Abstract
Mineral nutrition is a very important factor in the success of in vitro plant cultures. The aim was to compare the predictive capacity of the models obtained using a parametric technique such as multiple regression analysis with a nonparametric one such as artificial neural networks. These techniques were used for modeling the effect of total nitrogen concentration and the ratio nitrate: ammonium in the regeneration rate, oxidation rate, callus proliferation rate, number of buds per explant and buds-forming capacity index. Both the concentration of total nitrogen and the relationship between the concentrations of nitrate and ammonium influenced the morphogenetic responses. Optimal buds regeneration was in the range of 10–20 mM of the total nitrogen concentration and 1–2 of the nitrate: ammonium ratio. Higher concentrations of nitrogen produced an increase in the oxidation rate while the low nitrate: ammonium ratio favored the callus proliferation rate. Artificial neural network models presented a better precision to predict the different responses to the total content of nitrogen and the nitrate: ammonium rate, with higher coefficients of determination and correlation. They also presented a lower root mean square error for all the variables studied than the multiple regression analysis.
Key message
The use of artificial neural networks allows obtaining a better model of the effect of nitrogen on the organogenesis of Pinus taeda L. than traditional statistical techniques.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrogen is one of the main factors limiting the growth and development of plants among nutrients. Many essential compounds for the life of the plant are composed of this element, such as proteins, nucleic acids and chlorophyll, among others. Inorganic nitrogen, such as nitrate and ammonium, is incorporated into organic compounds that are used by the plant cell. In typical regeneration media, nitrogen is present in a greater proportion in the ionic forms of ammonium and nitrate (Ramage and Williams 2002) The optimal total nitrogen concentration (TNC) and nitrate: ammonium rate (NO3/NH4) in the medium depend on the type of tissue, the genetic material and the incubation conditions (Poothong and Reed 2016).
To understand the effect of a factor on a process it is necessary to create a model that allows predicting the value of the response from values of the independent variables. Multiple regression analysis (MRA) is a statistical technique for estimating the relationship between a dependent variable and independent variables, and formulates a linear relation equation between these variables (Uyanik and Guler 2013).
Another option that has been little used to model in vitro processes in plants are artificial neural networks (ANNs). It is a non-parametric technique, so it does not require assumptions such as normality and linearity, being able to detect non-linear effects that with other statistical techniques such as multiple regression could not be determined.
ANNs are computational models that manage to obtain and store information from processing units (artificial neurons) with multiple interconnections (Da Silva et al. 2017). Among the types of neural networks, there is a model called multilayer perceptrons that is characterized because the interconnection of neurons is created by feedback trained with the backpropagation algorithm. These networks learn to transform input data (independent variables) in a given response (dependent variable) (Panchal et al. 2011). The input layer is composed of input variables in separate neurons, while the output layer consists of the response variables. Between the input and output layer are the layers called hidden layers, which contain a variable number of interconnected neurons and a constant neuron related to the intercept synapses, which are not directly influenced by any input variable (Günther and Fritsch 2010).
The aim of this work was to compare the predictive capacity of the models obtained by multiple linear regressions and artificial neural networks on Pinus taeda in vitro organogenesis processes.
Materials and methods
Vegetal material consisted in mature zygotic embryos collected and isolated from clonal seed orchard of Pinus taeda L. (Livingston Parish) located at the geographical coordinates 27°59′ 0.4′S 55 58′ 6′W. Half strength Murashige and Skoog (1962) semisolid (agar 6.5 g L−1) medium with different TNC (5, 10, 20 and 30 mM) and NO3/NH4 (0.5, 1, 2 and 3) (Table 1), and supplemented with sucrose (30 g L−1), thidiazuron (0.45 mM), and 6-benzylaminopurine (0.44 mM) was used.
The isolated mature embryos were incubated under light (116 µmol m−2 s−1 PPFD, 14 h photoperiod) and temperature (27 ± 2 °C) controlled conditions. After 35 days of incubation the regeneration rate, oxidation rate, callus proliferation, number of buds originated per explant and bud-forming capacity (BFC) index were measured. BFC index was calculated as follows:
A factorial experimental design completely randomized with 16 treatments, five repetitions and an experimental unit of ten explants was used. MRA and ANNs were used to modeling the nitrogen effect, three repetitions were used for modeling and two to test the model. R 3.0.2 program (R Core Team 2013) with “MASS” (Venable and Ripley 2002) and “rsm” (Lenth 2009) packages were used for MRA and “neuralnet” (Fritsch et al. 2016) to build a multilayer perceptron neural network. In MRA, starting from a cubic model (formula 1), it was simplified using only the statistically significant terms (p < 0.05), when NAR is the NO3/NH4 and TNC is the total nitrogen concentration in mM. Box-Cox transformations were applied to all the variables because they did not comply with the assumptions for parametric analysis (normal distribution and homogeneous variance) (Box and Cox 1964).
Residuals vs fitted, normal Q-Q, scale-location and residuals vs leverage plots were used to diagnose the nature of the variables, such as normal distribution, homogeneity of variance and linearity.
Neural networks were made with a layer input with two neurons (one per independent variable), a hidden layer with three neurons and a neuron output (the response variable that was modeled) (Fig. 1). Previously the variables were transformed to a scale of between 0 for the minimum values and 1 for the maximum values.
To evaluate the predictive capacity of the models was used the coefficient of determination (R2), Pearson’s correlation coefficient (r) and the root mean square error (RMSE) using values that were not used to generate the models, which were calculated using the following formulas:
where Yi are the experimental values to evaluate the model, Yp is the corresponding data predicted, \({\overline {{\text{Y}}} _{\text{i}}}\) is the mean value of experimental data and n is the number of the experimental data.
Results and discussion
Both the TNC and the NO3/NH4 influenced the morphogenetic responses (Fig. 2). The numbers represent the treatments described in Table 1. It was observed that treatments with low TNC and high NO3/NH4 favored the formation of calluses, while high TNC and NO3/NH4 decreased the regeneration of the buds. Influences of TNC and NO3/NH4 on the induction and differentiation of plant cell cultures have been reported for some in vitro systems (Kovalchuk et al. 2018; Poothong and Reed 2016; Wada and Reed 2015).
Table 2 shows the regression coefficients, significance (based on a t-test), determination coefficients (R2) and adjusted R2 for the models obtained by MRA.
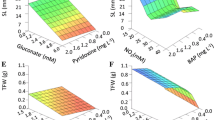
Optimal buds regeneration was in the range of 10–20 mM of TNC and 1–2 of NO3/NH4 in MRA and ANNs models (Figs. 3, 4).
Higher concentrations of nitrogen produced an increase in the oxidation rate (Figs. 5, 6), while the low NO3/NH4 favored the formation of calluses (Figs. 7, 8), to the detriment of the production of buds.
The number of buds per explant was greater in the range of 1–2 of NO3/NH4 in both models, and with TNC between 20 and 30 mM for MRA, while for ANNs it was greater between 10 and 20 mM (Figs. 9, 10).
BFC index was higher in the range of 10–20 mM of TNC and 1–2 of NO3/NH4 respectively in both models (Figs. 11, 12).
The relation NO3/NH4 present in the culture medium affects the activity of the growth regulators, and that the requirement of cytokinins for the meristematic activity is lower when the content of reduced nitrogen is reasonably high (George et al. 2008).
Residual vs fitted plot is used to detect nonlinearity and unequal error variations. Normal quantile–quantile graph (normal Q-Q) is a graphical technique to determine if the variable has a normal distribution. Scale-location plot shows the square root of the standardized residuals as a function of the fitted values. Residuals versus leverage plot help to identify influential data points in the model. Regeneration and oxidation rates transformed completely met the requirements of parametric analysis. This is observed in the residual vs fitted plots which results in a horizontal line close to 0 and in the distribution of the points in the normal Q-Q plot, which means that they have a normal distribution, homogeneity of variances and linearity. Transformed callus proliferation rate, number of buds and BFC index had no normal distribution, homogeneity of variance and linearity.
Table 3 shows the observed values, the values predicted by both models, coefficients of determination (R2), Pearson’s correlation coefficients (r) and root mean square errors (RMSE) for all the evaluated variables obtained with the test values. Models obtained by ANNs for the regeneration, callus proliferation, number of bud per explant and BFC index had high r (> 0.9) and R2 (> 0.8), while the oxidation rate showed a very low R2 for both models (< 0.3). For all the variables evaluated, r and R2 were higher while RMSE was lower in the models obtained by ANNs than those obtained from MRA. R2 is widely used to understand the sources of variation, since it represents the proportion of the variance explained by a given model (Nakagawa et al. 2017). On the other hand, the correlation is a measure of the association between two variables, which can be positive or negative. One of the ways to measure the correlation between variables is through the Pearson correlation coefficient (Emerson 2015). When the values of RMSE are smaller, the greater the prediction capacity of the model, because the difference between the values predicted by the model and the values observed in the experiment is smaller. This indicates that the models obtained by ANNs have better predictive capacity, since the predicted values are closer to those observed. It is also interesting to note that the models for regeneration and oxidation that comply with the assumptions of the parametric analysis have a similar prediction capacity with MRA and ANNs methodology, while the prediction capacity is notably greater in ANNs models for variables with a non-linear nature.
Biological processes, such as organogenesis, are non-linear in nature due to their complexity, since they depend on multiple factors and their interactions (Gallego et al. 2011). Various nonparametric analysis were used for mineral optimization in vitro cultures such as Chi-squared automatic interaction detection (CHAID) analysis (Akin et al. 2017), Classification and Regression Tree (CART) analysis (Kovalchuk et al. 2017) and Neurofuzzy logic (Alanagh et al. 2014).
Sarve et al. (2015) compared the prediction capacity of response surface models (RSM) and ANNs for the synthesis of biodiesel from sesame oil, who concluded that ANNs presented better prediction capacity with higher R2, and lower RMSE. Moreover, Astray et al. (2016) compared the models obtained by RSM and the ANN methodology to optimize the production of mixtures of oligosaccharides from sugar beet pulp. The ANNs models improved the RSM models between 5.58 and 61.78%.
Gago et al. (2010) came to the same conclusion using traditional statistical analysis and ANNs methodology in the proliferation of kiwis in vitro. ANNs methodology is easy to use and does not require assumptions such as traditional statistical analysis (regression analysis and ANOVA for example) and allows modeling using a limited number of experiments.
Other advantages offered by ANNs over traditional statistical analysis are the ability to process many types of data at the same time (continuous, discrete, binomial variables) that allows complex models and does not require a specific experimental design allowing the use of data generated previously (Gallego et al. 2011).
In conclusion, both TNC and NO3/NH4 influenced the morphogenetic responses and artificial neural network models presented a better precision to predict the different responses, with higher coefficients of determination and correlation. They also presented a lower root mean square error for all the variables studied.
Abbreviations
- ANNs:
-
Artificial neural networks
- BFC:
-
Bud-forming capacity
- MRA:
-
Multiple regression analysis
- NO3/NH4:
-
Nitrate: ammonium rate
- r:
-
Pearson’s correlation coefficient
- R2 :
-
Coefficient of determination
- RMSE:
-
Root mean squared error
- TNC:
-
Total nitrogen concentration
References
Akin M, Eyduran E, Reed BM (2017) Use of RSM and CHAID data mining algorithm for predicting mineral nutrition of hazelnut. Plant Cell Tissue Organ Cult 128(2):303–316
Alanagh EN, Garoosi GA, Haddad R, Maleki S, Landín M, Gallego PP (2014) Design of tissue culture media for efficient Prunus rootstock micropropagation using artificial intelligence models. Plant Cell Tissue Organ Cult 117(3):349–359
Astray G, Gullón B, Labidi J, Gullón P (2016) Comparison between developed models using response surface methodology (RSM) and artificial neural networks (ANNs) with the purpose to optimize oligosaccharide mixtures production from sugar beet pulp. Ind Crops Prod 92:290–299
Box GE, Cox DR (1964) An analysis of transformations. J R Stat Soc. Ser B (Methodol) 26:211–252
Da Silva IN, Spatti DH, Flauzino RA, Liboni LHB, dos Reis Alves SF (2017) Artificial neural networks. Springer International Publishing, Cham
Emerson RW (2015) Causation and Pearson’s correlation coefficient. J Vis Impair Blind 109(3):242–244
Fritsch S, Guenther F, Guenther MF (2016) Package ‘neuralnet’. The Comprehensive R Archive Network
Gago J, Martínez-Núñez L, Landín M, Gallego PP (2010) Artificial neural networks as an alternative to the traditional statistical methodology in plant research. J Plant Physiol 167(1):23–27
Gallego PP, Gago J, Landín M (2011) Artificial neural networks technology to model and predict plant biology process. In: Artificial neural networks-methodological advances and biomedical applications. InTech, London
George EF, Hall MA, De Klerk GJ (2008) The components of plant tissue culture media I: macro-and micro-nutrients. In: Plant propagation by tissue culture. Springer, Dordrecht, pp 65–113
Günther F, Fritsch S (2010) neuralnet: Training of neural networks. R J 2(1):30–38
Kovalchuk IY, Mukhitdinova Z, Turdiyev T, Madiyeva G, Akin M, Eyduran E, Reed BM (2017) Modeling some mineral nutrient requirements for micropropagated wild apricot shoot cultures. Plant Cell Tissue Organ Cult 129(2):325–335
Kovalchuk IY, Mukhitdinova Z, Turdiyev T, Madiyeva G, Akin M, Eyduran E, Reed BM (2018) Nitrogen ions and nitrogen ion proportions impact the growth of apricot (Prunus armeniaca) shoot cultures. Plant Cell Tissue Organ Cult 133(2):263–273
Lenth RV (2009) Response-surface methods in R, using rsm. J Stat Softw 32(7):1–17
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia plantarum 15(3):473–497
Nakagawa S, Johnson PC, Schielzeth H (2017) The coefficient of determination R2 and intra-class correlation coefficient from generalized linear mixed-effects models revisited and expanded. J R Soc Interface 14(134):20170213
Panchal G, Ganatra A, Kosta YP, Panchal D (2011) Behaviour analysis of multilayer perceptronswith multiple hidden neurons and hidden layers. Int J Comput Theory Eng 3(2):332
Poothong S, Reed BM (2016) Optimizing shoot culture media for Rubus germplasm: the effects of NH4+, NO3−, and total nitrogen. In Vitro Cell Dev Biol-Plant 52(3):265–275
R Core Team (2013) R: a language and environment for statistical computing. Vienna. http://www.R-project.org/. Accessed 8 Jun 2018
Ramage CM, Williams RR (2002) Mineral nutrition and plant morphogenesis. In Vitro Cell Dev Biol-Plant 38(2):116–124
Sarve A, Sonawane SS, Varma MN (2015) Ultrasound assisted biodiesel production from sesame (Sesamum indicum L.) oil using barium hydroxide as a heterogeneous catalyst: comparative assessment of prediction abilities between response surface methodology (RSM) and artificial neural network (ANN). Ultrason Sonochem 26:218–228
Uyanık GK, Güler N (2013) A study on multiple linear regression analysis. Proc-Soc Behav Sci 106:234–240
Venables WN, Ripley BD (2002) Modern Applied Statistics with S. 4th edn, Springer, New York. ISBN 0-387-95457-0
Wada S, Reed BM (2015) Trends in culture medium nitrogen requirements for in vitro shoot growth of diverse pear germplasm. In: VI International Symposium on production and establishment of micropropagated plants 1155 (pp. 29–36)
Acknowledgements
I thank the Botanical Institute of the Northeast (IBONE-UNNE-CONICET) for supporting this work and the forest company “Bosques del Plata S.A.” for supplying the seeds.
Author information
Authors and Affiliations
Contributions
BJO designed the experiment, executed it and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interests.
Additional information
Communicated by M. Paula Watt.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Barone, J.O. Use of multiple regression analysis and artificial neural networks to model the effect of nitrogen in the organogenesis of Pinus taeda L.. Plant Cell Tiss Organ Cult 137, 455–464 (2019). https://doi.org/10.1007/s11240-019-01581-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-019-01581-y