11248_2014_9824_MOESM1_ESM.tif
Supplemental Fig. 1 Expression of Hrip1 in transgenic Arabidopsis a The transcript and expression levels of Hrip1 in the 35S∷Hrip1 and rd29A∷Hrip1 expression lines, as analyzed by PCR, RT-PCR, and western blotting. Total RNA and protein were extracted from WT plants and Hrip1-expressing plants grown under normal or salt conditions for 8 h. Different expression lines were analyzed by western blotting. All lines were analyzed using an unquantified concentration of protein extract. The protein is 17 kDa. b Representative leaves photographed at 8 dpi for GUS detection of rd29A∷Hrip1∷GUS and 35S∷Hrip1∷GUS transgenic plants and WT under control and stress conditions. The plants were grown on MS agar containing 25 µg mL−1 kanamycin and then exposed to salt for 8 h; GUS activity is shown in rosette plants (TIFF 2767 kb)
11248_2014_9824_MOESM2_ESM.tif
Supplemental Fig. 2 Germination of transgenic Arabidopsis on MS medium and MS supplemented with 75 mM NaCl and 50 mM mannitol. a Germination of seeds on medium at three days after stratification. b Germination rate of transgenic Arabidopsis under salt and drought stresses. The results are presented as the means and standard errors from three independent experiments; three lines of each construct were used. A total of 60 seeds of each line were used for each experiment (TIFF 4447 kb)
11248_2014_9824_MOESM3_ESM.tif
Supplemental Fig. 3 Relative root length of transgenic Arabidopsis. Phenotypes on MS medium supplemented with different concentrations of NaCl or mannitol. The relative root lengths were measured on day 7 after stratification (n = 90 for each condition), and photographs were taken. The results are presented as the means and standard errors from three independent experiments. * and **and *** indicate significant differences at P < 0.05 (TIFF 4276 kb)
11248_2014_9824_MOESM4_ESM.tif
Supplemental Fig. 4 Expression of stress tolerance-related genes in WT and Hrip1-expressing plants, as analyzed by qRT-PCR. Total RNA was extracted from 21-d-old plants grown under normal or stress treatment for 0, 1.5 h, and 10.5 h. Actin was used as an internal control. Each column represents an average of three replicates, and the error bars indicate the standard deviation. Different * indicate significant differences in comparison with WT at P < 0.05. a Expression of DREB2A and P5CS2 with 200 mM NaCl. b Expression of LOS6 and NCED3 with 200 mM mannitol. c Expression of PR1 and BGL2 with Botrytis. d Expression of PR4 and Thi2.1 with Botrytis (TIFF 9421 kb)
Rights and permissions
About this article
Cite this article
Peng, XC., Qiu, DW., Zeng, HM. et al. Inducible and constitutive expression of an elicitor gene Hrip1 from Alternaria tenuissima enhances stress tolerance in Arabidopsis . Transgenic Res 24, 135–145 (2015). https://doi.org/10.1007/s11248-014-9824-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-014-9824-x