Abstract
Conventional Agrobacterium-mediated plant transformation often produces a significant frequency of transgenic events containing vector backbone sequence, which is generally undesirable for biotechnology applications. We tested methods to reduce the frequency of transgenic plants containing vector backbone by incorporating genes into the backbone that inhibit the development of transgenic plants. Four backbone frequency reduction genes, bacterial levansucrase (sacB), maize cytokinin oxidase (CKX), Phaseolus GA 2-oxidase (GA 2-ox), and bacterial phytoene synthase (crtB), each expressed by the enhanced CaMV 35S promoter, were placed individually in a binary vector backbone near the left border (LB) of binary vectors. In transformed soybean plants, the lowest frequency of backbone presence was observed when the constitutively expressed CKX gene was used, followed by crtB. Higher backbone frequencies were found among the plants transformed with the GA 2-oxidase and sacB vectors. In some events, transfer of short backbone fragments appeared to be caused by LB readthrough and termination within the backbone reduction gene. To determine the effect of the backbone genes on transformation frequency, the crtB and CKX vectors were then compared to a control vector in soybean transformation experiments. The results revealed that there was no significant transformation frequency difference between the crtB and control vectors, but the CKX vector showed a significant transformation frequency decrease. Molecular analysis revealed that the frequency of transgenic plants containing one or two copies of the transgene and free of backbone was significantly increased by both the CKX and crtB backbone reduction vectors, indicating that there may be a correlation between transgene copy number and backbone frequency.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Binary vectors used for Agrobacterium-mediated plant transformation usually consist of a transfer DNA (T-DNA) flanked by right border (RB) and left border (LB) 25 bp imperfect repeats. A plant selectable marker gene and at least one gene of interest are usually contained within the T-DNA. Vector sequences outside the T-DNA are referred to as the vector “backbone”, which contains genetic elements such as bacterial replication origins and antibiotic resistance genes for maintenance in both E. coli and Agrobacterium. In the process of Agrobacterium-mediated transformation, the Agrobacterium virulence protein virD2 cleaves one strand of the border sequences to initiate the production of a single stranded T-DNA (T-strand) with virD2 covalently attached to its 5′ end (Zupan et al. 2000). Both the RB and the LB can initiate T-strand formation, but the RB is more efficient at initiation, possibly due to the presence of one or more cis-active T-DNA transfer enhancer sequences adjacent to the RB (van Haaren et al. 1987). The LB is usually the site of T-strand termination (van Hareen et al. 1987, 1988). The backbone sequence can be introduced into cells when the T-strand is not terminated at the LB, or when T-strand initiates from the LB. (Kononov et al. 1997; Ramanathan and Veluthambi 1995; Wenck et al. 1997; van der Graaff et al. 1996, Huang et al. 2004). Depending on the plant species, Agrobacterium strains, and vectors used, 20–80% of the transgenic plants may contain the backbone sequence. (De Buck et al. 2000; Hanson et al. 1999; Olhoft et al. 2004; Rommens et al. 2004). Although backbone transfer can actually be exploited to generate useful transgenic plants (Huang et al. 2004), in most cases vector backbone sequences are not necessary for the utility of the transgenic plants, and may not be desirable for some commercial applications.
Methods have been tested to reduce the frequency of transfer of backbone sequences during plant transformation. In one approach, multiple tandem LB repeats were used to suppress the transfer of vector backbone in rice transformation (Kuraya et al. 2004). In another approach, a plant expression cassette is placed in the vector backbone, where the expression of the gene provides a phenotype if the gene, i.e. the vector backbone, is transferred into the plant cell. In one example, the phytotoxic gene barnase, which encodes a general RNase activity, was inserted into the binary vector backbone and used to produce backbone-free transgenic plants (Hanson et al. 1999). In this case, primary transgenic cells containing backbone sequences could not develop due to the lethal effect of barnase expression, while transgenic plants without the backbone sequence containing the barnase gene developed normally. However, transformation frequency (TF) decrease was observed in tobacco and tomato (Hanson et al. 1999). In the same way, the cytokinin synthesis gene ipt from Agrobacterium tumefaciens was inserted into a binary vector backbone for potato transformation, which produced an abnormal stunted phenotype in backbone containing events with less efficient for backbone event elimination (Rommens et al. 2004).
We tested the impact of placing non-lethal genes that interfere with plant development in a binary vector backbone in soybean transformation experiments to reduce the frequency of transgenic plants containing backbone sequences. We selected genes whose expression would be expected to disrupt normal plant development and/or cause severe aberrant phenotypes such as dwarfism, less elongated shoots and unexpanded leaves. Thus, only the primary transformed cells without such genes in the vector backbone can develop into normal shoots.
We tested four genes: levensucrase (sacB) from Bacillus subtilis (Steinmetz et al. 1985), maize cytokinin oxidase (CKX, Huang et al. 2003), Phaseolus GA 2-oxidase (2-ox, Thomas et al. 1999) and Erwinia phytoene synthase (crtB, Shewmaker et al. 1999). All four of these genes have been reported to produce pleiotropic phenotypes in various plant species when they were constitutively expressed (Fray et al. 1995; Huang et al. 2003; Ye et al. 2001; Werner et al. 2003), suggesting they may be useful for backbone frequency reduction.
The sacB gene in many bacteria such as Bacillus spp., Erwinia spp. etc, encodes levansucrase, which is responsible for levan (neutral polyfructan) synthesis using sucrose as a substrate (See review by Cairns 2003). Tobacco or ryegrass plants expressing the sacB gene with a vacuole targeting sequence driven by CaMV 35S promoter showed stunted phenotype (Ye et al. 2001). The native sacB gene without the vacuole targeting sequence could only be expressed in a tissue-specific manner. In corn, the sacB expressing kernels disturbed grain filling in later stage and resulted in shrunken seeds with very low germination frequency (Caimi et al. 1996). In potato, the expression of the native sacB in tubers leads to smaller tubers (Röber et al. 1996). These results suggested that expression of the native sacB gene inhibits plant cell development. Overexpression of the CKX in plants results in cytokinin deficiency which led to stunted shoots with smaller apical meristems in tobacco (Werner et al. 2001), and diminished activity of the vegetative and floral shoot apical meristems and leaf primordial in Arabidopsis (Yang et al. 2003; Werner et al. 2003). Constitutive expression of a phytoene synthase causes dwarfism in tomato (Fray et al. 1995) and both dwarfism and infertility in rice (Burkhardt et al. 1997). Overexpression of GA oxidases created severe dwarfism in many plant species due to significant drop of GA level (Huang et al. 1998; Sakamoto et al. 2003).
We present results here using these four genes in a binary vector backbone to reduce the frequency of transgenic soybean plants that contain vector backbone sequences, and thus increase the frequency of quality plants in transgenic soybean populations.
Materials and methods
Binary vector construction
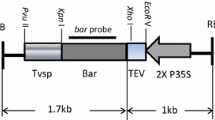
The four pleiotropic phenotype producing, or backbone reduction genes (BRGs), were cloned individually into an expression cassette with an enhanced CaMV 35S promoter (Kay et al. 1987) and a transcriptional terminator sequence from the nos gene (Depicker et al. 1982) according to standard methods (Sambrook et al. 1989). The resulting expression cassettes were further subcloned into a unique XhoI site adjacent to the LB region in the backbone of the control vector pMON83898 by blunt fragment ligation (Fig. 1). The T-DNA border regions were previously described by Horsch and Klee (1986). The RB region is a 357 bp DNA sequence that was originally isolated from A. tumefaciens plasmid pTiT37 (Depicker et al. 1982). The LB region is a 456 bp DNA sequence that was originally isolated from A. tumefaciens plasmid pTi15955 (Barker et al. 1983). The RK2 replicon oriV (Cross et al. 1986) is used for Agrobacterium maintenance and ori-322 for E. coli (Huang et al. 2004).
Five binary vectors used for soybean transformation. (1) pMON83898: the control vector with the selectable marker EPSPS-CP4 and gus genes. The XhoI site after the LB was used to insert the backbone reduction genes (BRGs). (2) pMON83907: the levensucrase (sacB) expression cassette was inserted into XhoI site of pMON83898. (3) pMON83908: the maize cytokinin oxidase (CKX) expression cassette was inserted into XhoI site of pMON83898. (4) pMON83912: the GA 2-oxidase (2-ox) expression cassette was inserted into XhoI site of pMON83898. (5) pMON83916: the Erwinia phytoene synthase (crtB) cassette was inserted into XhoI site of pMON83898. oriV: RK2 vegetative replication origin for vector maintenance in Agrobacterium; Ori-322: E. coli plasmid replication origin; aadA: spectinomycin resistance gene
The control vector pMON83898 contains the EPSPS-CP4 (Barry et al. 1992) gene conferring resistance to glyphosate driven by an FMV 35S promoter (Sanger et al. 1990) and terminated with a pea rbcS E9 polyadenylation signal (Schardl et al. 1987), as well as an intron-containing gus gene (Vancanneyt et al. 1990) driven by the enhanced CaMV 35S promoter (Kay et al. 1987) and terminated with the nos transcription terminator (Depicker et al. 1982).
Agrobacterium preparation and plant transformation
Agrobacterium tumefaciens strain ABI derived from the C58 strain (Koncz and Schell 1986) was used for soybean transformation. The binary vectors were transformed into the ABI strain by electroporation with the Bio-Rad Gene Pulser II according to the manufacturer’s instruction (Bio-Rad Laboratories, Hercules, CA). Soybean cultivar A3525 was used for Agrobacterium-mediated transformation as described (Martinell et al. 2002). Briefly, soybean seeds were imbibed for less than 14 hours at room temperature in Bean Germination Medium (BGM) (Martinell et al. 2002) and meristem explants were excised from imbibed seeds. The Agrobacterium strain containing each binary vector was grown in LB medium (Samsbrook et al. 1989) with 75 mg/l of spectinomycin and 50 mg/l of kanamycin in a gyratory shaker at 200 rpm for about 20–24 h at 28°C. The culture was spun down and directly re-suspended in inoculation medium (INO), which contains 2/5× of B5 macro elements and 1/10× of B5 micro elements (Gamborg et al. 1968), supplemented with 30 g/l glucose, 3.9 g/l MES, pH 5.4. The final optical density of Agrobacterium inoculum was adjusted to approximately 0.3 at OD660.
For each transformation experiment approximately 500 explants were placed in a Plantcon® (MP Biomedicals, Inc. Cat. No. 26-720-02) lid and mixed with 100 ml of Agrobacterium suspension and sonicated in a W-113 Sonicator for 20 s (Martinell et al. 2002). After sonication, the explants were co-cultured with Agrobacteria on a sterile filter paper within a Plantcon® for 2–4 days at 23°C. Explants were then transferred onto the surface of solid WPM medium (Lloyd and McCown 1980) purchased from Phytotech Lab (Shawnee Mission, KS, Cat. No. L449) with supplement of 20 g/l sucrose, 1.29 g/l calcium gluconate, 200 mg/l cefotaxime, 100 mg/l Ticarcillin, 200 mg/l carbenicillin, 12.68 mg/l glyphosate (Sigma, St. Louis, Missouri, Cat. No. PS 1051), and 4 g/l Agargel, pH 5.6. After 2 weeks, explants were transferred to the fresh WPM medium with the primary radicle inserted in the medium, and each Plantcon® contained about 25 explants.
Glyphosate resistant shoots, which were elongated with fully expanded trifolia, were identified 6–10 weeks post-inoculation. They were detached from original explants and transferred to Bean Rooting Medium (BRM) for rooting. The BRM contains 1/2× MS salts and 1× MS vitamins (Murashige and Skoog 1962) supplemented with 30 g/l sucrose, 100 mg/l cysteine, 0.1 mg/l IAA, 100 mg/l Ticarcillin and 4.2 mg/l glyphosate, pH 5.6 and solidified with 8 g/l washed agar. After 2–3 weeks on BRM, the rooted plantlets were transplanted to soil and grown in greenhouse for maturation and further analysis. The TF was defined as the number of EPSPS-CP4 positive plants/the number of initial explants.
Molecular analyses
Leaf samples of approximately 200 mg were collected from each plant. Total DNA was extracted as described by Dellaporta et al. (1983). The transgene copy numbers were estimated by Invader quantitative analysis using a probe specific to the EPSPS-CP4 gene according to the manufacturer’s instruction (http://www.twt.com/, Third Wave Technologies Inc. Madison, WI). The Invader DNA copy number assay was extensively validated previously by comparison with known copy samples and its accuracy is comparable to real time PCR (Taqman) or southern blot (Pielberg et al. 2003).
The presence of backbone sequence in transgenic plants was determined by Taqman assay using probes specifically for the oriV sequence or for LB sequence that is 3′ to the LB nicking site. In other experiments (data not shown) we tested the accuracy of the oriV Taqman assay results by the use of multiple probes across vector entire backbone in Southern blots. The Taqman assay result corresponded with presence of the entire backbone at least 95% of the time. Approximately 10 ng of genomic DNA was used for a qualitative Taqman reaction according to the manufacturer’s instructions (Applied Biosystems, Forster City, CA). The primers 5′ AACGCCTGATTTTACGCGAG 3′ (forward) and 5′ CAATACCGCAGGGCACTTATC 3′ (reverse), which amplify a 70 bp fragment and further detected with MGB (minor grove binding) Taqman probe 6-FAM-CCCACAGATGATGTGGAC, were used to analyze the oriV-containing backbone sequence. The LB primers 5′ GCACCCGGTGGAGCTT 3′ (forward) and 5′ TCTGCCTAACCGGCTCAGT 3′ (reverse), which amplify a 55 bp fragment, with the probe 6FAM-CATGTTGGTTTCTACGCAG were used to detect sequence 35 bp downstream from the LB nicking site.
The BRG presence was also analyzed by PCR. The following primer pairs were used to detect the BRG respectively: (1) 5′ GACTTCGGCAACATCACGTC 3′ (CKX forward) and 5′ AGGCTCTGGTTCACGAACAC 3′ (CKX reverse) to amplify a 712 bp CKX fragment in pMON83908; (2) 5′ GTCTCAGCCAGCATTGAACC 3′ (GA 2-ox forward) and 5′ ACCTGTAGAGCGTCACCAAC 3′ (GA 2-ox reverse) to amplify a 737 bp GA 2-ox fragment in pMON83912; (3) 5′ TAGCGTGCTGATGCTCTACAC 3′ (crtB forward) and 5′ AGGAGATGTAGTACGGCTCTG 3′ (crtB reverse) to amplify a 600 bp crtB fragment in pMON83916; (4) 5′ ACGGCACTGTCGCAAACTATC 3′ (sacB forward) and 5′ AGCTCAATCATACCGAGAGCG 3′ (sacB reverse) to amplify a 673 bp sacB fragment in pMON83907.
Experiment design and statistical analysis
The soybean transformation experiments were carried out in two sets: the first set including all four BRG constructs and the control vector was transformed individually to screen the most effective BRG genes and was designated as preliminary experiment in context, while the second set with two promising BRG constructs and the control vector was transformed in parallel with identical conditions and was designated as parallel experiment. Statistical analysis was performed with generalized linear model which appears to allow more flexibility in terms of analysis (http://www.ats.ucla.edu/STAT/sas/glimmix.pdf).
Results
Determination of BRG efficacy in preliminary experiments
The four BRG vectors and the control vector were used to transform soybean in multiple transformation experiments to determine the impact of the BRG on the frequency of plants containing vector backbone, as well as transgene copy number. In all BRG transformation experiments, no obviously aberrant shoots were observed. Normal shoots from the BRG containing vectors were harvested and rooted in the rooting medium in the same time frame as the control, indicating there is no delay with the BRG vector. Transgenic plants were screened for the presence or absence of the backbone sequence (Fig. 2). In preliminary experiments, the different construct transformations were conducted at different times (nonparallel experiments, Fig. 2). In the second set of experiments, all constructs were transformed at the same time under identical conditions (Fig. 3). The oriV sequence-specific probe (probe c, Fig. 2A) was used in a Taqman assay to detect the presence of backbone sequences that extend beyond the BRG gene. The BRG-specific fragment (probe b, Fig. 2A) was used in a PCR assay to detect the presence of backbone fragments that may not have extended beyond the BRG.
Molecular analysis of vector backbone frequency and copy number in transgenic soybean plants from preliminary experiments. (A) Three backbone assay probes in the vector backbone after the LB nicking site are indicated. a: LB 3′ Taqman probe detecting backbone sequence directly downstream of the LB nic site; b: BRG specific PCR primers detecting BRG presence; c: oriV Taqman probe detecting longer backbone sequence. (B) backbone frequency analysis in transgenic soybean plants transformed by the four BRG and the control vectors from the preliminary experiments. The vector backbone presence was analyzed by Taqman assay of oriV (probe c in A) and BRG specific PCR (probe b in A). (C) effect of BRGs in binary vector backbone on usable (1 or 2 copies, backbone-free) soybean transgenic plant frequencies. CK: control vector pMON83898; sacB: pMON83907; CKX: pMON83908; GA 2-ox: pMON83912; crtB: pMON83916. The percentage of backbone frequency in B or backbone-free frequency in C of transgenic soybean plants was indicated on the top of bars. *Statistically significant (P = 0.05)
Molecular analysis of vector backbone frequency and copy number in transgenic soybean plants from the in parallel transformation experiments. (A) Backbone frequency analysis in transgenic soybean plants transformed by the crtB or CKX BRG and the control vectors. The vector backbone presence was analyzed by Taqman assay of oriV (Fig. 2A probe c), BRG specific PCR (Fig. 2A probe b) or LB-specific Taqman probe (Fig. 2A probe a). (B) Effect of crtB or CKX BRGs in binary vector backbone on usable (1 or 2 copies, backbone-free) soybean transgenic plant frequencies. CK: control vector pMON83898; CKX: pMON83908; crtB: pMON83916. The percentage of backbone frequency in A or backbone-free frequency in B of transgenic soybean plants was indicated on the top of bars. * Indicating statistically significant (P = 0.05) from the control plasmid
As shown in Fig. 2B, the CKX, GA 2-ox or crtB constructs significantly reduced the frequency of transgenic plants with vector backbone when the oriV backbone fragment was probed. The CKX vector had the lowest frequency, with no oriV backbone-containing plants, followed by GA 2-ox (8.8%) and crtB vector (9.7%). The vector with sacB in backbone had the same backbone frequency as the control vector.
To determine if the shorter backbone fragments terminated before oriV were present, a second assay using PCR to test for the presence of the BRG (probe b, Fig. 2A) was conducted in the transgenic plants from the preliminary experiments. Transgenic plants containing BRG sequences but no oriV sequence were detected in transformants from three vectors (sacB, GA 2-ox and CKX). The frequencies observed for such plants were 13% (9 events out of 70) for the sacB vector, 15.6% (7 out of 45) for the GA 2-ox vector, 2.1% (2 events out of 92) for the CKX vector, and no such plants for the crtB vector after analyzing 31 plants (Fig. 2B). In the GA 2-ox and the sacB vector transformations, these additional plants with short vector backbone sequence significantly contributed to the total number of plants with backbone. The overall frequencies of the plants containing backbone with the oriV and/or BRG region were 24.4 and 33%, respectively, for GA 2-ox and sacB transformants. In addition, a few events that were BRG negative but oriV positive were also observed, albeit at lower frequencies: 3.4% (2/70) for the sacB, 4.4% (2/45) for the GA 2-ox and 3.2% (1/31) for the crtB construct.
Transgenic soybean plants from the preliminary experiments were subjected to the Invader assay to estimate the EPSPS-CP4 transgene copy number. As shown in Table 1, the frequencies of plants with low copy (1–2 copies) numbers for the EPSPS-CP4 transgene were 68.6% for sacB vector, 73.5% for the control, 75.6% for GA 2-ox, 74.7% for CKX and 83.9% for crtB.
The production of transgenic plants with low transgene copy number and absence of vector backbone sequences is important for basic research as well as many biotechnology applications. The most useful plants have 1–2 copies of the transgene, and are backbone-free. In the preliminary experiments, two BRG vectors, pMON83908 (CKX) and pMON83916 (crtB) showed frequencies of 74.7 and 83.9% respectively, of plants that meet that definition of useful plants, which was significantly higher (Fig. 2C) than the control vector with (63.3%). The other two BRG vectors, pMON83907 (sacB) and pMON83912 (GA 2-ox), had lower frequencies of useful plants due to significantly higher frequency of plants with backbone sequences, including both longer backbone fragments (oriV positive) and short backbone fragments (BRG positive but oriV negative) (Fig. 2C).
Determination of impact and BRG efficacy in parallel experiments
To further investigate the impact of the crtB and CKX BRG constructs on TF, frequency of plants with backbone sequences, and transgene copy number, we conducted additional transformation experiments, carrying out all steps for each construct in parallel, under identical conditions. Ten experiments were conducted with a total of approximately 5,000 explants for each vector. The average TF for crtB construct was 2.33 ± 0.76%, which was not significantly different from the TF of the control vector (2.80 ± 0.70%, P = 0.05). In contrast, the CKX vector resulted in a significant drop in TF (1.47 ± 0.31%, P = 0.05).
All plants generated from these experiments were assayed to determine the frequency of plants that contain vector backbone sequences. The oriV-specific probe (probe c, Fig. 2A) was used in the Taqman assay to detect the presence of long backbone sequences that extend into the oriV region. The probe specific for the BRGs (probe b, Fig. 2A) was used in PCR assays for detecting shorter backbone fragments that extended into the BRG, but terminated before oriV region. In these experiments, an additional probe, designed to detect backbone sequences adjacent to the LB nic site (probe a, Fig. 2A) was used in Taqman assay for detecting the presence of very short backbone fragments, beginning 18 bp from the nic site that may not have included the BRGs or the oriV region.
The results obtained from these experiments were consistent with the results from the preliminary transformation experiments. The frequency of plants with backbone sequences, determined with the oriV Taqman assay, decreased from 34.7% with the control vector to 4.2 and 6.8% with the CKX and the crtB BRG vectors, respectively (Fig. 3A). Only a few additional short LB 3′ events were observed in both crtB and CKX BRG constructs and the control vector (Fig. 3A).
Transgenic soybean plants from the parallel experiments were subjected to the Invader assay for estimates of the EPSPS-CP4 transgene copy number. Consistent results were observed with the CKX and crtB BRG constructs (Table 2) compared to the preliminary transformation experiments (Table 1), where the frequency of plants with 1 or 2 copies of the transgene for the crtB construct was very close to that in the control vector. However, the frequency of plants with a single copy of the transgenes (43.7%) was significantly higher for the CKX construct than the control and crtB constructs (P = 0.05).
In addition, in the latter comparison experiments, 1–2 copy, backbone-free plant frequencies for the CKX construct (67.6%) and the crtB construct (65.3%) were significantly higher than the control vector (52.3%) (Fig. 3B), which was consistent with previous non-parallel experiments.
Discussion
We tested the utility of inserting four pleiotropic phenotype-producing genes in the binary vector backbone to reduce the frequency of transgenic soybean plants that contain vector backbone sequences. All four BRG vectors produce transgenic soybean plants within the same timeframe. A binary vector containing the CKX gene was the most effective in reducing vector backbone frequency in soybean transformation. Cytokinin is an essential phytohormone for plant cell differentiation and plays a major role in plant morphogenesis. The CKX gene plays an opposite role in plant morphogenesis by catabolizing cytokine to fine tune the cytokine level for proper plant development (Armstrong 1994). Severe aberrant phenotypes were observed in CKX overexpressing plants (Werner et al. 2001, 2003; Yang et al. 2003). When the CKX gene is transferred into plant cells as part of the backbone sequences, it likely results in cytokinin deficiency in the transformed cells. Since the soybean transformation system used in our experiments is a shoot apical meristem-based organogenic system, which is highly dependent on endogenous cytokinin levels for shoot formation (Martinell et al. 2002), plants containing backbone sequence with the CKX expression cassette were presumably impaired in normal cell development by cytokinin depletion, and thus deficient in normal shoot formation.
The CKX binary vector resulted in unexpected TF decrease compared to the control vector. It seems unlikely that the TF drop with the CKX vector is due to the elimination of the events with the CKX backbone since the crtB BRG vector showed no significant TF drop even though the frequency of backbone-containing plants is the same as CKX vector. Frequently, many T-DNAs are transferred into multiple cells in the same meristem as we observed in gus transient expression assays (data not shown). It appeared that the CKX backbone T-DNAs not only degraded cytokinin in the transformed cells but also depleted cytokinin in the adjacent cells of the same meristem, which further inhibited shoot regeneration from the adjacent cells transformed with GOI alone. Evidence was shown that the CKX encodes a potentially secreted protein containing a putative transit peptide and glycosylation signal (Morris et al. 1999; Huang et al. 2003), which might indicate the CKX proteins were translocated into some adjacent cells.
The binary vector with the crtB gene in the backbone resulted in a TF equivalent to the control vector in soybean. It also resulted in a significant reduction in vector backbone frequency in transgenic soybean plants and thus an increase in the overall frequency of usable transgenic plants. Other studies also showed that constitutive expression of a phytoene synthase causes dwarfism in tomato (Fray et al. 1995) and both dwarfism and infertility in rice (Burkhardt et al. 1997). Phytoene synthase converts geranyl-geranyl diphosphate (GGDP) to phytoene and is a key enzyme for carotenoid biosynthesis. The precursor GGDP is also a key product for biosynthesis of gibberellins (GA), tocopherols, phytol, quinnones etc. GA is an important phytohormone for cell expansion and shoot elongation. Indeed, overexpression of a phytoene synthase in tomato resulted in 30-fold reduction in GA content in dwarf plants (Fray et al. 1995). It was proposed that the overexpression of the phytoene synthase depleted the GGDP precursor and thus down-regulated GA and phytol biosynthesis (Fray et al. 1995).
The importance of GA in shoot formation was further demonstrated with the GA 2-oxidase BRG construct. We observed a similar reduction in backbone frequency with this construct to the crtB construct if the plants with short backbone sequence (15.6% GA 2-ox PCR positive but oriV negative), i.e. those that probably did not contain an intact GA 2-ox gene, were excluded. The GA 2-oxidase oxidizes GA and thus regulates GA level to control plant cell development.
Unexpectedly, the use of the sacB gene in the binary vector backbone did not reduce the frequency of plants containing backbone sequences in this study. The high backbone frequency with the sacB BRG construct consisted of combinations of entire backbone (19%) and short backbone (12.7% sacB PCR positive but oriV negative). The sacB backbone-containing soybean transgenic plants could have resulted from low expression of the sacB gene. The localized regions of AT richness resembling plant introns, the presence of mRNA destabilizing ATTTA sequences and rare codon usage found in the sacB from Bacillus sp. were proposed to be associated with the low expression of the sacB gene in plants (Ye et al. 2001). It was suggested that codon optimization or another source of the sacB gene may improve the sacB gene expression in plants (Ye et al. 2001) and may thus enhance its utility for backbone reduction.
A significant number of transgenic plants with the short backbone containing the BRG but not oriV were recovered with the GA 2-oxidase and sacB constructs (12.7 and 15.6% for sacB and GA 2-ox construct, respectively), indicating that the T-DNA truncation occurred during T-DNA process or integration. The oriV sequences tested in the above experiments are positioned at 455 bp relative to the LB nic site in the control vector pMON83898 and at 3075 bp in the largest CKX BRG vector pMON83908. Since high frequency of transfer of short backbone sequences occurred in the same region of the two BRG plasmids, the results may suggest that T-DNA border-like sequences might be present in these regions. It was recently observed that plant genome derived border-like sequences could replace the RB and LB for Agrobacterium-mediated transformation (Rommens et al. 2004) and the Agrobacterium protein virD2 can recognize an oriT sequence from pTF-FC2 in an in vitro assay (Dube et al. 2004). Aligning the oriT and pTiC58 border sequences revealed a consensus TCCTG sequence adjacent to the nick site (Dube et al. 2004). Several TCCTG motifs are present between the BRG reverse primer and the oriV sequence, with one in nos transcriptional terminator that is part of all BRG cassettes. In addition, border nicking efficiency may be affected by flanking sequence AT content, and AT-rich upstream flanking is required for LB termination function (Rommens et al. 2004). Both sacB and GA 2-ox genes are AT-rich sequences containing 58.86 and 52.95% AT content, respectively, while CKX and crtB are GC-rich sequences with 30 and 34% AT content correspondingly. The upstream AT-rich sequences combined with the border-like consensus sequence may facilitate the termination of backbone transfer that occurs before oriV, especially for the sacB and GA 2-ox vectors. Whether there is indeed a cryptic border-like sequence present in these BRG cassettes remains to be determined. Removal of the potential border-like sequences in these BRG cassettes may eliminate events with short sequence transfer and increase the efficiency of overall backbone reduction.
Due to the significant short backbone presence in sacB and GA 2-ox BRG constructs, the rest backbone presence after the oriV backbone detection point may be questioned and more probes may be required for determining an ideal BRG for backbone frequency reduction. The short T-DNA presence is often due to T-DNA truncations occuring close to RB or LB during T-DNA process or integration. Middle T-DNA deletion is rare, which is very often associated with repeats by homologous recombination (Ye et al. unpublished observation) or potential cryptic border sequence, as indicated by plant derived T-DNA (Rommens et al. 2004). Intact over 20 kb backbone T-DNAs after LB are frequently integrated in the rice transformation with the single LB control vector (Kuraya et al. 2004). Our oriV detection represents at least 95% accuracy for entire backbone since multiple backbone probes including a probe before RB nicking site were extensively used to validate the detection in other earlier experiments (data not published).
The reliability of BRG strategy is not only dependent on the BRG expression, but it may also depend on the gene sequence context, which was not mentioned in previous studies (Hanson et al. 1999; Kuraya et al. 2004; Rommens et al. 2004). Furthermore, it is also dependent on the impact on TF. Potential negative effect on TF was observed in CKX gene and also in the lethal gene (Hanson et al. 1999).
Though there is minor difference of 1–2 copy frequency between the preliminary and the parallel experiments for CKX and crtB BRG constructs, the relative tendency of the 1–2 copy backbone-free frequency (Figs. 1 and 2) are consistently higher than the control, indicating that the backbone frequency reduction may help to increase the low copy usable plant frequency. The Invader copy number assay used in this research provides comparable accuracy to the real time Taqman or Southern blot within a 70–90% range for single copy transformants (Pielberg et al. 2003). The minor difference of single copy frequency between the preliminary and the parallel transformation experiments for CKX BRG construct (Tables 1 and 2) may be due to the assay discrepancy or transformation condition variation in the preliminary screening experiment, which was not performed in parallel.
In experiments with the control vector we observed a correlation between the presence of vector backbone and the presence of multiple copies of the transgene (data not shown). Transgenic plants with one or two copies of the transgene were approximately 90 and 80% backbone-free, respectively. The majority of backbone-containing plants contained multiple copies of the transgene. Thus, a reduction of the frequency of plants with vector backbone would be expected to increase the frequency of plants with low copy numbers of the transgene. This is indeed what we observed. The higher frequency of backbone-free plants with one or two copies in experiments using the CKX and crtB constructs is consistent with the fact that the multiple copy, backbone-containing events were efficiently eliminated in the two BRG constructs.
Previously the lethal gene, barnase, containing an intron was placed in the vector backbone for reducing backbone frequency in transgenic plants (Hanson et al. 1999). The Agrobacterium isopentenyl transferase (ipt) was also used in the vector backbone for backbone frequency reduction, which is dependent on the phenotype production with much less efficiency (Rommens et al. 2004). The CKX and crtB as BRGs as shown in this study provided an alternative method for vector backbone reduction in soybean and other crop transformation. Although the genes we used had the potential to form transgenic shoots with abnormal phenotypes, we did not use phenotypes as a screen for transgenic plants that did or did not contain vector backbone. The same criteria for advancement of putative transgenic shoots were used regardless of the construct. The CKX and crtB genes appear to block the development of shoots, so there was no requirement for additional time or resources to screen for backbone-free shoots based on phenotypes.
The use of non-lethal genes, such as CKX and crtB, to eliminate transgenic plants vector backbone sequences offers a potential advantage over the use of lethal genes. Depending on the gene used, a lethal gene may express in cells into which the vector backbone has been transformed, perhaps as part of an additional T-strand transferred to the plant cell, but has not and will not integrate. Such a potentially useful transformed cell would be eliminated by the use of a lethal gene, but may proliferate when a non-lethal gene is used. Indeed, transformation frequencies decreased in experiments that used the barnase gene (Table 1 in Hanson et al. 1999) for backbone reduction.
The use of multiple LB border elements has also been applied to reduce the frequency of vector backbone transfer in rice (Kuraya et al. 2004) to levels that are as low as the levels we have observed using a non-lethal gene in the backbone. We wanted to avoid possible complications arising from initiation of T-strands at the LB (Podevin et al. 2006) which could lead to transfer of short segments that initiate at the first LB and terminate at the second LB, which would be very difficult to detect.
Our strategies were designed to reduce the transfer of binary vector backbone in Agrobacterium-mediated transformation methods. Several other methods for delivery of DNA to plant cells exist, including direct DNA transfer methods using particle guns. Under optimal conditions these methods can efficiently produce transgenic plants with low copy number (Agrawal et al. 2005, also see Alterpeter et al. 2005 for recent review). Moreover, in the case of direct DNA transfer, it is relatively straightforward to purify transgene cassettes from vector backbone using, for example, gel purification prior to DNA delivery (Fu et al. 2000). Thus, the BRG strategy is not necessary for most direct DNA transfer methods.
References
Agrawal P, Kohli A, Twyman R, Christou P (2005) Transformation of plants with multiple cassettes generates simple transgene integration patterns and high expression levels. Mol Breed 16:247–260
Alterpeter A, Baisakh N, Bechy R, Bock B, Capell T, Christou P et al (2005) Particle bombardment and the genetic enhancement of crops: myths and realities. Transgenic Res 15:305–327
Armstrong DJ (1994) In: Mok DWS, Mok MC (eds) Cytokinins: chemistry, activity and function. CRC, Boca Raton, pp 139–154
Barker RF, Idler KB, Thompson DV, Kemp JD (1983) Nucleotide sequence of the T-DNA region from the Agrobacterium tumefaciens octopine Ti plasmid pTi15955. Plant Mol Biol 2:335–350
Barry G, Kishore G, Padgette S, Taylor M, Kolacz K, Weldon M, Re D, Eichholtz D, Fincher K, Hallas L (1992) In: Singh BK, Flores HE, Shannon JC (eds) Biosynthesis and molecular regulation of amino acids in plants. American Society of Plant Physiologists, pp 139–145
Burkhardt PK, Beyer P, Wünn J, Klöti A, Armstrong GA, Schledz M, von Lintig J, Potrykus I (1997) Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J 11:1071–1078
Caimi PG, McCole LM, Kerr PS (1996) Fructan accumulation and sucrose metabolism in transgenic maize endosperm expressing a Bacillus amyloliquefaciens sacB gene. Plant Physiol 110:355–363
Cairns AJ (2003) Fructan biosynthesis in transgenic plants. J Exp Bot 54:549–567
Cross MA, Warne SR, Thomas CM (1986) Analysis of the vegetative replication origin of broad-host-range plasmid RK2 by transposon mutagenesis. Plasmid 15:132–146
De Buck S, Wilde CD, Van Montague M, Depicker A (2000) T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol Breed 6:459–468
Dellaporta SL, Wood J, Hicks JB, (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Depicker A, Stachel S, Dhaese P, Zambryski P, Goodman HM (1982) Nopaline synthase: transcript mapping and DNA sequence. J Mol Appl Genet 1:561–573
Dube T, Kovalchuk I, Hohn B, Thomson JA (2004) Agrobacterium tumefaciens-mediated transformation of plants by the pTF-FC2 plasmid is sufficient and strictly dependent on the MobA protein. Plant Mol Biol 55:531–539
Fray RG, Wallace A, Fraser PD, Valero D, Hedden P, Bramley PM, Grierson D (1995) Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J 8:693–701
Fu XD, Duc LT, Fontana S, Bong BB, Tinjuangjun P, Sudhakar D, Twyman RM, Christou P, Kohli A (2000) Linear transgene constructs lacking vector backbone sequences generate low-copy-number transgenic plants with simple integration patterns. Transgenic Res 9:11–19
Gamborg OL, Miller RA, Ojima K (1968) Plant cell culture I. Nutrient requirement of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Hanson B, Engler D, Moy Y, Newman B, Ralston E, Gutterson N (1999) A simple method to enrich an Agrobacterium-transformed population for plants containing only T-DNA sequences. Plant J 19:727–734
Horsch RB, Klee HJ (1986) Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: Role of T-DNA borders in the transfer process. PNAS 12:4428–4432
Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM (1998) Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol 118:773–781
Huang S, Cerny RE, Qi Y, Bhat D, Aydt CM, Hanson DD, Malloy KP, Ness LA (2003) Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol 131:1270–1282
Huang S, Gilbertson L, Adams T, Malloy K, Reisenbigler E, Birr D, Snyder M, Zhang Q, Luethy M (2004) Generation of marker-free transgenic maize by regular two-border Agrobacterium transformation vectors. Transgenic Res 13:451–461
Kay R, Chan A, Daly M, McPherson J (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236:1299–1302
Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396
Kononov ME, Bassuner B, Gelvin SB (1997) Integration of T-DNA binary vector ‘backbone’ sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J 11:945–957
Kuraya Y, Ohta S, Fukuda M, Hiei Y, Murai N, Hamada K, Ueki T, Imaseki H, Komari T (2004) Suppression of transfer of non-T-DNA ‘vector backbone’ sequences by multiple left border repeats in vectors for transformation of higher plants mediated by Agrobacterium tumefaciens. Mol Breed 14:309–320
Lloyd G, McCown B (1980) Commercially feasible micropropagation of mountain laural (Kalmia latifolia) by use of shoot tip culture. Proc Int Plant Prop Soc 30:421–427
Martinell B, Julson LS, Emler CA, Huang Y, McCabe DE, Williams EJ (2002) Soybean Agrobacterium transformation method. US Patent # US6384301
Morris RO, Bilyeu KD, Laskey JG, Cheikh NN (1999) Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem Biophys Res Commun 255:328–333
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Olhoft PM, Flagel LE, Somers DA (2004) T-DNA locus structure in a large population of soybean plants transformed using the Agrobacterium-mediated cotyledonary-node method. Plant Biotechnol J 2:289–300
Pielberg G, Day AE, Plastow GS, Andersson L. (2003) A sensitive method for detecting variation in copy numbers of duplicated genes. Genome Res 13:2171–2177
Podevin N, De Buck S, De Wilde C, Depicker A. (2006) Insights into recognition of the T-DNA border repeats as termination sites for T-strand synthesis by Agrobacterium tumefaciens. Transgenic Res 15:557–571
Ramanathan V, Veluthambi K (1995) Transfer of non-T-DNA portions of the Agrobacterium tumefaciens Ti plasmid pTiA6 from the left terminus of TL-DNA. Plant Mol Biol 28:1149–1154
Röber M, Geider K, Müller-Röber B, Willmitzer L (1996) Synthesis of fructans in tubers of transgenic starch-deficient potato plant does not result in an increased allocation of carbohydrates. Planta 199:528–536
Rommens CM, Humara JM, Ye J, Richael C, Zhang L, Perry R, Swords K (2004) Crop improvement through modification of the plant’s own genome. Plant Physiol 135:421–431
Sakamoto T, Morinaka Y, Ishiyama K, Kobayashi M, Itoh H, Kayano T, Iwahori S, Matsuoka M and Tanaka H (2003) Genetic manipulation of gibberellin metabolism in transgenic rice. Nat Biotechnol 21:909–913
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbour Laboratory Press, New York
Sanger M, Daubert S, Goodman RM (1990) Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol 14:433–443
Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DF, Hunt AG (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61:1–11
Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY (1999) Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J 20:401–412
Steinmetz M, Le Coq D, Aymerich S, Gonzy-Treboul G, Gay P (1985) The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol Gen Genet 200:220–228
Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. PNAS 96:4698–4703
Vancanneyt G, Schmidt R, O’Connor-Sanchez A, Willmitzer L, Rocha-Sosa M (1990) Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Genet Genomics 220:245–250
van der Graaff E, den Dulk-Ras A, Hooykaas PJJ (1996) Deviating T-DNA transfer from Agrobacterium tumefaciens to plants. Plant Mol Biol 31:677–681
van Haaren MJ, Sedee NJ, Schilperoort RA, Hooykaas PJJ (1987) Overdrive is a T-region transfer enhancer which stimulates T-strand production in Agrobacterium tumefaciens. Nucleic Acids Res 15:8983–8997
van Haaren MJJ, Sedee NJA, Krul MJT, Schilperoort RA, Hooykaas PJJ (1988) Function of heterologous and pseudo border repeats in T-region transfer via the octopine virulence system of Agrobacterium tumefaciens. Plant Mol Biol 11:773–781
Wenck A, Czakó M, Kanevski I, Márton L (1997) Frequent collinear long transfer of DNA inclusive of the whole binary vector during Agrobacterium-mediated transformation. Plant Mol Biol 34:913–922
Werner T, Motyka V, Strand M, Schmülling T (2001) Regulation of plant growth by cytokinin. PNAS 98:10487–10492
Werner T, Motyka V, Laucou V, Smets R, van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15:2532–2550
Yang S, Yu H, Xu Y, Goh CJ (2003) Investigation of cytokinin-deficient phenotypes in Arabidopsis by ectopic expression of orchid DSCKX1. FEBS Lett 555:291–296
Ye X, Wu X, Zhao H, Frehner M, Nösberger N, Potrykus I, Spangenberg G (2001) Altered fructan accumulation in transgenic Lolium multiflorum plants expressing a Bacillus subtilis sacB gene. Plant Cell Rep 20:205–212
Zupan J, Muth TR, Draper O, Zambryski P (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J 23:11–28
Acknowledgments
The authors thank the Monsanto Middleton Soybean Transformation Team for transgenic plant production, the Middleton Trait Development team for greenhouse care, Drs. D. Somers and Y. Wan for critical reading manuscript and stimulating discussion, C. Lawson for helpful suggestions, and C. Marquez for statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ye, X., Williams, E.J., Shen, J. et al. Plant development inhibitory genes in binary vector backbone improve quality event efficiency in soybean transformation. Transgenic Res 17, 827–838 (2008). https://doi.org/10.1007/s11248-008-9169-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-008-9169-4