Abstract
The high-affinity potassium transporter (HKT) genes have crucial roles in the regulation of sodium and potassium transportation in many species, however little is known about maize HKT genes. In this study, we obtained two alternative splicing transcripts of ZmHKT1;1 gene. One was named as ZmHKT1;1a which has the intact coding sequence, and the other was named as ZmHKT1;1b which has a deficiency of the third exon and a retention of the second intron. The phylogenic tree analysis showed that both translation products of ZmHKT1;1a and ZmHKT1;1b belong to group I HKT proteins which prefer for Na+ transport than other cations. ZmHKT1;1a and ZmHKT1;1b showed different response to stress treatment in maize. Overexpressing ZmHKT1;1a or ZmHKT1;1b in transgenic tobacco plants conferred high salt tolerance by increasing root length and fresh weight of plants. When treated with high concentration of salt, transgenic tobacco plants manifested a trend of reduced Na+ content and increased K+ content in both shoot and root, suggesting that ZmHKT1;1 may involve in Na+ unloading and indirectly affect other transporter activity. It was also found that overexpression of ZmHKT1;1a and ZmHKT1;1b caused different expression of stress-related genes. The results in this study indicate that two alternative splicing variants of ZmHKT1;1 might be useful for the development of salt-tolerant transgenic crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinity is one of the major factors affecting agriculture and crop productivity. Deleterious effects of salinity on plants tissues are mainly divided into three aspects: osmotic stress, ion toxicity and nutrient deficiencies (Munns and Tester 2008). To cope with the negative effects of high sodium levels, plants have evolved a series of strategies, including sodium exclusion and/or sodium intracellular compartmentalization into the large cell vacuoles. Na+/H+ antiporters (NHXs) could mediated the sodium exclusion and the NHXs from several plant species have been shown to enhance salt tolerance (Apse et al. 1999; He et al. 2005; Patel et al. 2015; Mishra et al. 2015). By these strategies, plants could maintain Na+/K+ balance by limiting the over-accumulation of sodium in the cytosol (Hasegawa et al. 2000).
The high-affinity potassium transporter (HKT) genes, belonging to the Trk/Ktr/HKT transporter family, have been shown to play a crucial role in the regulation of sodium and potassium transportation in many species (Corratgé-Faillie et al. 2010). Since the first discovery of TaHKT2;1 in Triticum aestivum (Schachtman and Schroeder 1994), more and more HKT transporters from different species have been found. The HKT transporters have four pore domains (PDs), and HKTs are classified into two sub-families according to the amino acid difference in the first PD (Platten et al. 2006). Sub-family 1 HKTs exhibit a preference for sodium permeability; whereas members of sub-family 2 could select either sodium and/or potassium according to their external concentrations.
Both sub-family 1 and sub-family 2 HKTs have been shown to be able to enhance the salt tolerance of plants by overexpression in transgenic plants. Arabidopsis genome has only one HKT gene, AtHKT1;1, which can mediate the sodium exclusion from leaves under salt stress condition (Horie et al. 2009). It was shown that specifically expression of AtHKT1;1 in the mature root stele of Arabidopsis decreased the Na+ accumulation, whereas constitutively expression of AtHKT1;1 driven by 35S promoter led to a high accumulation of Na+ (Moller et al. 2009). When AtHKT1;1 was specifically expressed in the root cortical and epidermal cells, transgenic plants also had significantly improved Na+ exclusion (Plett et al. 2010). Transgenic barley plants overexpressing HvHKT2;1 could tolerate high concentration of salt (Mian et al. 2011). In these studies, the salt tolerance of transgenic plants are correlated with the significantly lower accumulation Na+ than WT plants.
Compared to only one HKT gene in dicot Arabidopsis, monocotyledon species have a much larger number of HKT genes. It was shown that wheat genome have five to eleven HKT genes (Huang et al. 2006) and there are at least nine HKT genes in rice (Garciadeblas et al. 2003). Recently, four sorghum HKT genes were identified (Wang et al. 2014). OsHKT1;5 was identified by map-based cloning of the SKC1 QTL locus in a segregating population, and the physiological analysis showed that OsHKT1;5 is involved in Na+/K+ homeostasis under salt stress (Ren et al. 2005). Another important crop HKT gene is TmHKT1;5 which was cloned from a Nax2 locus in the ancestral wheat germplasm, and field trials confirmed that the presence of TmHKT1;5 reduced leaf Na+ contents and increased wheat grain yield under salt stress condition (Munns et al. 2012). These two crop HKT genes are good candidates for improving salt tolerance of crops.
The characteristics and the function of maize HKT transporter is rarely known. In this study, we cloned two alternative splicing variants of maize HKT1;1. Both transcripts of ZmHKT1;1 was constitutively overexpressed in transgenic tobacco plants, which exhibited better growth status, significantly decreased sodium contents in shoots and roots compared to the WT plants under salt stress condition, indicating that ZmHKT1;1 could confer salt tolerance of transgenic tobacco plants.
Materials and methods
Maize growth, stress treatments and RNA isolation
Seeds from the maize inbred line B73 were germinated in pots containing sand, and the seedlings were grown in a growth chamber under a 16 h light and 8 h dark photoperiod at 22 °C for 3 weeks. For cold and heat treatment, the seedlings were transferred to a growth chamber at 4 and 42 °C, respectively. For ABA and salt treatment, seedlings were gently removed from the sand, transferred to water for 1 h and then transferred to 100 μM ABA, 250 mM NaCl, respectively. For dehydration treatments, seedlings were carefully removed from the sand, transferred to water for 1 h and then dehydrated on filter paper. Shoots were sampled at 0, 1, 3, 6, 12 and 24 h after treatment, respectively.
Total RNA was extracted using Trizol reagent (Invitrogen, USA) according to the manufacturer’s protocol. The cDNA was prepared with 5 μg total RNA with a TransScript II first-strand cDNA synthesis kit (TransGen, China).
Cloning of ZmHKT1;1a and ZmHKT1;1b and plasmid construction
Based on the maize cDNA sequence (GRMZM2G047616), the primers ZmHKT-infusion-F(5′-GGGACTCTTACCATGGATCCTCTCCGTTTTGGGTTTCTTG-3′) and ZmHKT-infusion-R (5′GGGGAAATTCGAGCTGGACACCCAGGTTGATCGAGCGAGTTT-3′) were designed to amplify maize HKT1;1 using cDNA from maize inbred line B73 as template. Two PCR fragments were obtained and cloned into plant expression vector pCAMBIA3301 by In-fusion technology.
Protein alignments and phylogenetic tree construction
Amino acids sequences were aligned using the Cluster X (http://www.clustal.org/) and phylogenetic tree was obtained by Cluster X and Mega (http://www.megasoftware.net/) software.
Quantitative real-time PCR assay
Quantitative real-time PCR (qRT-PCR) was performed using TransStart Green qPCR SuperMix kits (TransGen, China) according to the manufacturer’s instructions under following conditions: initial denaturation at 95 °C for 2 min, 40 cycles at 95 °C for 10 s, 56 °C for 20 s and 72 °C for 30 s. At the end of the qRT-PCR cycles, the products were subjected to melt curve analysis to verify the specificity of PCR amplification. Three independent experiments were performed. Relative expression levels were calculated using the \(2^{{ - {\Delta \Delta }C_{\text{t}} }}\) formula with β-tubulin as a housekeeping gene (Livak and Schmittgen 2001). The used primers were shown in Supplemental Table 1.
Tobacco transformation
Plant expression plasmids were transferred into competent cells of the A. tumefaciens strain EHA105 through freeze–thaw treatment, respectively. The transformed A. tumefaciens colonies were selected on YEB-agar plates containing 100 mg mL−1 of kanamycin and 125 mg mL−1 of streptomycin. The positive colonies were identified by PCR amplification of the inserted genes and used for the tobacco transformation as previously described (Horsch et al. 1985). The transgenic plants were confirmed by PCR amplification of the interested gene.
Salt tolerance evaluation of transgenic plants
Heterozygous tobacco seeds of the T1 generation were germinated on MS medium containing 10 mg L−1 phosphinothricin and grown for 7 days at 100 μmol m−2 s−1 with a 16 h light/8 h dark period. The living seedlings with a similar size were transferred onto MS medium in plates containing different amounts of NaCl and grown vertically for another 1 week. The alive seedlings were also transferred to MS medium in plastic boxes containing different concentration of NaCl, and grown for another 1 month. The injury was observed and the fresh weight and main root length of transgenic plants were measured.
Na+ and K+ content analysis
The transgenic plants grown on the MS medium in plastic boxes containing different concentration of NaCl were used for measure the Na+ and K+ contents. The measurements were determined in shoot and root according to the described method with some modification (Shukla et al. 2012). Around 0.2 g tobacco plant tissues were digested in 5 ml of a mixed solution of nitric acids and perchloric acid (4:1). The solution was boiled on a hot plate at 175 °C until it turned colorless. The digested samples were then analyzed using a flame emission photometric method for Na+ and K+ concentration. Values from the flame photometer were converted into Na+ and K+ content values.
Results
Cloning of two alternative splicing variants of ZmHKT1;1
The BLAST analysis was performed in MaizeGDB using wheat OsHKT1;5 sequence and a full-length cDNA (GRMZM2G047616) was found to have high identity with OsHKT1;5. This maize gene was named as ZmHKT1;1 which contains two exons and one intron. When specific primers were designed to clone it, two ZmHKT1;1 cDNA fragments with different size were amplified and sequenced. One was named as ZmHKT1;1a which contains 1482 bp in the open reading frame (ORF) and encodes a protein with 493 amino acids, and this cDNA is identical to the predicted sequence in MaizeGDB. The other was named as ZmHKT1;1b which contains 1308 bp in the ORF and encodes a truncated protein with 435 amino acids. Compared with ZmHKT1;1a, the intron was not spliced in ZmHKT1;1b, and the termination codon in the intron leads to the translation of a truncated protein (Fig. 1a). The deduced amino acid sequences of ZmHKT1;1a and ZmHKT1;1b were aligned with the known HKT proteins. As other HKT proteins, ZmHKT1;1a has four membrane-pore-membrane (MPM) motifs, whereas the fourth MPM motif of ZmHKT1;1b was destroyed due to the pre-termination of translation (Fig. 1b).
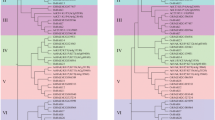
To understand the phylogenetic relationship of ZmHKT1 and those from other species, a phylogenic tree of ZmHKT1;1 and other 18 known HKT proteins was constructed (Fig. 2). HKT transporters can be classified into two groups (Horie et al. 2009), and the phylogenetic tree clearly showed that ZmHKT1;1a and ZmHKT1;1b belong to group I (Fig. 2). So these two ZmHKT1;1 variants might be Na+ transporters because group I HKT proteins prefer for Na+ transport than other cations (Horie et al. 2009).
The phylogenetic relationship of maize ZmHKT1;1 with HKTs from other plant species. The GenBank access numbers of the HKTs are: AAF68398 (AtHKT1;1), AAF97728 (EcHKT1;1), AAD53890 (EcHKT1;2), ABK58096 (HvHKT1;5), AAK52962 (McHKT1;1), AAO73474 (McHKT1;1), CAD37183 (OsHKT1;1), CAD37185 (OsHKT1;3), CAD37197 (OsHKT1;4), BAB93392 (OsHKT1;5), ABG33947 (TaHKT1;5-B1), ABG33948 (TaHKT1;5-B2), ABG33945 (TaHKT1;5-D), ABK41858 (TmHKT1;4-A1), ABK41857 (TmHKT1;4-A2), ABG33946 (TmHKT1;5), ABG339460 (TtHKT1;5-B1), ABG33941 (TtHKT1;5-B2), CAJ01327 (HvHKT2;1), BAB61791 (OsHKT2;2), CAD37187 (OsHKT2;3), CAD37199 (OsHKT2;4), AAA52749 (TaHKT2;1), DAA54361.1 (ZmHKT1;1a)
ZmHKT1;1a and ZmHKT1;1b showed different response to stress treatment in maize
The expression pattern of ZmHKT1;1a and ZmHKT1;1b under stress condition was investigated. One of the primers for ZmHKT1;1b was designed in the intron sequence which existed in ZmHKT1;1b, leading to the specific amplification of ZmHKT1;1b not ZmHKT1;1a. The primer specific for the amplification of ZmHKT1;1a was designed in the junction region of exon 2 and exon 3, leading to the specific amplification of ZmHKT1;1a (Fig. 1). When treated with 250 mM NaCl, the expression of ZmHKT1;1b in maize seedlings had 15 fold of increase 6 h after treatment, whereas the transcriptional level of ZmHKT1;1a was induced 12 h after treatment with threefold of increase (Fig. 3a). ZmHKT1;1b also showed faster response to heat stress than ZmHKT1;1a. 12 h after heat treatment, more than tenfold increase of ZmHKT1;1b was observed compared to the twice fold of increase of ZmHKT1;1a (Fig. 3c). The transcriptional level of both ZmHKT1;1a and ZmHKT1;1b were increased by ABA treatment, and 9- and 4-fold increase of transcriptional level were observed for ZmHKT1;1a and ZmHKT1;1b, respectively (Fig. 3b). 24 h after cold treatment, ZmHKT1;1a had 200 fold of transcript increase and ZmHKT1;1b had 62 fold of transcript increase (Fig. 3d). In dehydration treated maize seedlings, the transcriptional level of ZmHKT1;1a was not affected, whereas the transcriptional level of ZmHKT1;1b was decreased (Fig. 3e). These results clearly showed that ZmHKT1;1a and ZmHKT1;1b had different response to stress treatment in maize.
Real-time PCR analysis of the expression of ZmHKT1;1a and ZmHKT1;1b in response to different stress treatment. a Treatment with 250 mM NaCl; b treatment with 100 μM ABA; c heat treatment at 42 °C; d cold treatment at 4 °C; e dehydration treatment. The relative transcriptional level was analyzed using \(2^{{ - {\Delta \Delta }C_{\text{t}} }}\) method, and the transcriptional level at 0 h was normalized as 1.00. The experiments were performed in triplicate, and values are the mean ± SE of three samples
Overexpression of ZmHKT1;1 increased the salt tolerance of transgenic tobacco plants
To further study the function of two alternative splicing ZmHKT1;1 variants, transgenic tobacco plants overexpressing ZmHKT1;1a and ZmHKT1;1b, respectively, were obtained. Both genes were controlled by the CaMV 35S promoter and cloned into the plasmid pCAMBIA3301 to construct the vectors for plant transformation (Supplemental Fig. 1). The vectors were introduced into tobacco plants (Nicotiana tabacum var. Samsum) via Agrobacterium-mediated transformation. Transgenic tobacco plants were confirmed by PCR and RT-PCR analysis, and two transgenic lines (1–7 and 1–13) overexpressing ZmHKT1;1a and two transgenic tobacco lines (2–3 and 2–8) overexpressing ZmHKT1;1b were chosen for further analysis (Supplemental Fig. 1).
T2 progeny tobacco seeds were germinated on MS medium using 10 mg L−1 phosphinothricin as a selective reagent to eliminate non-transgenic plants. The germinated transgenic tobacco seedlings were moved on to the MS medium containing 0, 200 and 300 mM NaCl, respectively, and grown vertically for another 7 days. On the medium without NaCl, all transgenic plants grew similarly to the WT plants (Fig. 4a). Treatment with NaCl inhibited the growth of WT plants and the transgenic plants (Fig. 4a). The fresh weight (FW) and main root length (MRL) were measured to assess the inhibition of plant growth by NaCl. When treated with 200 and 300 mM NaCl, significantly higher fresh weight and root length were observed for transgenic plants overexpressing ZmHKT1;1a or ZmHKT1;1b, than those of WT plants (Fig. 4b, c). These results clearly showed that the over-expression of ZmHKT1;1a or ZmHKT1;1b in plants could offer high salt tolerance.
The growth of transgenic tobacco seedlings on MS medium containing different concentration of NaCl. a Photograph of transgenic tobacco seedlings. Heterozygous tobacco seeds of the T1 generation were germinated on MS medium containing 10 mg L−1 phosphinothricin and grown for 7 days at 100 μmol m−2 s−1 with a 16 h light/8 h dark period. The living seedlings with a similar size were transferred to MS medium in plates containing 0, 200, 300 mM of NaCl, respectively, and grown vertically for another 1 week. b The fresh weight of transgenic plants. c The main root length of transgenic plants. 1–7 and 1–13 are two independent transgenic lines expressing ZmHKT1;1a; 2–3 and 2–8 are two independent transgenic lines expressing ZmHKT1;1b. Data are shown as the average ± SE of 15 plants. Experimental data was analyzed by t test and the asterisks in columns mean significant difference from WT plants at *P < 0.05 or **P < 0.01 level
To further confirm the salt tolerance of transgenic tobacco plants over-expressing ZmHKT1;1a and ZmHKT1;1b, the germinated T2 transgenic tobacco seedlings were moved on to the MS medium containing 0, 200 and 300 mM NaCl in plastic boxes, respectively, and grown for another 4 weeks. Transgenic plants showed better salt tolerance than non-transgenic plants (Fig. 5a), and had significantly higher fresh weight than non-transgenic plants (Fig. 5b). We also put tobacco leaf discs in the liquid 1/4 MS medium containing different concentration of NaCl, and transgenic plants showed more salt tolerance than WT plants (Supplemental Fig. 2).
The growth of transgenic tobacco plants under salt treatment for 4 weeks. a Photograph of transgenic tobacco plants. Heterozygous tobacco seeds of the T1 generation were germinated on MS medium containing 10 mg L−1 phosphinothricin and grown for 7 days at 100 μmol m−2 s−1 with a 16 h light/8 h dark period. The alive seedlings were transferred onto MS medium in plastic boxes containing different concentration of NaCl, and grown for another 1 month. b The fresh weight (FW) of transgenic plants. 1–7 and 1–13 are two independent transgenic lines expressing ZmHKT1;1a; 2–3 and 2–8 are two independent transgenic lines expressing ZmHKT1;1b. Data are shown as the average ± SE of five samples each of which contains three plants. Experimental data was analyzed by t test and the asterisks in columns mean significant difference from WT plants at *P < 0.05 or **P < 0.01 level
Transgenic tobacco plants accumulated less Na+ and more K+ under salt treatment
Transgenic plants were treated with NaCl, and the accumulation of Na+ and K+ in shoot and root was measured. When plants grew under normal condition, there was no difference of Na+ and K+ concentration between WT plants and transgenic plants, indicating that overexpression of ZmHKT1;1a and ZmHKT1;1b did not affect the cation accumulation. Treatment with 200 mM NaCl led to significantly increase of Na+ and K+ contents in both WT plants and transgenic plants (Fig. 6). However, the transgenic plants had significantly less Na+ contents than that in WT plants (Fig. 6a, c), indicating that overexpression of ZmHKT1;1 reduced the accumulation of Na+ under salt stress condition. The similar results were observed when plants grew in hydroponic culture with different concentration of NaCl (Supplemental Fig. 3). Compared to the decreased Na+ contents, transgenic plants accumulated more K+ in both shoots and roots than WT plants which were treated with NaCl (Fig. 6b, d), indicating that the increased K+ could play roles in protecting plant cells from Na+ stress.
Na+ and K+ contents in shoots (a, b), and roots (b, d) of transgenic plants. Heterozygous tobacco seeds of the T1 generation were germinated on MS medium containing 10 mg L−1 phosphinothricin and grown for 7 days at 100 μmol m−2 s−1 with a 16 h light/8 h dark period. The alive seedlings were transferred onto MS medium in plastic boxes containing different concentration of NaCl, and grown for another 1 month. 1–7 and 1–13 are two independent transgenic lines expressing ZmHKT1;1a; 2–3 and 2–8 are two independent transgenic lines expressing ZmHKT1;1b. Data are shown as the average ± SE of three replicates. Experimental data was analyzed by t test and the asterisks in columns mean significant difference from WT plants at P < 0.05 level
Overexpression of ZmHKT1;1 affected the expression levels of some stress-related genes
To further investigate the mechanism of salt tolerance of transgenic plants, qRT-PCR was performed to compare the expression of eleven stress-related genes (Fig. 7; Supplemental Table 2). It was observed that overexpression of ZmHKT1;1b decreased the transcription levels of NtGPX, NtSPS, NtERD10B, NtRub-SS, NtAPX2, NtCAX3, NtNHX4, NtAPX1 and AY554170 in transgenic tobacco leaves, whereas overexpression of ZmHKT1;1a only decreased the expression of NtCAX3 and NtERD10B. When the plants were treated with 300 mM NaCl, higher expression levels of NtGPX in transgenic plants overexpressing ZmHKT1;1a or ZmHKT1;1b than that in WT plants was observed. The transgenic plants overexpressing ZmHKT1;1b also had higher NtSPS levels than WT plants when treated with 300 mM NaCl. These results indicate that ZmHKT1;1b might be more effective than ZmHKT1;1a to confer salt tolerance of transgenic plants.
The relative expression changes of stress-related genes in ZmHKT1;1a and ZmHKT1;1b transgenic tobaccos. 7-day old alive seedlings were transferred onto MS medium in plastic boxes containing different concentration of NaCl, and grown for another 1 month. 1–7 and 1–13 are two independent transgenic lines expressing ZmHKT1;1a; 2–3 and 2–8 are two independent transgenic lines expressing ZmHKT1;1b. The relative transcriptional level was analyzed using \(2^{{ - {\Delta \Delta }C_{\text{t}} }}\) method, and the transcriptional level of WT plants was normalized as 1.00. Data are shown as the average ± SE of three independent replicates. Experimental data was analyzed by t test and the asterisks in columns mean significant difference from WT plants at *P < 0.05 or **P < 0.01 level
Discussion
HKT genes, i.e. rice OsHKT1;5 (Ren et al. 2005) and wheat TmHKT1;5 (Munns et al. 2012), play important roles in the salt tolerance of crops. To clone the maize orthologue gene of HKT1;5, we performed BLAST analysis in MaizeGDB using rice OsHKT1;5 sequence and a full-length cDNA (GRMZM2G047616) was found to have high identity with OsHKT1;5. When specific primers were designed to clone it, two alternative splicing variants of ZmHKT1 (named as ZmHKT1;1a and ZmHKT1;1b) were amplified. Compared with ZmHKT1;1a, the intron was not spliced in ZmHKT1;1b, and the termination codon in the intron leads to the translation of a truncated protein. It is known that HKT genes contain two introns, and sub-family 1 have significantly larger introns than sub-family 2 (Platten et al. 2006). It has been shown that in salt sensitive rice line IR29, the presence of K+, Cs+ or Rb+ caused incomplete splicing of OsHKT1 intron, leading to larger transcript which contains one intron and encodes a dysfunctional HKT protein (Golldack et al. 2002). This kind of incomplete splicing might be due to the stress treatment. There was a alternative splicing of the rice OsHKT1;4 gene, and the spliced variants could be translated into a truncated protein due to the presence of translation stop codon in the second intron (Cotsaftis et al. 2012). It was suggested that the alternative splicing of OsHKT1;4 might control the horizontal axis in the shoot system (Cotsaftis et al. 2012).
Only two maize HKT orthologous sequences (ZmHKT1;1 and ZmHKT2;1) could be retrieved from maize genomes, and we could not amplify the sequence of ZmHKT2;1 from the cDNA of salt-treated plants (data not shown). So, the alternative splicing of ZmHKT1;1 could provide maize plant with more HKT copy, which can increase the salt tolerance of maize. Generally, sorghum has high salt tolerance than maize, might due to that there are four HKT members in sorghum compared to the two genes in maize (Wang et al. 2014). We noticed that both alternative splicing variants of ZmHKT1;1 exist under normal condition. However, ZmHKT1;1b showed faster response to salt and heat stress than ZmHKT1;1a. In dehydration treated maize seedlings, the transcriptional level of ZmHKT1;1a was not affected, whereas the transcriptional level of ZmHKT1;1b was decreased (Fig. 3e). These results clearly showed that ZmHKT1;1a and ZmHKT1;1b had different response to stress treatment in maize. The transcripts of these two variants also showed different levels in different maize tissues (Supplemental Fig. 4).
To further study the function of the alternative splicing variants of ZmHKT1;1 genes, we performed transgenic tobacco plants. When treated with high concentration of NaCl, transgenic plants overexpressing ZmHKT1;1a or ZmHKT1;1b had significantly higher fresh weight and root length than WT plants (Figs. 4, 5), indicating the high salt tolerance of transgenic plants. It has been reported that transgenic plants overexpressing AtHKT1;1 which was driven by constitutive promoter were relatively more salt sensitive than WT plants, whereas transgenic plants overexpressing the same gene specifically in root stele cells accumulated less salt in the shoot and became more salt tolerant (Moller et al. 2009). However, transgenic barley plants overexpressing HvHKT2;1 which was driven by strong constitutive ubiquitin promoter had increased salt tolerance (Mian et al. 2011). In our experiments, both ZmHKT1;1a and ZmHKT1;1b were controlled by CaMV 35S promoter which is a strong constitutive promoter, and overexpression of both genes could confer plants with high salt tolerance, indicating that not all HKT genes need to be cell type specifically expressed in transgenic plants to improve the salinity tolerance. It has also been shown that the reduction of TaHKT2;1 expression increased the salt tolerance of plants (Laurie et al. 2002). These results suggests that HKT proteins might play roles with multiple mechanism.
Exclusion of Na+ from shoots and photosynthetic tissues is a mechanism of salt tolerance (Moller and Tester 2007). HKTs detoxify the salt stress mainly by decreasing the accumulation of Na+ and it has been confirmed that the main role of AtHKT1;1 in transgenic plants is to avoid the accumulation of excessive Na+ in the plant shoots (Almeida et al. 2013). When treated with salt, the transgenic plants overexpressing ZmHKT1;1a or ZmHKT1;1b had significantly less Na+ contents in transgenic plants than that in WT plants (Fig. 6), indicating that overexpression of ZmHKT1;1 reduced the accumulation of Na+ under salt stress condition. The increased K+ content could protect plant cells from Na+ stress. Many studies have shown that increasing the K+/Na+ ratio is crucial for Na+ tolerance in plants, and maintaining high K+/Na+ ratio in shoots is highly correlated with salinity tolerance (Hauser and Horie 2010). So, it is not surprising that transgenic plants overexpressing ZmHKT1;1 accumulated more K+ in both shoots and roots than WT plants (Fig. 6).
Most HKTs have eight transmembrane domains and four highly conserved P-loops. It was shown that the glycine residue in the first P-loop is crucial for K+ selectivity and transport (Maser et al. 2002). Four TaHKT2;1 mutants in the second and third P-loop reduced the low affinity Na+ uptake, indicating the important roles of certain residues in the second and third P-loops (Rubio et al. 1995, 1999). The E464Q mutation in the fourth P-loop lowered the affinity of HKT1 for Na+, confirming the fourth P-loop plays important role in the binding and transport of Na+ (Diatloff et al. 1998). The fourth P-loop of ZmHKT1;1b was destroyed due to the pre-termination of translation. We did not observe obvious difference of salt-tolerant level, Na+ or K+ contents between transgenic plants overexpressing ZmHKT1;1a and plants overexpressing ZmHKT1;1b, indicating that the un-integrity of the fourth P-loop did not affect its function of salt detoxification when it was overexpressed. However, we did not know whether the transgenic plants expressing both genes still have the similar salt tolerance when ZmHKT1;1a and ZmHKT1;1b were driven by their native promoter.
Although there was no difference of the salt tolerance of transgenic tobacco plants overexpressing ZmHKT1;1a or ZmHKT1;1b, we found that overexpression of these two variants caused different expression of stress-related genes (Fig. 7). It was observed that overexpression of ZmHKT1;1b decreased the transcriptional levels of more stress-related genes in transgenic tobacco leaves, compared to the plants overexpressing of ZmHKT1;1a. Alternative splicing of pre-mRNAs can increase the diversity in the proteomic world, and various environmental stresses affected the alternative splicing (Graveley 2001). Overexpression of ZmHKT1;1b which has intron retention might led to the stress simulation of transgenic plant cell, so more stress-related genes were induced. ZmHKT1;1b plays role in the detoxification of salt stress with somewhat difference with ZmHKT1;1a due to the lack of the fourth P-loop, which need further investigation.
References
Almeida P, Katschnig D, de Boer AH (2013) HKT transporters—state of the art. Int J Mol Sci 14(10):20359–20385
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by over expression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 85:1256–1258
Corratgé-Faillie C, Jabnoune M, Zimmermann S, Véry A-A, Fizames C, Sentenac H (2010) Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cell Mol Life Sci 67(15):2511–2532
Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M (2012) A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 7(7):e39865
Diatloff E, Kumar R, Schachtman DP (1998) Site directed mutagenesis reduces the Na+ affinity of HKT1, an Na+ energized high affinity K+ transporter. FEBS Lett 432:31–36
Garciadeblas B, Senn ME, Banuelos MA, Rodriguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34(6):788–801
Golldack D, Su H, Quigley F, Kamasani UR, Munoz-Garay C, Balderas E, Popova OV, Bennett J, Bohnert HJ, Pantoja O (2002) Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J 31(4):529–542
Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17:100–107
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Hauser F, Horie T (2010) A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ 33(4):552–565
He C, Yan J, Shen G, Fu L, Holaday AS, Auld D, Blumwald E, Zhang H (2005) Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant Cell Physiol 46:1848–1854
Horie T, Hauser F, Schroeder JI (2009) HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci 14(12):660–668
Horsch R, Fry J, Hoffman N, Eichholz D, Rogers S, Fraley R (1985) A simple and general method for transferring genes into plants. Science 227(4691):1229–1231
Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R (2006) A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 142(4):1718–1727
Laurie S, Feeney KA, Maathuis FJ, Heard PJ, Brown SJ, Leigh RA (2002) A role for HKT1 in sodium uptake by wheat roots. Plant J 32(2):139–149
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408
Maser P, Hosoo Y, Goshima S, Horie T, Eckelman B, Yamada K, Yoshida K, Bakker EP, Shinmyo A, Oiki S, Schroeder JI, Uozumi N (2002) Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci USA 99(9):6428–6433
Mian A, Oomen RJ, Isayenkov S, Sentenac H, Maathuis FJ, Very AA (2011) Over-expression of an Na+-and K+-permeable HKT transporter in barley improves salt tolerance. Plant J 68(3):468–479
Mishra S, Alavilli H, Lee B, Panda SK, Sahoo L (2015) Cloning and characterization of a novel vacuolar Na+/H+ antiporter gene (VuNHX1) from drought hardy legume, cowpea for salt tolerance. Plant Cell Tissue Organ Cult 120:19–33
Moller IS, Tester M (2007) Salinity tolerance of Arabidopsis: a good model for cereals? Trends Plant Sci 12(12):534–540
Moller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21(7):2163–2178
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30(4):360–364
Patel MK, Joshi M, Mishra A, Jha B (2015) Ectopic expression of SbNHX1 gene in transgenic castor (Ricinus communis L.) enhances salt stress by modulating physiological process. Plant Cell, Tissue Organ Cult 122:477–490
Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin H-X, Luan S (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11(8):372–374
Plett D, Safwat G, Gilliham M, Skrumsager Moller I, Roy S, Shirley N, Jacobs A, Johnson A, Tester M (2010) Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PLoS ONE 5(9):e12571
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37(10):1141–1146
Rubio F, Gassmann W, Schroeder JI (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270:1660–1663
Rubio F, Schwarz M, Gassmann W, Schroeder JI (1999) Genetic selection of mutations in the high affinity K+ transporter HKT1 that define functions of a loop site for reduced Na+ permeability and increased Na+ tolerance. J Biol Chem 274(11):6839–6847
Schachtman DP, Schroeder JI (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370:655–658
Shukla PS, Agarwal PK, Jha B (2012) Improved salinity tolerance of Arachis hypogaea (L.) by the interaction of halotolerant plant-growth-promoting rhizobacteria. J Plant Growth Regul 31(2):195–206
Wang TT, Ren ZJ, Liu ZQ, Feng X, Guo RQ, Li BG, Li LG, Jing HC (2014) SbHKT1;4, a member of the high-affinity potassium transporter gene family from Sorghum bicolor, functions to maintain optimal Na+/K+ balance under Na+ stress. J Integr Plant Biol 56(3):315–332
Acknowledgments
This work is financially supported by the National Basic Research Program of China (2014CB138202) and the Agricultural Science and Technology Innovation Program (ASTIP) of CAAS.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ren, Z., Liu, Y., Kang, D. et al. Two alternative splicing variants of maize HKT1;1 confer salt tolerance in transgenic tobacco plants. Plant Cell Tiss Organ Cult 123, 569–578 (2015). https://doi.org/10.1007/s11240-015-0861-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-015-0861-9