Abstract
To develop doubled haploid (DH) mapping populations of hexaploid Brassica, 10 F1 hybrids derived from crosses between allohexaploid Brassica parents were evaluated in this study. The allohexaploid Brassica parents were selfed progenies of unique interspecific crosses between Brassica rapa (genome AA) × B. carinata (BBCC), B. nigra (BB) × B. napus (AACC), and a complex cross between B. juncea (AABB), B. napus and B. carinata, with relatively stable chromosome number (2n = 54). Hexaploid status and chromosome behavior during meiosis I in four promising F1 hybrids were assessed using microscopy and flow cytometry, and progeny were obtained following microspore culture. Hybrids H11-2 and H16-1 demonstrated higher amenability for embryo generation, plantlet regeneration, and frequency of production of DH microspore-derived progeny of hexaploid DNA content (6x) compared to hybrids H08-1 and H24-1. A total of 370 6x DH progeny were selected out of 693 plantlets from H11-2, 241/436 from H16-1, 23/54 from H08-1, and 21/56 from H24-1. DH progenies of hybrids H11-2 and H16-1 were then designated as promising mapping populations of a new hexaploid Brassica species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassica crops play important roles in agriculture, from highly diverse vegetables to oilseed crops. Natural polyploidisation among the three diploid species (Brassica oleracea, B. nigra and B. rapa) produced three allotetraploid agricultural species (B. carinata, B. juncea, and B. napus), as described in the Triangle of U (U 1935). The diploid Brassica species contain three distinct genomes: Brassica rapa (AA genome), B. nigra (BB genome) and B. oleracea (CC genome) (U 1935). Each species is genetically diverse and has its own specific adaptations. For example, B. rapa is very genetically diverse and has many useful traits, such as its strong resistance to humidity and cold tolerance and its short growth period in subtropical climates, but relatively low grain yield and disease susceptibility limits its application (Kimber and McGregor 1995).
Polyploidisation is an important feature of plant evolution and speciation (Levin 2002; Bennett 2004; Soltis and Soltis 1995). More than 70 % of angiosperms have gone through one or more genome duplication events during the process of evolution (Masterson 1994). The three Brassica diploid species naturally combined in pairwise fashion to form three allotetraploid species, namely B. juncea (AABB), B. carinata (BBCC) and B. napus (AACC). However, no allohexaploid Brassica species are known in nature. Recently, it was proposed that an allohexaploid Brassica species could be formed and tested for tolerance to a range of environmental conditions (Chen et al. 2011), based on the observation that many allopolyploid species are more widely adaptable than their lower ploidy parent species (Murphy et al. 1995). For example, bread wheat (Triticum aestivum), a hexaploid crop with A, B and D genomes, is more widely adapted than its lower ploidy relatives, and is now grown from the tropics to the extreme latitudes of agricultural production (Dubcovsky and Dvorak 2007; Leitch and Leitch 2008). Oats are also a hexaploid crop species (Avena sativa L., genome AACCDD) (Feldman 2001). Chen et al. (2011) hypothesized that an allohexaploid Brassica would also have greater buffering capacity against the shock of climate change and adaptation to a wider range of heat-stressed, drought-stressed, toxic or saline land than their diploid or tetraploid relatives.
Recently, unique allohexaploid Brassica lines have been synthesized independently in Australia and China through different interspecific hybridization approaches. Two allohexaploid Brassica populations were developed in China from interspecific crosses between B. rapa and B. carinata, and these populations were advanced 4–5 generations with selection for increasing stability (Li et al. 2005a, b; Tian et al. 2010; Ge et al. 2009). In Australia, allohexaploid Brassica was produced through the cross between B. nigra and B. napus (Pradhan et al. 2010) and also through crosses among the allotetraploid species B. juncea, B. napus and B. carinata (Mason et al. 2012). These allohexaploid Brassica from different sources provide invaluable germplasm to breed a diverse but stable hexaploid species. Hexaploid Brassica showed instability in chromosome number during selfing, and progenies were selected for “normal” chromosome pairing and segregation over several selfing generations (Tian et al. 2010). This is a similar issue to that experienced in the synthetic allohexaploid crop triticale (AABBRR), the stability of which was improved greatly in later generations following artificial allopolyploidisation from different genetic backgrounds (Larter and Gustafson 1980).
We chose diverse parental Brassica material with hexaploid chromosome status from various sources in China and Australia (Li et al. 2005a, b; Tian et al. 2010; Ge et al. 2009; Pradhan et al. 2010; Mason et al. 2012) for intercrossing and selection of potentially stable hexaploid Brassica populations. The current research was undertaken to establish DH populations for future genetic mapping of Brassica hexaploids.
Microspore culture is important for genetic and genomic research on Brassica species (Xu et al. 2007; Ferrie and Mollers 2011). While genetic variation exists within the allotetraploid B. napus for production of DH progeny through microspore culture, various techniques have improved embryo recovery and diploidisation from microspore culture (Ahmadi et al. 2012; Takahira et al. 2011). In some diploid Brassica species such as B. oleracea, it is more difficult to obtain embryos (Winarto and da Silva 2011). Colchicine is most widely used for chromosome doubling (Zhou et al. 2002a, b; Mohammadi et al. 2012). DH populations derived from microspore culture have been widely used to establish genetic linkage maps of allotetraploid B. carinata and B. napus (Guo et al. 2012; Raman et al. 2012). Ferreira et al. (1994) used a DH population from microspore culture to establish a RFLP linkage map of B. napus. Yu et al. (2009) constructed an SSR-based genetic linkage map of diploid B. rapa using a DH population. This is the first report of microspore culture on hexaploid Brassica to establish DH populations for genetic mapping.
Materials and methods
Plant materials
Allohexaploid Brassica parents were derived from several interspecific crosses between B. rapa (AA) and B. carinata (BBCC) (Li et al. 2005a, b; Ge et al. 2009), B. nigra (BB) and B. napus (AACC) (Pradhan et al. 2010) and complex crosses among the allotetraploid species B. juncea (AABB), B. napus and B. carinata (Mason et al. 2012) (Table 1).
A total of 18 plants were selected for crossing from 4 different sources of allohexaploid plants with 30 % or higher pollen viability and the expected number of 54 chromosomes in somatic tissue as determined by microscopic examination. Each selected plant was numbered and crossed in various combinations with other selected plants. Several flowers on each plant were emasculated and pollinated in each cross. Crosses yielded a total of 0–66 hybrid seeds. From 22 successful crosses, 100 F1 hybrid plants were grown together with 18 parental lines in the glasshouse at The University of Western Australia in 2010 to evaluate hybrid vigour and agronomic suitability. One F1 seed per cross was used to grow as a mother plant for microspore culture.
The parent allohexaploids and their F1 hybrid plants were grown in the glasshouse for chromosome counting and pollen viability assessment (Table 1). From 100 F1 hybrids formed among these sources of Brassica allohexaploids, 10 hybrids with diverse pedigree, high parental chromosome number, relatively high pollen viability and improved seed-setting ability were chosen for preliminary observation in microspore culture. Cuttings were made from these 10 F1 hybrids and were grown in a phytotron at 18 °C/13 °C (day/night) with a 16 h photoperiod at a light intensity of approximately 500 μmol m−2 s−1 for a second round of microspore culture, chromosome counting and pollen viability (Table 1).
Cytological observations
Chromosome number at mitotic metaphase
The chromosome numbers of 10 F1 Brassica hybrid plants and microspore-derived (MD) progenies were determined in somatic tissue on the young ovaries of new buds, at mitotic metaphase. At least 3 young ovaries per F1 were collected, treated with 2 mM 8-hydroxyquinoline for 4 h and then fixed in a mixture of ethanol: acetic acid (3:1) overnight, transferred to fresh fixative mixture and stored in freezer until use. Cells were checked for chromosome number following the method of Li et al. (Larter and Gustafson 1980; Li et al. 1995). Tissue was hydrolyzed in 1 N HCl at 60 °C for about 8 min, squashed in a drop of 10 % modified carbol fuchsin and the chromosome number was recorded at mitotic metaphase in 3–5 somatic cells. The highest number was chosen as the final chromosome number.
Chromosome pairing during meiosis
Meiosis was observed in pollen mother cells (PMC) in young anthers from the terminal inflorescence buds, following a modification of Li et al. (1995). More than 3 young buds (approximately 1.5 mm in length) were collected on ice between 9 a.m. and 10 a.m. in the morning from each hybrid plant in the phytotron, fixed in Carnoy’s solution [ethanol: acetic acid (3:1)] (Larter and Gustafson 1980; Li et al. 1995) overnight, transferred into fresh fixative solution every day until the solution became clear and stored in a freezer until use. The anthers were hydrolyzed in 1 N HCl at 60 °C for about 2 min and squashed gently in a drop of 10 % modified carbol fuchsin. A total of 100 PMCs were observed for each hybrid plant under the microscope.
Pollen viability
Pollen viability was measured by staining pollen grains from three young open flowers with 1 % acetocarmine. Approximately 600 pollen grains from anthers of three flowers were counted from each F1 hybrid plant. Normal pollen grains were large, round and densely stained red and easily distinguished from small, non-stained dead pollen and immature microspore stage cells (Li et al. 1995).
Microspore isolation and regeneration
In a preliminary study, about 80 flower buds were collected from cuttings of 10 F1 hybrids in the phytotron, placed on ice and brought to the laboratory where buds with length of 2–4 mm were selected and sterilized in 1 % hypochlorite for 15 min. The sterilized buds were transferred to a 100 μm sterile filter in the laminar flow and washed with 800 ml sterile water. After washing, the buds were transferred to a sterile beaker and 5 ml 1/2 B5-13 (half strength B5 Gamborg medium from Austratec TM plus 13 % sucrose in Millipore water, pH to 5.8). The microspores were released into the solution by gently squeezing buds using a glass rod. Then the solution with microspores were filtered through a 44 μm filter into a sterile 50 ml tube and adjusted to 30 ml with 1/2 B5-13 media. The microspores were centrifuged for 5 min at 1,200 rpm, the supernatant was discarded and resuspended in 30 ml fresh 1/2 B5-13 medium and centrifuged again for 5 min at 1,200 rpm. The supernatant was discarded again and the microspores were resuspended in 30 ml NLN-13 solution (NLN medium powder from Austratec plus 13 % sucrose in Millipore water, pH to 5.8).
Microspore suspensions were prepared following the method of Cousin and Nelson (2009) and Nelson et al. (2009), transferred to fresh NLN-13 medium and adjusted to a density of 4 × 105 mL−1. The microspore suspension was divided into 10 mL lots, to which were added 100 μL 1 % activated charcoal and 0.5 % colchicine in Petri dishes. All the Petri dishes were sealed and wrapped with aluminium foil. Microspores were induced by a heat treatment at 32.5 °C for three days, after which 10 mL NLN-13 medium was added to each dish, and the plates were then placed in a culture room at 25 °C. Two weeks later, the plates were put on a shaker (60 rpm) for 1 week, foil was removed, and they were placed in light (60 μmol m−2 s−1) on a shaker for one more week. The dishes were transferred to 4 °C for storage and cold treatment to enhance embryo development, following the method of Zhang et al. (2006). Two weeks later, young embryos with good quality were transferred to 90 mm Petri dishes (25 embryos per dish) and counted.
Four F1 hybrids were chosen for further manipulation of plantlets in culture as described by Takahira et al. (2011). Regenerated shoots with roots were transferred into soil in pots with a clear plastic cover to increase humidity for 1 week in the phytotron.
Flow cytometry analysis
Approximately 5 mg of fresh young leaf material was collected from microspore-derived (MD) plantlets in the phytotron, chopped by razor and prepared for flow cytometry following the conventional method, using lettuce as a control, as described by Nelson et al. (2009). The stained nuclei samples were analyzed using a BD FACSCanto II (BD Biosciences, San Jose, CA, USA) flow cytometer with a 488 nm laser. The genome size of samples was estimated through the use of a reference standard (lettuce) with known genome DNA content of 5.95 pg (Nelson et al. 2009). In addition, control Brassica samples were included, including allotetraploid B. napus. The absolute DNA amount of a sample was calculated based on the values of the G1 peak means: Sample 2C DNA content = [(sample G1 peak mean)/(standard G1 peak mean)] × [standard 2C DNA content (pg DNA)].
Sample data were acquired using BD FACSDiva V6.1.1 (BD Biosciences San Jose, CA, USA), which is the operating software on the FACSCanto II. Experimental data were analyzed and CV calculations performed using FlowJo V7.2.5 (Tree Star Inc., Ashland, OR, USA) flow cytometry analysis software. Samples were classified as hexaploid, triploid, mixed or other types of ploidy.
Statistical analysis
The data were analyzed using a statistical package, SPSS version 16.0 (SPSS, Chicago, IL, USA). The variation among F1 hybrids in pollen viability, production of normal cells, four different abnormal cell types during meiosis, embryo production and also plant regeneration in microspore culture, was evaluated by analysis of variance (ANOVA) followed by Tukey’s Honest Significant Differences test. All statistical analysis data had three repeats. Results were considered significant at P ≤ 0.05.
Results
Ten hexaploid F1 hybrids were selected for these studies based on diverse pedigree and stable and high chromosome numbers in parent lines, relatively high pollen viability and high seed yield of the F1 hybrids in glasshouse, and high rates of embryogenesis, plantlet regeneration rate and chromosome doubling ability in microspore culture (Table 1). The grandparents of the hexaploid hybrids included 5 of the 6 Brassica species in U’s triangle (U 1935) (Table 1), which improved the likelihood of finding high genetic diversity between parents in the A, B and C genomes.
Chromosome behavior and pollen viability in hexaploid F1 hybrids
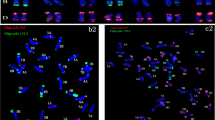
Chromosome number of parent plants used in crossing, and 10 hexaploid F1 hybrid plants, were close to or precisely 54 (Table 1; Fig. 1). Pollen viability in 10 F1 hybrids ranged from a low of 20.8 % in H02-4 to a high of 95.7 % in H04-6 (Table 1; Fig. 2).
Chromosome behavior during meiosis and microspore isolation from four hexaploid hybrids
We chose four hexaploid F1 hybrids (H08-1, H11-2, H16-1 and H24-1) for further cytogenetic studies and microspore culture, based on the hexaploid status and genomic diversity of the parents, high chromosome number, relatively high pollen viability, high seed yield in the F1 hybrids in the glasshouse, and preliminary microspore culture results (Table 1).
The four hexaploid F1 hybrids were examined for chromosome pairing at diakinesis of metaphase I in PMCs. Five different kinds of PMCs were observed during meiosis I and II, which included normal cells, cells with lagging chromosomes, triads, polyads and cells with unequal division (Fig. 3). The percentage of cells with “normal” pairing was highest in F1 hybrid H11-2 (70 %) and lowest in H16-1 (36 %) (Table 2). Several cells showed univalents or multivalents (Fig. 3), possibly as a result of homoeology between chromosomes in the A, B and C genomes. These abnormal chromosomes were not on the equatorial plate at metaphase I, or they moved towards opposite poles at anaphase I.
Chromosome pairing in PMCs of four hexaploid F1 hybrids. a Diakinesis PMCs with 27 II in H24-1. b PMC of H24-1 with a lagging bivalent at anaphase I. c Diakinesis PMC of H08-1 showing multivalents. d PMC of H24-1 showing normal segregation at anaphase I. e PMC of H11-2 showing triad at anaphase II. f PMC with a chromosome bridge. g PMC of H16-1 showing polyad. h PMC of H08-1 with unequal segregation at anaphase I
The four hexaploid F1 hybrids showed a range of embryo generation ability and plantlet regeneration rate during microspore culture. The number of embryos per Petri dish (an average of 3 dishes per hybrid isolated on three different days) varied significantly between hybrids. The embryos with good quality were transferred into solid medium to generate plantlets. Hybrid H11-2 had the highest number (394) of embryos per dish and also had the highest number of plantlets (693 plantlets) and plantlet regeneration rate (86.8 %) (Table 3). Hybrid H16-1 had 172 embryos per dish, the second highest number of plantlets (463 plantlets) and plantlet regeneration rate (72.7 %) (Table 3). Hybrid H08-1 had the second highest number (286) of embryos per dish, only 54 plantlets and plantlet regeneration rate (50.5 %) (Table 3). Hybrid H24-1 had 64 embryos per dish, only 56 plantlets and the lowest plantlet regeneration rate (11.2 %) (Table 3).
Ploidy level in microspore-derived plants using flow cytometry
The ploidy level of all MD plantlets from the four hexaploid F1 hybrids was estimated from nuclear DNA content measured in flow cytometry (Nelson et al. 2009). The peak of nuclear DNA content (PI_DNA-A) by flow cytometry for B. napus was found to be at approximately 50,000 units, with and without lettuce, which had a peak of PI_DNA-A at approximately 125,000 units (Figs. 4a, b). The PI_DNA-A of hexaploid MD progeny and parents was about 75,000 units (Fig. 4c) and triploid MD progeny was about 37,500 units (Fig. 4d). Based on flow cytometry tests, MD plantlets in the four populations were categorized as hexaploid, triploid, mixed or “other” (unidentified) ploidy level (Table 4). In these experiments, DH progeny were hexaploid and haploid progeny were triploid, since the parents were hexaploid and the chromosome numbers of MD progenies were checked (Fig. 5). Hybrids H11-2 and H16-1 gave the largest DH populations for genetic mapping (370 and 241 DH progeny, respectively), whereas H08-1 and H24-1 each resulted in less than 25 DH progeny (Table 4).
Examples of flow cytometry results. a Brassica napus (the peak of nuclear DNA content is 50,000 units), b B. napus (the peak of nuclear DNA content is 50,000 units) + lettuce (the peak of nuclear DNA content is 125,000 units), c microspore-derived (MD) hexaploid plant H16-1-002 + lettuce (the peak of nuclear DNA content is 75,000 units), and d MD triploid plant H16-1-011 + lettuce (the peak of nuclear DNA content is 37,500 units)
DH progenies of hybrids selected as candidates for mapping population
The hexaploid hybrid H11-2 showed the highest “normal” gamete production, embryo generation ability and plantlet regeneration rate (Table 3), the highest doubling frequency (Table 4) and also formed the highest number of hexaploid DH progeny (Table 4). H16-1 produced the second highest number of hexaploid DH progeny and also showed lower embryo generation ability, the second highest number of plantlet regeneration rate but lowest “normal” gamete production. H08-1 had the lower number of hexaploid DH progeny, the second highest embryo generation ability, lower plantlet regeneration rate and lower doubling frequency. H24-1 had the lowest embryo production per dish and plantlet regeneration rate (Table 3), lowest doubling frequency (Table 4), and lowest number of hexaploid DH progeny formed (Table 4). High correlation was found between embryo number per Petri dish and plantlet regeneration rate (r2 = 0.82), and between embryo number per Petri dish and doubling frequency (r2 = 0.91) for the four hybrids used in microspore culture (H08-1, H11-2, H16-1, and H24-1). Based on these findings, DH progenies of two hybrids H11-2 and H16-1 were proposed as candidates for allohexaploid mapping populations.
Discussion
The most common method of producing artificial Brassica allohexaploids is crossing an allotetraploid and a diploid species with a complementary genome, and then doubling the chromosome complement of the F1 (a sterile triploid) through the use of colchicine (Li et al. 2005a, b; Tian et al. 2010; Ge et al. 2009; Pradhan et al. 2010; Mason et al. 2012). Many polyploid plants have been produced via chromosome doubling technology including Brassica (Meng et al. 1998; Li et al. 2004), potato (Johnstone 1939; Saisingtong et al. 1996) and tobacco (Takashima et al. 1995). Synthetic hexaploid wheat has been generated by crossing tetraploid durum wheat (2n = 4x = 28, AABB) with diploid Aegilops tasschii Coss (2n = 2x = 14, DD) to generate genetic diversity for modern wheat breeding (Dreisigacker et al. 2008). The hexaploid triticale (AABBRR) is a successful synthetic allopolyploid derived through crossing tetraploid wheat (AABB) and rye (RR) (Ammar et al. 2004). The stability of triticale was highly improved after the lines from different genetic backgrounds were intercrossed and selected for genomic stability. Triticale has been widely used in agricultural production (Comai 2000).
In our research, a number of crosses were made between allohexaploid Brassica parents from diverse genetic backgrounds. All the lines used as parents in this study had close to the expected hexaploid number of 54 chromosomes and around 50 % normal pollen grains, and were derived from various genetic backgrounds including almost all the species of the Brassica U triangle (Table 1). Four hexaploid hybrid plants were chosen for microspore culture to establish future DH mapping populations of hexaploid Brassica.
Chromosome stability is the major factor for the establishment and persistence of polyploidy in plants. Many artificial allopolyploids, such as Triticum, Gossypium and Arabidopsis, show homologous chromosome pairing and stable chromosome number during meiosis after several generations of selfing (Comai 2000; Liu et al. 2001; Zhang et al. 2004). New synthetic allohexaploid wheat hybrids inherited meiotic irregularities which were progenitor-dependent (Mestiri et al. 2010). We used a range of sources of allohexaploid Brassica, which were relatively genetically stable after several generations of selfing (Ge et al. 2009), as parents in our crossing experiments.
Cytological observation of chromosome behavior during meiosis in allohexaploid parents demonstrated abnormal chromosome pairing and segregation, such as laggards, triads, polyads, unequal cell division, chromosome bridges and chromosome loss. The appearance of a chromosome bridge and lagging chromosomes indicated that pairing of homoeologous chromosomes in different genomes may lead to chromosome rearrangements such as inversions or exchange (Busso et al. 1987). Meng et al. (1998) and Rahman (2001) produced a pentaploid line (AABCC) through intercrossing the artificial hexaploid (AABBCC) and B. napus (AACC). They found some B. napus (2n = 38) from the selfing progenies of this pentaploid line. This showed that the B genome chromosomes are lost during selfing and rarely pair with A and C genome chromosomes (Rahman 2001).
Recently, Tian et al. (2010) proved by in situ hybridization that the lagging chromosomes during meiosis of a Brassica hybrid pentaploid were mostly either B genome or A/C pairs. Nearly 50 % of the PMCs had trivalents and quadrivalents, which revealed partial homology among the A, B and C genomes. The bivalents most likely belonged to the B genome. In our research, we observed abnormal chromosome behavior during meiosis in hexaploid Brassica parents and DH progeny that might be due to pairing between homologous and non-homologous regions.
It is possible that, given the occurrence of giant pollen cells (Fig. 3), some of the 6x progeny were products of unreduced gametes rather than colchicine-induced doubled haploids, as was found in microspore-derived progeny of an unbalanced ABCC hybrid reported by Nelson et al. (2009). Molecular marker characterisation of the 6x progeny would distinguish between DH (all homozygous loci) and unreduced gametes (combination of heterozygous and homozygous loci).
F1 hybrids H11-2 and H08-1 had high embryo production, as well as high “normal” gamete production. The parents of both hybrids included an allohexaploid parent plant derived from B. rapa × B. carinata and a parent plant derived from (B. juncea × B. napus) × B. carinata (Table 1). The F1 hybrids H24-1 and H16-1 had lower embryo production. The parents of these two hybrids included an allohexaploid parent plant derived from B. rapa × B. carinata and a parent plant derived from B. napus × B. nigra (Table 1). It seems that the F1 hybrids H11-2 and H08-1 from B. rapa × B. carinata and (B. juncea × B. napus) × B. carinata were much easier to obtain embryos than the other cross. B. nigra in the parentage may have reduced the ability of H24-1 and H16-1 to form embryos in microspore culture, as this species has been recalcitrant in microspore culture (Govil et al. 1986; Hetz and Schieder 1989). The more successful F1 hybrids H11-2 and H08-1 included parentage from all three allotetraploid Brassica species plus diploid B. rapa. Less successful F1 hybrids H24-1 and H16-1 were derived from two allotetraploids and two diploid Brassica species.
We found two F1 hybrids (H11-2 and H16-1) that generated several hundred allohexaploid DH progenies as shown by flow cytometry. Both of these hybrids have different sources of A, B and C genomes from their grandparents, and are therefore genetically diverse and ideal for genetic mapping of various traits including chromosome pairing stability in a new Brassica hexaploid species.
Abbreviations
- ANOVA:
-
Analysis of variance
- DH:
-
Doubled haploid
- MD:
-
Microspore-derived
- PMC:
-
Pollen mother cells
- PI:
-
Propidium iodide
- RFLP:
-
Restriction fragment length polymorphism
- SSR:
-
Simple sequence repeats
References
Ahmadi B, Alizadeh K, da Silva JAT (2012) Enhanced regeneration of haploid plantlets from microspores of Brassica napus L. using bleomycin, PCIB, and phytohormones. Plant Cell Tiss Org Cult 109:525–533
Ammar K, Mergoum M, Rajaram S (2004) The history and evolution of triticale. In: Mergoum M, Macpherson HG (eds) Triticale improvement and production (FAO plant production and protection paper 179). Food and Agriculture Organization of the United Nations, Rome, pp 1–9
Bennett MD (2004) Perspectives on polyploidy in plants-ancient and neo. Biol J Linn Soc 82:411–423
Busso C, Attia T, Robbelen G (1987) Trigenomic combinations for the analysis of meiotic control in the cultivated Brassica species. Genome 29:331–333
Chen S, Nelson MN, Chèvre AM, Jenczewski E, Li ZY, Mason AS, Meng JL, Plummer JA, Pradhan A, Siddique KHM, Snowdon RJ, Yan GJ, Zhou WJ, Cowling WA (2011) Trigenomic bridges for Brassica improvement. Crit Rev Plant Sci 30:524–547
Comai L (2000) Genetic and epigenetic interactions in allopolyploid plants. Plant Mol Biol 43:387–399
Cousin A, Nelson M (2009) Twinned microspore-derived embryos of canola (Brassica napus L.) are genetically identical. Plant Cell Rep 28:831–835
Dreisigacker S, Kishii M, Lage J, Warburton M (2008) Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Aust J Agric Res 59:413–420
Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316:1862–1866
Feldman M (2001) The origin of cultivated wheat. In: Bonjean AP, Angus WJ (eds) The World wheat book. Lavoisier Publishing, Paris, pp 1–56
Ferreira ME, Williams PH, Osborn TC (1994) RFLP mapping of Brassica napus using doubled-haploid lines. Theor Appl Genet 89:615–621
Ferrie AMR, Mollers C (2011) Haploids and doubled haploids in Brassica spp. for genetic and genomic research. Plant Cell Tiss Org Cult 104:375–386
Ge XH, Wang J, Li ZY (2009) Different genome-specific chromosome stabilities in synthetic Brassica allohexaploids revealed by wide crosses with Orychophragms. Ann Bot 104:19–31
Govil S, Babbar SB, Gupta SC (1986) Plant regeneration from in vitro cultured anthers of black mustard (Brassica nigra Koch). Plant Breed 97:64–71
Guo SM, Zou J, Li RY, Long Y, Chen S, Meng JL (2012) A genetic linkage map of Brassica carinata constructed with a doubled haploid population. Theor Appl Genet 125:1113–1124
Hetz E, Schieder O (1989) Plant regeneration from isolated microspores of black mustard (Brassica nigra). Science for plant breeding: XII Eucarpia congress, Göttingen, pp 10–15
Johnstone FE (1939) Chromosome doubling in potatoes induced by colchicine treatment. Am J Potato Res 16:288–304
Kimber DS, McGregor DI (1995) Brassica oilseeds: production and utilization. CAB International, Wallingford
Larter EN, Gustafson JP (1980) Triticale. In: Fehr WR, Hadley HH (eds) Hybridization of crop plants. American Society of Agronomy & Crop Science Society of America, Madison, pp 681–694
Leitch AR, Leitch IJ (2008) Genomic plasticity and the diversity of polyploid plants. Science 320:481–483
Levin DA (2002) The role of chromosomal change in plant evolution. Oxford University Press, New York
Li Z, Liu HL, Luo P (1995) Production and cytogenetics of intergeneric hybrids between Brassica napus and Orychophragmus violaceus. Theor Appl Genet 91:131–136
Li M, Qian W, Meng WJ, Li Z (2004) Construction of novel Brassica napus genotypes through chromosomal substitution and elimination using interploid species hybridization. Chromosom Res 12:417–426
Li MT, Li ZY, Zhang CY, Qian W, Meng JL (2005a) Reproduction and cytogenetic characterization of interspecific hybrids derived from crosses between Brassica carinata and B. rapa. Theor Appl Genet 110:1284–1289
Li MT, Zhang CY, Li ZY, Meng JL (2005b) The analysis of the biological characters in hexaploid hybrids derived from Brassica carinata and Brassica rapa. Acta Agron Sin 31:1579–1585
Liu B, Brubaker CL, Mergeai G, Cronn RC, Wendel JF (2001) Ployploid formation in cotton is not accompanied by rapid genomic changes. Genome 44:321–330
Mason A, Yan GJ, Cowling WA, Nelson M (2012) A new method for producing allohexaploid Brassica through unreduced gametes. Euphytica 186:277–287
Masterson J (1994) Stomatal size in fossil plants-evidence for polyploidy in majority of angiosperms. Science 264:421–424
Meng JL, Shi SW, Gan L, Li ZY, Qu XS (1998) The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B. campestris (AA) and B. carinata (BBCC) with B. napus. Euphytica 103:329–333
Mestiri I, Chague V, Tanguy AM, Huneau C, Huteau V, Belcram H, Coriton O, Chalhoub B, Jahier J (2010) Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol 186:86–101
Mohammadi PP, Moieni A, Ebrahimi A, Javidfar F (2012) Doubled haploid plants following colchicine treatment of microspore-derived embryos of oilseed rape (Brassica napus L.). Plant Cell Tiss Org Cult 108:251–256
Murphy EV, Zhang Y, Zhu W, Biggs J (1995) The human glioma pathogenesis-related protein is structurally related to plant pathogenesis-related proteins and its gene is expressed specifically in brain tumors. Gene 159:131–135
U N (1935) Genomic analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Nelson MN, Mason A, Castello MC, Thomson L, Yan GJ, Cowling WA (2009) Microspore culture preferentially selects unreduced (2n) gametes from an interspecific hybrid of Brassica napus L. × Brassica carinata Braun. Theor Appl Genet 119:497–505
Pradhan A, Plummer JA, Nelson MN, Cowling WA, Yan GJ (2010) Successful induction of trigenomic hexaploid Brassica from a triploid hybrid of B. napus L. and B. nigra (L.) Koch. Euphytica 176:87–98
Rahman MH (2001) Production of yellow-seeded Brassica napus through interspecific crosses. Plant Breeding 120:463–472
Raman H, Raman R, Nelson MN, Aslam MN, Rajasekaran R, Wratten N, Cowling WA, Kilian A, Sharpe AG, Schondelmaier J (2012) Diversity array technology markers: genetic diversity analyses and linkage map construction in rapeseed (Brassica napus L.). DNA Res 19:51–65
Saisingtong S, Schmid JE, Stamp P, Buter B (1996) Colchicine mediated chromosome doubling during anther culture of maize (Zea mays L.). Theor Appl Genet 92:1017–1023
Soltis DE, Soltis PS (1995) The dynamic nature of polyploidy genomes. Proc Natl Acad Sci USA 92:8089–8091
Takahira J, Cousin A, Nelson MN, Cowling WA (2011) Improvement in efficiency of microspore culture to produce doubled haploid canola (Brassica napus L.) by flow cytometry. Plant Cell Tiss Org Cult 104:51–59
Takashima H, Hasegawa H, Nakamura A (1995) A simple method for chromosome doubling in tobacco anther culture. Breed Sci 45:107–110
Tian E, Jiang Y, Chen L, Zou J, Liu F, Meng JL (2010) Synthesis of a Brassica trigenomic allohexaploid (B. carinata × B. rapa) de novo and its stability in subsequent generations. Theor Appl Genet 121:1431–1440
Winarto B, da Silva JAT (2011) Microspore culture protocol for Indonesian Brassica oleracea. Plant Cell Tiss Org Cult 107:305–315
Xu L, Najeeb U, Tang GX, Gu HH, Zhang GQ, He Y, Zhou WJ (2007) Haploid and doubled haploid technology. Adv Bot Res 45:181–216
Yu SC, Zhang FL, Yu RB, Zou YM, Qi JN, Zhao XY, Yu YJ, Zhang DS, Li L (2009) Genetic mapping and localization of a major QTL for seedling resistance to downy mildew in Chinese cabbage (Brassica rapa ssp pekinensis). Mol Breed 23:573–590
Zhang LQ, Liu DC, Yan ZH, Lan XJ, Zheng YL, Zhou YH (2004) Rapid changes of microsatellite flanking sequence in the allopolyploidization of new synthesized hexaploid wheat. Sci China Ser C 47:553–561
Zhang GQ, He Y, Xu L, Tang GX, Zhou WJ (2006) Genetic analyses of agronomic and seed quality traits of doubled haploid population in Brassica napus through microspore culture. Euphytica 149:169–177
Zhou WJ, Hagberg P, Tang GX (2002a) Increasing embryogenesis and doubling efficiency by immediate colchicine treatment of isolated microspores in spring Brassica napus. Euphytica 128:27–34
Zhou WJ, Tang GX, Hagberg P (2002b) Efficient production of doubled haploid plants by immediate colchicine treatment of isolated microspores in winter Brassica napus. Plant Growth Regul 37:185–192
Acknowledgments
This work was supported partly by the Science and Technology Department of Zhejiang Province (2012C12902-1, 2011R50026-5), the National Natural Science Foundation of China (31000678), and the Australia-China Special Fund, jointly managed by the Department of Innovation Industry Science and Research (Australia) and the Ministry of Science and Technology, National Natural Science Foundation (China). I.A. Astarini was the recipient of an Indonesian government Program of Academic Recharging (PAR). We thank Anouska Cousin for helping in microspore culture procedures.
Author information
Authors and Affiliations
Corresponding author
Additional information
X. X. Geng and S. Chen: equal first authors.
Rights and permissions
About this article
Cite this article
Geng, X.X., Chen, S., Astarini, I.A. et al. Doubled haploids of novel trigenomic Brassica derived from various interspecific crosses. Plant Cell Tiss Organ Cult 113, 501–511 (2013). https://doi.org/10.1007/s11240-013-0292-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-013-0292-4