Abstract
Soil salinity is a major abiotic stress that seriously affects crop productivity worldwide. One of the mechanisms that allow plants to withstand salt stress is vacuolar sequestration of Na+, through a Na+/H+ antiporter. We isolated a new vacuolar Na+/H+ antiporter gene (VuNHX1) from a drought hardy grain legume, cowpea (Vigna unguiculata L.). The cDNA is 1,981 bp, with an open reading frame of 1,629 bp encoding a predicted protein of 542 amino acids with a deduced molecular mass of 59.6 kDa. VuNHX1 displays a conserved amiloride binding domain (84LFFIYLLPPI93) in third transmembrane (TM3) region. Phylogenetic and bioinformatic analysis indicated VuNHX1 belonging to Class-I clade of plant NHX exchangers with high similarity with legume Na+/H+ antiporters. To assess its role in Na+ exchange, we performed complementation studies using the salt sensitive yeast mutant strain AXT3. The results showed that VuNHX1 complemented for the loss of yeast NHX1 under NaCl, KCl and LiCl stress in the salt sensitive phenotype of the yeast strain AXT3. The expression profiles revealed significant induction of VuNHX1 in cowpea seedlings under salt, cold and dehydration stress. Both expression analysis and ion estimation under salt stress indicated the VuNHX1 expression preferentially in roots than in leaves. Overexpression of VuNHX1 in transgenic Arabidopsis conferred enhanced salt tolerance in transgenic Arabidopsis lines while the wild type plants exhibited growth retardation. This study shows that VuNHX1 is a potential gene for salt tolerance, and can be used in future for developing cisgenic salt tolerant cowpea and transgenic salt tolerant crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinity is a major abiotic stress that limits crop growth and productivity worldwide (Kronzucker and Britto 2011). In saline soil, Na+ is the most predominantly toxic ion. The excess accumulation of Na+ in the cytosol leads to disruption of ion homeostasis, hyper-osmotic stress, inhibition of cytoplasmic enzyme activities, production of reactive oxygen species, and causes adverse effect on photosynthesis and cellular metabolism (Hasegawa et al. 2000; Zhu 2001). Therefore, salt toxicity is mainly attributed to Na+ specific damage to the plant cytoplasm (Zhao et al. 2007). In order to avoid Na+ toxicity, the plant cell may either exclude the ions from the cell by the plasmamembrane Na+/H+ antiporter, or store them inside the vacuole by pumping the Na+ to the vacuole via vacuolar Na+/H+ antiporters (Zhu 2003). These Na+(K+)/H+ exchangers (also known as NHX-type cation/proton antiporters) function cooperatively to prevent Na+ uptake, increase efflux of Na+ from cell and compartmentalize Na+ into vacuole (Reguera et al. 2014). Pumping sodium ions into vacuole via vacuolar Na+/H+ antiporters facilitates the reduction in the toxic levels of sodium in the cytosol, and the increase in the vacuolar osmotic potential with the concomitant increase in negative water potential which favors enhanced cellular water uptake and tissue water retention under high soil salinity (He et al. 2005). In addition to the known role of Na+/H+ antiporters in ion homeostasis, recent findings indicated their involvement in other key cellular processes, not necessarily limited to salinity stress response, such as calcium signaling, sulfur metabolism, cell structure and cell growth, as well as intracellular vesicular trafficking and protein targeting (Apse et al. 2003; Sottosanto et al. 2004), Berry ripening (Hanana et al. 2007), and recessive mutation in vacuolar Na+/H+ antiporter gene leading to purple flower of Japanese morning glory (Yamaguchi et al. 2001). Current evidence suggests that halophytes and glycophytes share many fundamental transport and osmotic solute biosynthetic mechanisms for ion compartmentalization and osmotic adjustment (Hasegawa 2013). Vacuolar Na+/H+ antiporter gene of Arabidopsis (AtNHX1) was the first plant homologue of yeast Na+/H+ exchanger to be cloned (Apse et al. 1999) and the transcript level of AtNHX1 was up-regulated by NaCl and abscisic acid (Shi and Zhu 2002). Vacuolar NHX1 gene was identified in several glycophytes including rice (Fukuda et al. 1999), calico flower (Zhang et al. 2008), rape seed (Wang et al. 2003), cotton (Wu et al. 2004), hybrid tea rose (Kagami and Suzuki 2005), maize (Zorb et al. 2005), wheat (Brini et al. 2005), and halophytes including ice plant (Chauhan et al. 2000), gmelin’s saltbush (Hamada et al. 2001), beetroot (Xia et al. 2002), seepweed (Ma et al. 2004) and umari keerai (Jha et al. 2011). Over-expression of vacuolar antiporter genes resulting in enhanced salt tolerance in different plant species (Apse et al. 1999; Chen et al. 2008; He et al. 2005; Ohta et al. 2002; Wu et al. 2005; Xue et al. 2004; Zhang and Blumwald 2001; Zhang et al. 2001) suggests their important role in salt tolerance and ion homeostasis.
Cowpea (Vigna unguiculata L. Walp.) is an important grain legume that serves both as food and fodder for resource poor populace in semi-arid tropics (Singh 2005; Timko et al. 2007). It is cultivated in different parts of the world, viz. Africa, India, USA and Australia, under a wide range of climatic conditions such as semiarid to sub-humid and is well adapted to heat and drought (Hall 2012). In plants, there exists extensive overlap and crosstalk between salinity and drought stress-responsive signaling pathways. The distinctive character of cowpea to withstand drought makes it a valuable system for mining abiotic stress responsive genes and understanding the molecular mechanisms of abiotic stress tolerance. However, there has been no attempt to clone and understand the function of NHX1 in cowpea. Moreover, salinity and limited water availability impose severe constraints on cowpea production. Consequently, development of cowpea cultivars endowed with enhanced tolerance to salinity and dehydration stress possibly through overexpression of native NHX1 by cis-genic approach offers both environment and consumer acceptable way for sustainable production on hostile soil.
In this study, we report cloning and molecular characterization of a vacuolar Na+/H+ antiporter gene from cowpea (VuNHX1), a drought tolerant grain legume. We investigated the VuNHX1 expression patterns under salinity and other abiotic stresses, functional validated in salt sensitive yeast AXT3 mutant and over-expressed in Arabidopsis with intention to reveal the possible role of VuNHX1 in salt tolerance.
Materials and methods
Plant material and stress treatment
Cowpea plants (Vigna unguiculata L. cv. Pusa Komal) were grown in a growth chamber in hydroponic system consisting of Hoagland’s solution under conditions of 16 h photoperiod at 25 °C and 70 % relative humidity. Ten days old seedlings were subjected to different abiotic stress treatments such as salt, dehydration and cold for different time periods (0, 6, 12, and 24 h). Salt stress was applied by adding NaCl to the hydroponic medium to a final concentration of 200 mM. For dehydration stress, the seedlings were drawn out of the hydroponic system and their roots were exposed to air in a room at 27 °C. The cold stress treatment consisted of incubating the seedlings at 4 °C.
Isolation of VuNHX1 cDNA
A pair of degenerate PCR primers, FP: 5′-GGKTTTCARGTRAAAAAGAAGCA-3′ and RP: 5′-GTRCTSGTGATCATRAYTGCATT-3′ were designed for amplification of partial cDNAs of Na+/H+ antiporter analogue gene from cowpea based the conserved regions of vacuolar Na+/H+ antiporter from legume species. Total RNA was isolated using RNeasy Plant Mini Kit (Qiagen, Venlo, Limburg, Netherlands). The first strand cDNA synthesis and RT-PCR were performed using Revert Aid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) with the pair of degenerate primers. The amplified product was cloned into TA cloning vector, pTZR/T (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced. The cDNA synthesized using gene specific primer (GSP: 5′-GCACCCAAAGTTATGACAGCAC-3′) and superscript™ II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) was purified using a SNAP column for efficient removal of unincorporated dNTPs and remaining GSP. Terminal transferase (TdT) enzyme generated a homopolymeric tail by incorporating dCTPs to 3′ end of the purified cDNA. The dC-tailed cDNA was amplified using abridged anchor (AAP: 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′) and gene specific (GSP1: 5′-TGTACCAATAGCACCAAACAACATGATG-3′) primers. A nested PCR was performed with abridged universal anchor (AUAP: 5′-GGCCACGCGTCGACTAGTAC-3′) and gene specific (GSP2: 5′-GTCATGAAGTTAACAAAAAACTGC-3′) primers using the first PCR product as template. Similarly, the 3′ untranslated sequence was obtained by using “Rapid Amplification of cDNA Ends Kit” (Version E, Life Technologies/Invitrogen, Carlsbad, CA, USA). The cDNA was synthesized using a dT-adapter primer (AP: 5′-GGCCACGCGTCGACTAGTAC(T)17-3′). The 3′-RACE PCR was performed with abridged universal anchor (AUAP: 5′-GGCCACGCGTCGACTAGTAC-3′) and gene specific (GSP3:5′-GCTGTATATTGGAAGGCACTCT-3′) primers. The full length of VuNHX1 was amplified using forward (FLP: 5′-ATGGTCTTTGAAATCAGTTCTGTTGTTTC-3′) and reverse (RLP: 5′-TCAACGCCATTGATGACCATTACGTTC-3′) primers, cloned into TA cloning vector pTZR/T (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced (GenBank accession no.: JN641304.2).
In-silico analysis
Multiple sequence alignment and phylogenetic analysis were performed using Clustal W (Thompson et al. 1997). Based on the neighbor-joining (NJ) algorithm, an unrooted phylogenetic tree was constructed using MEGA4: Tree Explorer software (Tamura et al. 2007). The transmembrane prediction was performed with TMpred software (Hofmann and Stoffel 1993) and secondary structure prediction was made using SOPMA (Self-Optimized Prediction Method with Alignment) server (Geourjon and Deleage 1995). Post-translational modification of VuNHX1 was predicted by searching for conserved motifs of N- and O-glucosylation and N-myristoylation sites using ScanProsite (Gattiker et al. 2002).
Yeast strain and vector construction
Saccharomyces cerevisiae strains used in tolerance assays were wild-type W303-1B (MATα ade2-1 can1-100 his3-11, 15 leu2-3,112 trp1-1 ura3-1) and mutant strain AXT3 (∆ ena1-4::HIS3 ∆nha1::LEU2 ∆nhx1::TRP1, ura3-1). Yeast strains were grown in YPD (1 % Yeast extract, 2 % peptone and 2 % glucose), YPGal (1 % Yeast extract, 2 % peptone and 2 % galactose), SC (0.67 % Yeast nitrogen base and 2 % glucose) and APGal synthetic minimal media (10 mM arginine, 8 mM phosphoric acid, 2 mM MgSO4, 1 mM KCl, 0.2 mM CaCl2, 2 % galactose, trace vitamins, and minerals; pH 4.0) supplemented with appropriate amino acids as indicated.
The VuNHX1 ORF was digested from the pTZR/T-VuNHX1 vector by KpnI/BamHI and sub-cloned into the KpnI/BamHI site of the yeast expression vector pYES2.0 (Invitrogen, Carlsbad, CA, USA).
Functional assays using the yeast mutant
The yeast mutant AXT3 was transformed with pYES2.0-VuNHX1 and pYES2.0 as a control using Lithium acetate method (Gietz et al. 1992) and selected on SC ura− medium. All yeast strains were grown at 30 °C for 16–18 h to reach OD600-1.0 in liquid YPGlu (Glu/Ura−) media. Cultures were normalized to an OD600-0.006 and inoculated into liquid APGal (Gal/Ura−) media (Rodriguez-Navarro and Ramos 1984) supplemented with different concentrations of NaCl (0, 50, 75, and 100 mM), KCl (0, 0.5, 1.0, and 1.5 M), and LiCl (0, 15, 20 and 25 mM). Growth status was compared after culturing strains at 30 °C for 2 days. Similarly, 10, 100 and 1,000 fold serial dilutions of yeast cultures (OD600-1.0) were spotted on solid APGal media supplemented with or without 75 mM NaCl, 0.5 M KCl, and 50 mM LiCl and further, on solid YPGal media supplemented with or without 50 µg/ml hygromycin-B antibiotic. Growth status was compared after culturing the strains at 30 °C for 3 days.
Intracellular measurement of Na+ and K+ distribution in yeast mutant
Yeast strains were grown in liquid APGal media, pH 4.0 supplemented with or without 70 mM NaCl, and harvested at an OD600 of 0.3–0.4. Cells were centrifuged at 3,000g/3 min, washed twice in ice-cold 10 mM MgCl2, 10 mM CaCl2 and 1 mM HEPES buffer and resuspended in the same buffer. The relationship between cell density (Absorbance at OD600) and yeast dry weight was determined. Total intracellular ion was determined by addition of HCl to a final concentration of 0.4 % and incubated at 95 °C for 20 min. After removal of cell debris the supernatant was measured for presence of total Na+ and K+. To differentiate Na+ and K+ concentration in cytoplasmic and vacuolar region, cells were grown and washed as above and resuspended in 2 % cytochrome c, 18 µg/ml antimycin, 1 mM HEPES, 10 mM MgSO4, 10 mM CaCl2, and 5 mM 2-Deoxy d-Glucose solution. Cytochrome c is known to selectively permeabilize the plasma membrane of cells. After 20 min incubation at room temperature, cells were washed thrice with the same solution without cytochrome c. Cytoplasmic ion content was determined by pooling the supernatants. The remaining vacuolar ions were extracted with addition of HCl in a final concentration of 0.4 % and incubated at 95 °C for 20 min (Venema et al. 2003). The Na+ and K+ distribution in the cytoplasmic and vacuolar fractions were measured in Flame Photometer (Systronics, Bhopal, MP, India).
Gene copy determination by Southern analysis
For gene copy number analysis in cowpea, genomic DNA (20 µg) of cowpea was digested separately with restriction endonucleases EcoRI, HindIII and PstI. Digested DNA was electrophoretically fractionated on a 1 % agarose gel and blotted onto Zeta-Probe nylon membrane (Bio-Rad, Hercules, CA, USA). The blot was hybridized with DIG-labeled 0.9 kb PCR product, corresponding to the coding region of VuNHX1 using the primers, forward (SF: 5′-GCTGTATATTGGAAGGCACTCT-3′) and reverse (RF: 5′-TCAACGCCATTGATGACCATTACGTTCAG-3′). Southern hybridization was carried out using hybridization solution containing 50 % formamide, 5 X SSC, 5 X Denhardt’s solution, 0.05 M sodium phosphate pH 6.5, 0.1 % SDS, 10 % Dextran sulfate, 0.1 mg/ml sheared denatured salmon-sperm DNA and 20 ng/ml probe at 42 °C for 18 h. Washing and detection was performed according to instructions of the DIG Labeling and Detection system (Roche Diagnostics, Mannheim, Germany).
Expression analysis by semi-quantitative PCR and determination of Na+ and K+ content
Cowpea seedlings subjected to different treatments were used for expression analysis by semi-quantitative PCR. Total RNA was isolated using RNeasy Plant Mini Kit (Qiagen, Venlo, Limburg, Netherlands). The first strand cDNA synthesis was performed using Revert Aid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) and the products were subsequently used for PCR using gene specific primers (RFP: 5′-GCTGTATATTGGAAGGCACTCT-3′ and RRP: 5′-CAATGTCCAAGGCATCCATACC-3′). The PCR conditions were as follows: 95 °C for 10 min followed by 28 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s and finally 72 °C for 10 min. Housekeeping VuNSR9 primers (FN: 5′-GCACAGTTTGGGTATATTG-3′ and RN: 5′-GAGTAAAACTGGCAAAAATTAG-3′) were used as an internal control. The PCR conditions for housekeeping gene were as follows: 95 °C for 10 min followed by 28 cycles at 95 °C for 30 s, 49 °C for 30 s, 72 °C for 30 s and finally 72 °C for 10 min. Entire experiments were performed in triplicate. Leaves and roots of untreated and salt-treated cowpea seedlings were harvested at different time intervals (0, 6, 12, 18, 24, and 48 h). The samples were dried, digested with concentrated HNO3 at 90 °C for 30 min and centrifuged at 12,000 rpm for 10 min. The suspension was diluted with sterile water and analyzed for Na+ and K+ content in Flame photometer (Systronics, Bhopal, MP, India).
Generation of transgenic Arabidopsis plants
The CaMV35S::VuNHX1::35ST (2.3 kb) was digested from the pRT101VuNHX1 (4.9 kb) vector by PstI and subcloned into the PstI site of plant binary vector pCAMBIA2301 (11.6 kb) to result the plant binary construct pCAMBIA2301-35S::VuNHX1 (13.9 kb). Further, a RD29A promoter region (0.898 kb) of AtRD29A (DQ071887.1) was amplified from A. thaliana genomic DNA using forward (RD29F: 5′-CTGAAATTTCTGCAAGAATC-3′) and reverse (RD29R: 5′-TCCAATAGAAGTAATCAAACC-3′) primers and cloned into TA cloning vector pGEMT easy (3.015 kb) (Promega, Madison, WI, USA). The RD29A promoter was digested as EcoRI fragment from pGEMTRD29A (4.2 kb) and cloned into EcoRI site of plant binary construct pCAMBIA2301-35S::VuNHX1 (13.9 kb) by replacing the 0.4 kb CaMV35S promoter in 35SP::VuNHX1::35STer cassette resulting pCAMBIA2301-RD29A::VuNHX1 (14.4 kb). The plant binary constructs, pCAMBIA2301-35S::VuNHX1 and pCAMBIA2301-RD29A::VuNHX1 were mobilized to A. tumefaciens strain GV3101 by electroporation at 1,250 V with capacitance of 25 mF and resistance of 400 ohm.
Arabidopsis thaliana (ecotype Columbia-0) plants were transformed using floral dipping method (Clough and Bent 1998). The T1 transgenic plants were screened on ½ MS medium (Duchefa, Haarlem, Netherlands) supplemented with 50 mg/l kanamycin (Duchefa, Haarlem, Netherlands). The transgenic selections were continued until T4 generation to obtain homozygous transgenic lines (35S::VuNHX1 or RD29A::VuNHX1).
Analysis of transgenic Arabidopsis by real-time PCR
Total RNA extraction from wild-type and T4 transgenic lines and First-strand cDNA synthesis were carried out as per the procedures described previously. The products were subsequently used as templates for real-time PCR analysis using gene specific forward primer (VrRTF: 5′-TGATCCAATCCATCGTCCAA-3′) and 35S poly-A reverse primer (TerparR: 5′-GCGAAACC CTATAAGAACCCTAATTCC-3′) for amplification of a 0.28 kb fragment of VuNHX1::35S poly-A. The AtUbiquitin was used as internal control using UBQ1FP: 5′-AGAGCTG TCAACTGCAGGAAGAA-3′ and UBQ1RP- 5′-ACAAGAAAAACAAACCCTATCAAAGG primers to amplify a 150 bp fragment. Real-time PCR was performed using USB VeriQuest™ SYBR Green qPCR Master Mix (2X) (Affymetrix, Santa Clara, CA, USA) on a 7500 Real-Time PCR System (Applied Biosystem, Foster City, California, USA) according to manufacturer’s instructions with a final primers concentration of 200 nM for both genes. The experiment was repeated twice independently with three replicates. The expression values relative to the standard curve was calculated for each sample. The relative expression of VuNHX1 in wild-type (WT) and transgenic Arabidopsis lines was estimated by normalizing expression values of VuNHX1 with that of housekeeping AtUBQ1.
Effect of salinity on transgenic 35S::VuNHX1 and RD29A::VuNHX1 Arabidopsis lines
In order to evaluate the effect of salinity on seed germination, T4 seeds of wild-type (WT) and homozygous transgenic Arabidopsis were surface sterilized and vernalized for 3 days at 4 °C on ½ MS medium (Murashige and Skoog 1962) supplemented with or without 150 mM NaCl and transferred for 7 days to a growth chamber maintained at 22 °C and 60 % relative humidity with a 16 h photoperiod.
Wild type (WT) and T4 homozygous transgenic Arabidopsis seeds were grown on ½ MS medium (Murashige and Skoog 1962) for 5 days and thereafter transferred to ½ MS medium supplemented with NaCl (150 and 200 mM). The corresponding root length was measured in WT and transgenic lines (35S:: VuNHX1 and RD29A::VuNHX1) after 7 days of NaCl treatment and the difference was calculated. The emergence of lateral roots in each case was also studied. Mean data was collected from ten replicates (n = 10) for wild-type (WT) and T4 transgenic Arabidopsis lines.
To test the effect of salinity on mature plants, germinated seedlings were grown initially on ½ MS medium for 5 days and subsequently transferred to soilrite for 2 weeks. The WT and T5 transgenic lines (35S:: VuNHX1 and RD29A::VuNHX1) were subjected to salt stress for 2 weeks by watering them with ½ MS nutrient liquid media supplemented with 200 mM NaCl. The leaves were harvested and the Na+ and K+ content was estimated using method described previously.
For measurement of chlorophyll content, leaf samples were homogenized in 95 % ethanol, lysate was centrifuged at 3,000 rpm for 10 min and absorbance was recorded for the extract at wavelength of 648 and 664 nm (Lichtenthaler 1987).
Lipid peroxidation was measured as the amount of malondialdehyde (MDA) determined by the thiobarbituric acid (TBA) reaction. Briefly, 0.2 g of fresh leaf samples were homogenized with 5 ml of 0.25 % TBA containing 10 % TCA (tricloroacetic acid). The homogenate was boiled for 30 min at 95 °C and centrifuged at 10,000g for 10 min. Absorbance values were recorded at 532 nm and values corresponding to non-specific absorption at 600 nm were subtracted (Heath and Packer 1968).
For colorimetric estimation of proline, leaf samples (0.5 g) were homogenized with 5.0 ml of sulfosalicylic acid (3 %). 2 ml of homogenate was incubated with 2 ml glacial acetic acid and 2 ml ninhydrin reagent at a ratio of 1:1:1 in boiling water bath at 100 °C for 30 min. After cooling, 4 ml toluene was added to the reaction mixture, mixed vigorously and absorbance was measured at 520 nm (Bates et al. 1973). Mean data was collected from three replicates (n = 3) for wild-type (WT) and T4 kanamycin selected transgenic Arabidopsis lines.
Statistical analysis
Statistical comparison between the variances was determined by ANOVA (Analysis of variance) and significant differences between mean values were determined by Bonferroni analysis. Statistically significant mean values were denoted as different letters (P ≤ 0.05).
Results
Molecular characterization of VuNHX1 and sequence analysis
The full-length cDNA of VuNHX1 was obtained by RT-PCR and RACE method. The 1,981 bp cDNA of VuNHX1 contained an open reading frame (ORF) of 1,629 bp which encoded a polypeptide of 542 amino acid residues with an estimated molecular mass of 59.60 kDa and isoelectric point of 6.76. The sequence of VuNHX1 cDNA has been deposited at GenBank under accession number JN641304.2.
Multiple sequence alignment revealed a higher degree of homology between amino acid sequences of VuNHX1 and vacuolar Na+/H+ antiporter of other higher plants (Online Resource 1). The highest identity was found to be 97.42 % with mungbean (Vigna radiata), an Asiatic grain legume, 92.44 % with soybean (Glycine max), 88.1 % with Peashrub (Caragana korshinskii), 87.45 % with narrow-leaved bird’s-foot trefoil (Lotus tenuis), 87.25 % with alfalfa (Medicago sativa), 86.9 % with chickpea (Cicer arietinum), 86.51 % with white clover (Trifolium repens), and 84.84 % with Goat’s rue (Galega orientalis) (Online Resource 1).
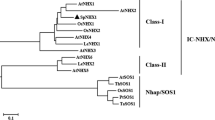
Recent phylogenetic analysis of the Na+/H+ exchanger (NHE) family had revealed two distinct subgroups corresponding to plasma membrane (PM-NHE) and intracellular transporters (IC-NHX) (Brett et al. 2005a). The two subgroups were distinct from one another in ion selectivity, kinetic properties, inhibitor sensitivity, and physiological role. In our work, a phylogenetic analysis of a number of Na+/H+ antiporters was generated by CLUSTAL W and MEGA 4 software program (Online Resource 2 a). The phylogenetic analysis showed that VuNHX1 formed a clade with class-I type intracellular Na+/H+ (IC-NHX) exchangers, more closely related to legume NHX homologs, and distinct from class-II type IC-NHX exchangers as well as from the cluster of plasma membrane Na+/H+ transporters (PM-NHE) (Online Resource 2 a). These results suggested that the product of VuNHX1 localized on the tonoplast.
A hydropathy plot generated by TMPred software indicated that VuNHX1 with highly hydrophobic N-terminal end consisting of 11 putative hydrophobic regions (Online Resource 2 b), which was different from 9 transmembrane domains in AtNHX1 and OsNHX1 (Fukuda et al. 1999; Yamaguchi et al. 2003), and a longer hydrophilic C-terminal end (Online Resource 2 b). The amiloride binding motif, 84-LFFIYLLPPI-93 in VuNHX1 is highly conserved among eukaryotic Na+/H+ exchangers (Online Resource 1). As shown in Online Resource 1, VuNHX1 was highly conserved in transmembrane domains (TM 1, 3, 5, 7, 8, 10) with other reported legume NHXs. However, comparatively low conservation was observed in case of transmembrane domains (TM 2, 4, 6, 9, 11) and the hydrophilic C-Terminal end. Secondary structure prediction by SOPMA indicated occurrence of alpha helix (49.45 %), extended strand (14.21 %), beta turn (3.69 %), and random coil (32.66 %) as represented in Online Resource 3. The putative post-translational modification sites predicted by ScanProsite software are as follows: 2 N-glycosylation sites, 14 phosphorylation sites, 9 N-myristoylation sites, and 1 Leucine Zipper site (Online Resource 4).
Functional characterization of VuNHX1 using yeast mutants
In order to assess whether VuNHX1 had a role in Na+/H+ exchange, we used a yeast mutant AXT3 lacking the potential plasma membrane antiporters, ENA 1–4 and NHA1 required for Na+ efflux, as well as the vacuolar antiporter NHX1 for compartmentation of toxic Na+ in vacuole. Therefore, the lack of these transporters in AXT3 cells renders them sensitive to Na+ due to the absence of effective protective machinery. Yeast cells transformed with the empty vector pYES2.0 and the construct pYES2.0-VuNHX1 were compared for their ability to grow under salt stress, LiCl and KCl. As shown in Fig. 1 a, wild type and mutant yeast cells transformed with either construct displayed normal growth on APGal liquid media, under normal physiological conditions. However, a statistically significant difference (P ≤ 0.05) in growth pattern was observed in W303-1B wild-type and AXTYES2.0 cells under stress conditions. Under salt stress at 50 mM NaCl, no significant difference in growth was observed between AXTYES2.0 and AXTVuNHX1 cells. However, with increase in NaCl concentration (75 and 100 mM NaCl) a marked difference was observed in their growth. The AXTVuNHX1 cells showed 1.85 and 2.87 times better survival efficiency as compared to AXTYES2.0 cells, under 75 and 100 mM NaCl stress respectively (Fig. 1a). Similarly, pronounced difference under increase in external [K+] between AXTYES2.0 and AXTVuNHX1 cells was observed at 1 M KCl unlike, 0.5 and 0.75 mM KCl stress. AXTYES2.0 cells were able to survive at a similar rate as that of AXTVuNHX1 cells under 15 and 20 mM LiCl stress (Fig. 1a). But, significant growth difference was observed at 25 mM LiCl suggesting the efficient survival of AXTVuNHX1 than AXTYES2.0 cells owing to the cation/proton antiporter activity of VuNHX1 (Fig. 1a). The yeast complementation assay was performed on solid APGal minimal medium and similar results were obtained as observed in liquid media supplemented with different concentrations of Na+, K+ and Li+. The survival efficiency of AXT3 strains expressing VuNHX1 was greater than AXT3 cells transformed with empty pYES2.0 vector (Fig. 1b). Wild-type strain W303-1B was taken as control in each case. Heterologous expression of VuNHX1 under GAL1-inducible promoter restored salt tolerance in AXTVuNHX1 cells at 50 µg/l hygromycin-B (Fig. 1b). Hygromycin-B, a cationic protein synthesis inhibitor is known to confer hypersensitivity in Δnhx1 and Δnhx1 Δnha1 mutants, owing to its increased uptake by cells under changes in plasma membrane electrical potential but, no effect on wild-type and Δnha1 mutants with intact ScNHX1 (Ali et al. 2004; Brett et al. 2005b). Therefore, the growth sensitivity of AXTVuNHX1 cells under a higher concentration of hygromycin-B was least affected indicating the functional complementation of ScNHX1 by heterologous expression of VuNHX1 as compared to AXTYES2.0 cells lacking effective salt tolerance machinery.
a Heterologous expression of VuNHX1 in yeast mutant. Wild type (W303-1B) and ∆ ena1-4 ∆nha1 ∆nhx1 mutant (AXT3) strains used for the complementation assay were transformed with pYES2.0 vector only and pYESVuNHX1 recombinant vector and labeled as AXTYES2.0 and AXTVuNHX1, respectively. Cation sensitivity assay of transformed yeast strains under various concentrations of NaCl (0, 50, 75, 100 mM), KCl (0, 0.5, 0.75, 1.0 M), and LiCl (0, 15, 20, 25 mM). Saturated seed cultures for each strain was diluted to an OD600 of 0.006 and inoculated to liquid APGal medium (pH 5.5) supplemented with or without above mentioned concentrations of NaCl, KCl, and LiCl. Growth was observed at 30 °C after 3 days and absorbance recorded at 600 nm. Data are means of 3 independent events (n = 3) and SE are plotted in the graph. Statistically significant values at P ≤ 0.05 are indicated as different letters using Bonferroni analysis. b 101-, 102-, and 103-fold serial dilutions of saturated seed cultures of each strain were spotted onto APGal media (pH 5.5) supplemented with or without 75 mM NaCl, 50 mM LiCl, 1.0 M KCl and YPGal media (pH 5.5) supplemented with or without 50 µg/ml Hyg. The plates were incubated at 30 °C for 3 days
Na+ and K+ distribution in yeast mutants
The total intracellular ion distribution in the yeast cells (W303-1B, AXTTES2.0, AXTVuNHX1) was determined. Under normal physiological and salt stress condition, no significant difference was observed in total [Na+] in yeast cells, moreover, AXTYES2.0 cells showed relatively higher value than W303-1B and AXTVuNHX1 cells. However, statistically significant difference (P ≤ 0.05) was observed in total [K+] in yeast cells. The W303-1B and AXTVuNHX1 exhibited 2.8 and 1.9 times higher accumulation of total K+, respectively than AXTYES2.0 cells (Fig. 2). This could be due to the activity of antiporters other than Na+/K+/H+ Nhx1 in vacuole involved in K+ transport. Further, ion analysis studied in vacuolar and cytoplasmic region for understanding of distribution of ions in yeast strains revealed no significant difference in vacuolar Na+ accumulation under normal physiological condition (Fig. 2). However, K+ accumulation was found significantly higher in W303-1B (2.7 times) and AXTYESVuNHX1 (2.1 times) as compared to AXTYES2.0 (Fig. 2). Under salt stress, AXTYES2.0 cells exhibited 2.1 and 1.9 times lower vacuolar Na+ than wild-type and ∆ena ∆nha1 ∆nhx1 mutant expressing VuNHX1 (Fig. 2). This could be attributed to the fact that AXTYYES2.0 cells lacked vacuolar antiporter ScNHX1 involved in sequestration of cations. Further, under salt stress the vacuolar K+ accumulation was 2.4 and 1.94 times higher in W303-1B and AXTVuNHX1 as compared to AXTYES2.0 cells. The comparatively higher K+ in AXTVuNHX1 than AXTYES2.0 cells indicated the possible role of VuNHX1 in compartmentalizing K+ along with Na+ in vacuole. Higher [Na+] was observed in the cytoplasmic region of AXTYES2.0 cells as compared to W303-1B and AXTVuNHX1 cells owing to lack of plasma membrane and vacuolar antiporters involved in exclusion and compartmentalization of excess toxic ions.
Total intracellular ion estimation in yeast strains. Wild type (W303-1B) and ∆ ena1-4 ∆nha1 ∆nhx1 mutant (AXT3) strains used for the complementation assay were transformed with pYES2.0 vector only and pYESVuNHX1 recombinant vector and labeled as AXTYES2.0 and AXTVuNHX1, respectively. Yeast cells were grown in APG medium (pH 4.0) with 1 mM KCl supplemented in presence (stressed, S) or absence of 70 mM NaCl (unstressed, US) and harvested at a cell density of 0.3. Total intracellular, vacuolar and cytoplasmic Na+ and K+ content was determined as described in the materials and methods section. Data are means of 3 independent events (n = 3) and SE are plotted in the graph. Statistically significant values at P ≤ 0.05 are indicated as different letters using Bonferroni analysis
Copy number analysis of VuNHX1
The copy number of NHX gene in the cowpea genome was determined by Southern blot analysis. The cowpea genomic DNA was digested separately with three restriction enzymes, EcoRI, HindIII and PstI and then blotted onto membrane and hybridized to a probe specific to 3′-region of VuNHX1 CDS that lacked all three restriction sites. The results indicated presence of a single copy of NHX1 in cowpea genome as revealed from a single hybridization signal in HindIII digested genomic DNA (Fig. 3). The occurrence of two and three signals in case of EcoRI and PstI digested genomic DNA respectively could be due to presence of multiple sites of EcoRI and PstI in VuNHX1.
Expression analysis of VuNHX1 under abiotic stress
To understand the potential role of VuNHX1 in abiotic stress, the expression of this gene was assessed under different stress conditions. Cowpea seedlings were subjected to salt, dehydration or cold treatment for various time periods (0, 6, 12, and 24 h), total RNA was extracted from control and treated seedlings, and used for semi-quantitative RT-PCR assays. The results showed varied response of VuNHX1 under salt, cold and dehydration stress at whole plant level. Under salt stress, a gradual increase in the transcript level was observed after 6 h, in leaves and roots of treated cowpea seedlings (Fig. 4a). The expression of VuNHX1 in cowpea seedlings subjected to salt stress was significantly upregulated (Fig. 4b) indicating the association of NHX1 with salt stress response. The expression of VuNHX1 was also found upregulated under dehydration stress with strong induction at 6 and 12 h of osmotic stress (Fig. 4b), and the results were in agreement with the fact that high saline condition known to mimic osmotic stress in plants. Interestingly, a 3.75 fold increase in VuNHX1 expression was observed under cold stress at 6 h and the expression decreased by 2.2 fold with further increase in cold stress period relative to the seedlings under normal condition (Fig. 4b).The increased expression level under cold stress could be accounted for the cross-talk between NHX1 and stress components involved in low temperature stress.
a Semi-quantitative RT-PCR for studying expression patterns of VuNHX1 under salt stress (200 mM NaCl). Total RNA was isolated from leaves and roots of 10 days old cowpea seedling under 200 mM NaCl treatment at time intervals of 0, 6, 12, and 24 h. b Semi-quantitative RT-PCR for studying expression patterns of VuNHX1 under different abiotic stress conditions such as salt, cold and drought stress. Total RNA was isolated from 10 days old cowpea seedling under 200 mM NaCl, Cold (4 °C), and dehydration treatment at time intervals of 0, 6, 12, and 24 h. PCR fragments of 260 and 292 bp size corresponding to VuNHX1 and NSR9 gene were fractionated electrophoretically on 2 % agarose gel stained with 10 mg/ml ethidium bromide. c Total intracellular ion measurement in leaves and roots of mid stage cowpea seedlings. Na+ and K+ content in leaves and roots of unstressed and salt stressed cowpea seedlings harvested at time intervals of 0, 6, 12, 24, and 48 h was measured using flame photometer. Values indicate mean ± SE (n = 3). Statistically significant values at P ≤ 0.05 are indicated as different letters using Bonferroni analysis
Ion accumulation in salt stressed cowpea seedlings
The Na+ and K+ content in leaves and roots of untreated and salt-treated cowpea seedlings were measured after different time intervals. As shown in Fig. 4c, under normal physiological condition, the Na+ content was almost similar in both leaves as well as roots of cowpea seedlings. Under salt stress, the Na+ accumulation was significantly increased in roots than leaves and the increase was concomitant with increase in exposure time (Fig. 4c). Significant difference (P ≤ 0.05) in Na+ content was observed after 6 h of salt stress in roots and 18 h of stress in leaves relative to unstressed condition (Fig. 4c). The Na+ accumulation was increased by 9.3-fold in roots and 6.1-fold in leaves, after 48 h of salt treatment (Fig. 4c). However, no significant difference was observed in [K+] in leaves under salt stress, unlike in roots after 6 h of salt stress. As compared to control condition, 2.9- and 33.7-fold higher Na+/K+ values were observed in case of leaves and roots, respectively, at 48 h of salt stress. The restriction of movement of Na+ to leaves could possibly the reason for enhanced Na+ accumulation at 48 h of salt stress in cowpea seedlings, and a higher K+/Na+ ratio of 2.1 in leaves as compared to 0.12 in roots. Maintenance of higher K+ in leaves is essential for cellular and ionic homeostasis to attain unhindered growth under hypersalinity and hyperosmotic condition.
Growth of transgenic Arabidopsis overexpressing VuNHX1 under salt stress
To validate the function of VuNHX1 in model plant, Arabidopsis during salt stress, independent T4 homozygous Arabidopsis lines expressing VuNHX1 either through constitutive CaMV35S promoter (Fig. 5a) or stress-responsive AtRD29A promoter (Fig. 5b), were subjected to salt stress. The difference in their growth and survival was monitored. The effect of salt stress on germination was studied with Arabidopsis lines (WT; 35S::VuNHX1, line #4 and RD29A::VuNHX1, line #13) after one week exposure to 150 mM NaCl. Wild-type and transgenic lines exhibited no apparent difference in growth under normal physiological condition (Fig. 5c). However, under salt stress, the transgenic lines (#4 and # 13) exhibited significantly better growth and survival as compared to WT (Fig. 5c).
T-DNA region (7.6 kb) of a pCAMBIA2301-35S::VuNHX1 and b pCAMBIA2301-RD29A::VuNHX1 (14.4 kb). Restrcition enzyme PstI and EcoRI used for cloning 35SP::VuNHX1::35STer cassette (2.3 kb) and RD29A::VuNHX1::35STer cassette (2.8 kb) into plant binary vector pCAMBIA 2301 is also highlighted. LB left border, RB right border; 35SPromoter, cauliflower mosaic virus 35S promoter; RD29A promoter, stress indicible AtRD29A promoter; CaMV 35S poly-A, cauliflower mosaic virus 35S terminator; nos poly-A, nopaline transferase terminator; nptII, neomycin phosphotransferase; intron-gus-A, intron interrupted β-glucuronidase; VuNHX1, Vigna unguiculata NHX1. c Study on effect of salt stress (150 mM NaCl) on germination efficiency of wild-type WT (col-0) and transgenic Arabidopsis 35S:VuNHX1 (#4) and RD29A::VuNHX1 (#13) lines
The physiological growth parameters (root length and lateral root development) were monitored in WT and independent T4 homozygous transgenic Arabidopsis lines expressing VuNHX1 constitutively (Line #4, 35S::VuNHX1) and in inducible manner (Line #13, RD29A::VuNHX1) that were subjected to 150 or 200 mM NaCl. The difference in root length was measured after 7 days of salt stress. Under control unstressed condition, no significant difference was observed between WT and transgenic Arabidopsis lines (Fig. 6a). However, a significant difference (P ≤ 0.05) in root length was observed in transgenic Arabidopsis lines under 150 and 200 mM NaCl stress as compared to WT plants. Under salt stress, root growth inhibition was significantly lower in transgenic Arabidopsis lines (35S::VuNHX1 and RD29A::VuNHX1) than WT (Fig. 6a, b). Lateral root development was affected by salinity stress. Under 200 mM salt stress, significantly (P ≤ 0.05) higher lateral root number was observed in transgenic lines (#4 and #13) as compared to WT (Fig. 6b).
a Study of root growth inhibition in wild type (WT, Col-0) and transgenic Arabidopsis lines (Line #4, 35S::VuNHX1 and Line #13, RD29A::VuNHX1) upon salt stress. Excessive NaCl-induced root growth inhibition in Col-0 wild-type (WT) plants was observed as compared to transgenic lines. b Root length (cm) and lateral root number was measured after exposure of WT and transgenic lines to 150 and 200 mM NaCl stress for 1 week. Values indicate mean ± SE (n = 10). Statistically significant values at P ≤ 0.05 are indicated as different letters using Bonferroni analysis. c Salt tolerance assay in wild type (WT, Col-0) and transgenic Arabidopsis plants expressing VuNHX1 constitutively (Line #4, 35S::VuNHX1) and inducibly (Line #13, RD29A::VuNHX1) subjected to 200 mM NaCl treatment for 2 weeks. d Relative transgene expression level of VuNHX1 in transgenic Arabidopsis lines. No transgene expression was observed in WT. A 0.28 kb fragment of VuNHX1::35SployA and 0.150 kb fragment of AtUBQ1 was amplified in case of transgenic Arabidopsis lines
The effect of salt stress on mature Arabidopsis plants was studied by subjecting the mature WT and transgenic lines (Line #4, 35S::VuNHX1 and Line #13, RD29A::VuNHX1) to salt stress (200 mM NaCl) and growth and physiological parameters were recorded. Under salt stress, plant growth was found inhibited in WT with symptoms of salinity induced leaf senescence whereas, transgenic plants (35S::VuNHX1 and RD29A::VuNHX1) exhibited better survival and comparatively less salinity induced leaf senescence (Fig. 6c). Further, real-time PCR analysis indicated a 1.34-fold higher expression of VuNHX1 in line #13 (RD29A::VuNHX1) as compared to line #4 (35S::VuNHX1) (Fig. 6 d) indicating the stress inducible expression of VuNHX1 provided enhanced salt tolerance in comparison to constitutive expression.
The leaves from WT and transgenic lines (#4 and #13) were analyzed for Na+ and K+ accumulation, chlorophyll, malondialdehyde (MDA) and proline content, after subjecting the plants to salt stress for 2 weeks. Under normal physiological condition, no difference in Na+ content was observed between WT and transgenic lines however, transgenic lines #4 and #13 accumulated 1.06 and 1.34 times higher Na+ than WT, respectively after salt stress. Transgenic line #4 exhibited 1.93 times and line #13 1.66 times higher accumulation of K+ than WT. Consequently, transgenic lines #4 and #13 maintained a 1.8 and 1.2 fold higher K+/Na+ ratio, respectively than WT (Fig. 7). The maintenance of K+ homeostasis under salinity is essential for regulation of plant growth and survival.
Na+ and K+ content (μmoles/g DW) was estimated in leaves of salt stressed (200 mM NaCl) and unstressed (0 mM NaCl) WT and transgenic lines (Line #4, 35S::VuNHX1 and Line #13, RD29A::VuNHX1), as described in “Materials and methods” section. Values indicate mean ± SE (n = 3). Statistically significant values at P ≤ 0.05 are indicated as different letters using Bonferroni analysis
The salt stress apparently penalized the photosynthesis machinery in WT as evident from 1.9 fold lower chlorophyll content in WT as compared to transgenics (Fig. 8). The MDA content was found 1.2 fold lower in transgenics as compared to WT under salt stress (Fig. 8) indicating lower lipid peroxidation in Arabidopsis overexpressing VuNHX1. A 1.53 fold higher proline content in transgenic line #4 than WT revealed better osmoprotection under salt stress in transgenic Arabidopsis overexpressing VuNHX1. In general, the transgenic line (#4) expressing VuNHX1 under constitutive promoter exhibited higher proline accumulation than transgenic line (#13) that expressed VuNHX1 under stress inducible promoter (Fig. 8).
Physiological analysis of WT (Col-0) and transgenic Arabidopsis plants expressing VuNHX1 constitutively (Line #4, 35S::VuNHX1) and inducibly (Line #13, RD29A::VuNHX1) upon salt stress. Changes in chlorophyll, MDA and proline content were estimated and analyzed as explained in “Materials and methods” section. Values indicate mean ± SE (n = 3). Statistically significant values at P ≤ 0.05 are indicated as different letters using Bonferroni analysis
Discussion
In this report, a vacuolar Na+/H+ antiporter gene, VuNHX1 was isolated from cowpea, a dry land grain legume with wide adaptability to varied climatic conditions. Cowpea is known as a drought hardy crop and has been reported to have moderate tolerance to salinity (Murillo-Amador et al. 2006; Duzdemir et al. 2009). Sequence analysis revealed that it encoded an AtNHX1-like antiporter protein, and consequently named as VuNHX1. The VuNHX1 is the first orthologue of AtNHX1 isolated from cowpea. Phylogenetic analysis of predicted VuNHX1 protein revealed it as Na+/H+ antiporter located in the vacuole and clearly distinct from those located at the endosomal or plasma membrane. The results indicated that VuNHX1 was evolutionary closer and shared a high homology with legume NHX1. The topological analysis showed VuNHX1 comprised of 11 transmembrane domains, in addition to the putative amiloride binding motif at TM3, common to vacuolar Na+/H+ antiporters. These results suggest that VuNHX1 corresponds to a Na+/H+ antiporter localized in the vacuole. The protein homology and topology suggested that VuNHX1 might have similar function and regulation role in salt tolerance as that of dicot glycophyte plants. Southern analysis of cowpea genomic DNA revealed presence of a single copy of VuNHX1, unlike the earlier reports of occurrence of multiple copies NHX1 in other plants (Wang et al. 2003; Qingxia et al. 2009).
The role of VuNHX1 was assessed by complementation of yeast wild type and mutant AXT3 strains. Yeast endosomal Na+/H+ exchanger shares functional similarity with plant Na+/H+ exchanger, and thereby provide an excellent platform to validate the function of plant Na+/H+ exchangers in salt tolerance (Darley et al. 2000; Quintero et al. 2000). Plasma membrane and endosomal Na+/H+ antiporters in yeast are involved in ion homeostasis, and cellular pH maintenance. Therefore, ∆ena∆nha1∆nhx1 mutant shows high sensitivity to increased level of Na+ and K+ at low pH condition. Additionally, the ∆nhx1 mutant is sensitive to hygromycin-B, a cationic aminoglycoside antibiotic due to defective sequestration in vacuolar lumen (Brett et al. 2005b). Heterologous expression of plant endosomal Na+/H+ antiporters, AtNHX1 (Hernandez et al. 2009), MzNHX1 (Qingxia et al. 2009), MsNHX1 (Bao-Yan et al. 2008), TSNHX1 (Gouiaa et al. 2012), AeNHX1 (Qiao et al. 2007), CmNHX1 (Wang et al. 2011), ThNHX1 (Wu et al. 2009), PgNHX1 (Rajagopal et al. 2007) in mutant AXT3 have earlier allowed to complement its salt sensitive phenotype. Expression of VuNHX1 in ∆ena∆nha1∆nhx1 mutant enhanced its growth abilities under low osmolarity media and in presence of high concentrations of Na+, K+, and Li+ as compared to AXT3 mutant harboring only pYES2.0. Suppression of hygromycin sensitivity further supported the role of VuNHX1 in functional complemention of yeast vacuolar NHX exchanger, ScNHX1. The total Na+ and K+ content in AXT3 cells transformed with pYES2.0 and pYESVuNHX1 were in accordance with the function of VuNHX1. The results suggested that the role of VuNHX1 is probably limited to vacuolar sequestration of alkali cations for establishing ion homeostasis. Similar findings were reported for OsNHX1 expressed in ∆ena∆nha1∆nhx1 mutant (Kinclova-Zimmermannova et al. 2004). Moreover, VuNHX1 expression in AXT3 showed enhanced K+ distribution within both cytoplasmic and vacuolar fractions which was in accordance with the suggested vacuolar cation/proton activity.
In this study, it was observed that VuNHX1 expression is responsive to salt stress and upregulation of VuNHX1 was higher in roots than in leaves. Differential distribution of Na+ in plants is a deciding factor for enhanced salt tolerance (Tester and Davenport 2003). Higher expression in roots was reported for ZmNHX (Zorb et al. 2005), ThNHX1 (Wu et al. 2009) and AlNHX1 (Zhang et al. 2008) whereas, lower expression reported for TrNHX1 (Tang et al. 2010), MsNHX1 (Bao-Yan et al. 2008) and DmNHX1 (Zhang et al. 2012). In cowpea, in addition to salt stress, dehydration and cold stress also induced the expression of VuNHX1. However, down-regulation of NHX has been reported under cold stress in P. glaucum (Rajagopal et al. 2007) and T. halophila (Wu et al. 2009) with no expression in A. thaliana (Shi and Zhu 2002). In cowpea, the upregulation of VuNHX1 in response to dehydration and cold stress is indicative of possible cross-talks between various abiotic stress. Up-regulation of NHX1 under dehydration stress has been reported for EgNHX1 (Baltierra et al. 2012), GmNHX1 (Li et al. 2006), and OsNHX1 (Fukuda et al. 2004). Cowpea under salt stress accumulated higher Na+ in roots than shoots and the content increased with increase in stress period. The results were in agreement with the semi-quantitative expression data indicating that higher Na+ were sequestered in root vacuoles and thereby restricting their movement to the aerial part that maintained a higher K+/Na+ ratio. The VuNHX1 upregulation and higher sequestration of Na+ in roots was indicative to the salt stress response associated with NHX1 in cowpea.
Inhibition of seed germination, plant growth and induced senescence are accelerated by salt stress (Lee and Zhu 2010). In our results, transgenic Arabidopsis expressing VuNHX1 through stress inducible promoter (Line #13) or constitutive promoter (Line #4) exhibited comparable degree of salt tolerance. The transgenic lines (#4 and #13) showed significantly higher germination rate, root length and lateral root emergence than WT. The higher Na+ displayed in leaves of transgenic lines (#4 and 13) was possibly due to the efficient compartmentalization of Na+ by VuNHX1. The transgenic lines showed reduced leaf senescence, chlorophyll and MDA content indicating an active protection against salt and oxidative stress. The higher K+ accumulation in transgenic plants under salt stress could be due to Na+/K+/H+ activity of VuNHX1. The maintenance of higher K+/Na+ ratio in transgenic plants was essential for ionic homeostasis under salt stress. Our results clearly demonstrated that transgenic Arabidopsis plants expressing VuNHX1 had better survival under salt stress.
In summary, we have isolated a novel vacuolar Na+/H+ antiporter gene from cowpea for the first time, which allowed a yeast mutant strain lacking Scnhx1 to suppress its Na+ sensitive phenotype. Additionally, we showed that VuNHX1 expression increased significantly in response to salinity, dehydration as well as cold stress, even though most studies associate its role to salt stress. Moreover, VuNHX1 stable expression in A. thaliana plants subjected to high salt concentration improved their performance in comparison to wild type plants. These results suggested VuNHX1 overexpression in cowpea through cisgenic and in other legumes by transgenic approach might confer enhanced tolerance to salinity, and possibly to other stress through cross talk triggering.
References
Ali R, Brett CL, Mukherjee S, Rao R (2004) Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J Biol Chem 279:4498–4506
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Apse MP, Sottosanto JB, Blumwald E (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J 36:229–239
Baltierra Q, Castillo M, Gamboa MC et al (2012) Molecular characterization of a novel Na+/H+ antiporter cDNA from Eucalyptus globules. Biochem Biophys Res Commun 430:535–540
Bao-Yan AN, Yan L, Jia-Rui LI et al (2008) Expression of a vacuolar Na+/H+ Antiporter gene of alfalfa enhances salinity tolerance in transgenic Arabidopsis. Acta Agron Sin 34:557–564
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Brett CL, Donowitz M, Rao R (2005a) The evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol 288:223–239
Brett CL, Tukaye DN, Mukherjee S, Rao R (2005b) The yeast endosomal Na+(K+)/H+ exchanger Nhx1 regulates cellular ph to control vesicle trafficking. Mol Biol Cell 16:1396–1405
Brini F, Gaxiola RA, Berkowitz GA, Masmoudi K (2005) Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump. Plant Physiol Biochem 43:347–354
Chauhan S, Forstoefel N, Ran Y, Quigley F, Nelson DE, Bohnert HJ (2000) Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crsytallinum. Plant J 24:511–522
Chen LH, Zhang B, Xu ZQ (2008) Salt tolerance conferred by overexpression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1 in common buckwheat (Fagopyrum esculentum). Transgenic Res 17:121–132
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Darley CP, van Wuytswinkel OCM, van der Woude K, Mager WH, de Boer AH (2000) Arabidopsis thaliana and Saccharomyces cerevisiae NHX1 genes encode amiloride sensitive electroneutral Na+/H+ exchangers. Biochem J 351:241–249
Duzdemir O, Unlukara A, Kurunc A (2009) Response of cowpea (Vigna unguiculata) to salinity and irrigation regimes. N Z J Crop Hort Sci 37:271–280
Fukuda A, Nakamura A, Tanaka Y (1999) Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim Biophys Acta 1446:149–155
Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45:146–159
Gattiker A, Gasteiger E, Bairoch A (2002) ScanPROSITE: a reference implementation of a PROSITE scanning tool. Appl Bioinformatics 1:107–108
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11:681–684
Gietz D, St. Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20:1425
Gouiaa S, Khoudi H, Leidi EO et al (2012) Expression of wheat Na+/H+ antiporter TNHXS1 and H+-pyrophosphatase TVP1 genes in tobacco from a bicistronic transcriptional unit improves salt tolerance. Plant Mol Biol 79:137–155
Hall AE (2012) Phenotyping cowpeas for adaptation to drought. Front Physiol 3:00155
Hamada A, Shono M, Xia T, Ohta M, Hayashi Y, Tanaka A, Hayakawa T (2001) Isolation and characterization of a Na+/H+ antiporter gene from the halophyte Atriplex gmelini. Plant Mol Biol 46:35–42
Hanana M, Cagnac O, Yamaguchi T, Hamdi S, Ghorbel A, Blumwald E (2007) A grape berry (Vitis vinifera L.) cation/proton antiporter is associated with berry ripening. Plant Cell Physiol 48:804–811
Hasegawa PM (2013) Sodium (Na+) homeostasis and salt tolerance of plants. Environ Exp Bot 92:19–31
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Bio 51:463–499
He CX, Yan JQ, Shen GX, Fu LH, Holaday AS, Auld D, Blumwald E, Zhang H (2005) Expression of an arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant Cell Physiol 46:1848–1854
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetic and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198
Hernandez A, Jiang X, Cubero B, Nieto PM, Bressan RA, Hasegawa PM, Pardo JM (2009) Mutants of the Arabidopsis thaliana cation/H+ antiporter AtNHX1 conferring increased salt tolerance in yeast: the endosome/prevacuolar compartment is a target for salt toxicity. J Biol Chem 284:14276–14285
Hofmann K, Stoffel W (1993) A database of membrane spanning proteins segments. Biol Chem 374:166
Jha A, Joshi M, Yadav NS, Agarwal PK, Jha B (2011) Cloning and characterization of the Salicornia brachiata Na+/H+ antiporter gene SbNHX1 and its expression by abiotic stress. Mol Biol Rep 38:1965–1973
Kagami T, Suzuki M (2005) Molecular and functional analysis of a vacuolar Na+/H+ antiporter gene of Rosa hybrida. Genes Genet Syst 80:121–128
Kinclova-Zimmermannova O, Flegelova H, Sychrova H (2004) Rice Na+/H+-antiporter Nhx1 partially complements the alkali-metal-cation sensitivity of yeast strains lacking three sodium transporters. Folia Microbiol 49:519–525
Kronzucker HJ, Britto DT (2011) Sodium transport in plants: a critical review. New Phytol 189:54–81
Lee BH, Zhu JK (2010) Phenotypic analysis of Arabidopsis mutants: germination rate under salt/hormone-induced stress. Cold Spring Harb Protoc 2010:pdb-prot4969
Li WYF, Wong FL, Tsai SN, Phang TH et al (2006) Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ 29:1122–1137
Lichtenthaler HK (1987) Chlorophyll fluorescence signatures of leaves during the autumnal chlorophyll breakdown. J Plant Physiol 131:101–110
Ma XL, Zhang Q, Shi HZ, Zhu JK, Zhao YX, Ma CL, Zhang H (2004) Molecular cloning and different expression of a vacuolar Na+/H+ antiporter gene in Suada salsa under salt stress. Biol Plant 48:219–225
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Murillo-Amador B, Troyo-Dieguez E, Garcia-Hernandez JL et al (2006) Effect of NaCl salinity in the genotypic variation of cowpea (Vigna unguiculata) during early vegetative growth. Sci Hortic 108:423–431
Ohta M, Hayashi Y, Nakashima A, Hamada A, Tanaka A, Nakamura T, Hayakawa T (2002) Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett 532:279–282
Qiao WH, Zhao XY, Li W et al (2007) Overexpression of AeNHX1, a root-specific vacuolar Na+/H+ antiporter from Agropyron elongatum, confers salt tolerance to Arabidopsis and Festuca plants. Plant Cell Rep 26:1663–1672
Qingxia Z, Xuefeng X, Wang Y, Tianzhong L, Jin K, Zhenhai H (2009) Isolation and preliminary function analysis of a Na+/H+ antiporter gene from Malus zumi. Afr J Biotechnol 8:4774–4781
Quintero FJ, Blatt MR, Pardo JM (2000) Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett 471:224–228
Rajagopal D, Agarwal P, Tyagi W et al (2007) Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Mol Breed 19:137–151
Reguera M, Bassil E, Blumwald E (2014) Intracellular NHX-type cation/H+ antiporters in plants. Mol Plant 7:261–263
Rodriguez-Navarro A, Ramos J (1984) Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol 159:940–945
Shi H, Zhu JK (2002) Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol 50:543–550
Singh BB (2005) Cowpea [Vigna unguiculata (L.) Walp. In: Singh RJ, Jauhar PP (eds) Genetic resources, chromosomal engineering and crop improvement, vol 1. CRC Press, Boca Raton, pp 117–162
Sottosanto JB, Gelli A, Blumwald E (2004) DNA array analyses of Arabidopsis thaliana lacking a vacuolar Na+/H+ antiporter: impact of AtNHX1 on gene expression. Plant J 40:752–771
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tang R, Li C, Xu K et al (2010) Isolation, functional characterization, and expression pattern of a vacuolar Na+/H+ antiporter gene TrNHX1 from Trifolium repens L. Plant Mol Biol Rep 28:102–111
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgin DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Timko MP, Ehlers JD, Roberts PA (2007) Cowpea. In: Kole (ed) Genome mapping and molecular breeding in plants, vol 3, pulses, sugar and tuber crops. Springer, Berlin pp 49–67
Venema K, Belver A, Marin-Manzano MC, Rodgriguez-Rosales MP, Donaire JP (2003) A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J Biol Chem 278:22453–22459
Wang J, Zuo K, Wu W, Song J, Sun X, Lin J, Li X, Tang K (2003) Molecular cloning and characterization of a new Na+/H+ antiporter gene from Brassica napus. DNA Seq 14:351–358
Wang S, Zhang YD, Perez PG et al (2011) Isolation and characterization of a vacuolar Na+/H+ antiporter gene from Cucumis melo L. Afr J Biotechnol 10:1752–1759
Wu CA, Yang GD, Meng QW, Zheng CC (2004) The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol 45:600–607
Wu YY, Chen QJ, Chen M, Chen J, Wang XC (2005) Salt-tolerant transgenic perennial ryegrass (Lolium perenne L.) obtained by Agrobacterium tumefaciens-mediated transformation of the vacuolar Na+/H+ antiporter gene. Plant Sci 169:65–73
Wu C, Gao X, Kong X et al (2009) Molecular cloning and functional analysis of a Na+/H+ antiporter gene ThNHX1 from a halophytic plant Thellungiella halophila. Plant Mol Biol Rep 27:1–12
Xia T, Apse MP, Aharon GS, Blumwald E (2002) Identification and characterization of a NaCl-inducible vacuolar Na+/H+ antiporter in Beta vulgaris. Physiol Plant 116:206–212
Xue ZY, Zhi DY, Xue GP, Zhang H, Zhao YX, Xia GM (2004) Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 167:849–859
Yamaguchi T, Fukada-Tanaka S, Inagaki Y, Saito N, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Iida S (2001) Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant Cell Physiol 42:451–461
Yamaguchi T, Apse MP, Shi H, Blumwald E (2003) Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci USA 100:12510–12515
Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19:765–768
Zhang HX, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98:12832–12836
Zhang GH, Su Q, An LJ, Wu S (2008) Charcterization and expression of a vacuolar Na+/H+ antiporter gene from the monocot halophyte Aeluropus littoralis. Plant Physiol Biochem 46:117–126
Zhang H, Liu Y, Xu Y, Chapman S, Love AJ, Xia T (2012) A newly isolated Na+/H+ antiporter gene, DmNHX1, confers salt tolerance when expressed transiently in Nicotiana benthamiana or stably in Arabidopsis thaliana. Plant Cell Tissue Organ Cult 110:189–200
Zhao JS, Zhi DY, Xue ZY, Liu H, Xia GM (2007) Enhanced salt tolerance of transgenic progeny of tall fescue (Festuca arundinacea) expressing a vacuolar Na+/H+ antiporter gene from Arabidopsis. J Plant Physiol 164:1377–1383
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Zorb C, Noll A, Noll A, Karl S, Leib K, Yan F, Schubert S (2005) Molecular characterization of Na+/H+ antiporters (ZmNHX) of maize (Zea mays L.) and their expression under salt stress. J Plant Physiol 162:55–65
Acknowledgments
We express our sincere thanks to Prof. Edward Blumwald and Dr. Olivier Cagnac for the yeast strains, W303 and AXT3 respectively. We also thank Dr. Luciana LoureiroPenha for providing pYES2.0 vector for yeast expression analysis, Department of Civil Engineering, IIT Guwahati for use of Flame Photometry. LS is grateful to DBT (Department of Biotechnology, Government of India) for its support through various Grants (BT/PR10818/AGR/02/591/2008 and BT/01/NE/PS/08) for legume improvement program. BhL is grateful to Rural Development Administration, Republic of Korea for its support by Next-Generation BioGreen 21 Program (PJ009104). SM is grateful to MHRD for Research Fellowship. SKP is grateful to DBT (Department of Biotechnology, Government of India) for its support.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mishra, S., Alavilli, H., Lee, Bh. et al. Cloning and characterization of a novel vacuolar Na+/H+ antiporter gene (VuNHX1) from drought hardy legume, cowpea for salt tolerance. Plant Cell Tiss Organ Cult 120, 19–33 (2015). https://doi.org/10.1007/s11240-014-0572-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-014-0572-7