Abstract
Ethylene is a critical signal that influences cut flower opening and senescence in tree peony (Paeonia suffruticosa), which is a traditional ornamental plant in China. Arabidopsis ETHYLENE-INSENSITIVE3 (EIN3) acts as a key transcription factor of the ethylene signaling pathway, suggesting a possible role for its homologues in regulation of cut flower postharvest development. In this study, three EIN3 homologous genes including PsEIL1, PsEIL2, and PsEIL3 have been isolated from petals of tree peony. Deduced amino acid sequences of conserved domains of PsEILs share high similarities with the Arabidopsis EIN3 protein. Real-time reverse transcription-polymerase chain reaction (RT-PCR) analysis indicated that three PsEILs were differentially expressed in various tissues of tree peony, and none was flower specific. During cut flower postharvest development, PsEIL1 transcript in petals accumulated at a relatively low level at the opening stage and reached the highest level of mRNA accumulation at senescence; PsEIL2 and PsEIL3 transcripts were gradually increased and peaked at full opening stage followed by a decline when petals wilted. However, these PsEIL genes exhibited differential responses to ethylene and 1-methycyclopropene (1-MCP) treatments. Compared with the control, the mRNA level of PsEIL1 was not influenced by either ethylene or 1-MCP treatment, whereas both PsEIL2 and PsEIL3 transcripts were significantly increased by exogenous ethylene, and only PsEIL3 was strongly inhibited by 1-MCP. These results suggest that PsEIL transcripts are spatiotemporally regulated, and the transcriptional regulation of PsEIL3 may play an important role in ethylene-mediated cut flower opening and senescence in tree peony.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The gaseous phytohormone ethylene plays a crucial regulatory role in diverse aspects of plant growth and development (Abeles et al. 1992; Bleecker and Kende 2000; Lavee et al. 2010; Lu et al. 2011), including flower opening and senescence (Reid et al. 1989; Reid and Wu 1992; Shibuya 2012). The climacteric rise in ethylene influences flower senescence of ethylene-sensitive flowers, which is associated with changes in gene expression (Borochov and Woodson 1989; Lawton et al. 1989). In this event, the ethylene perception and signal transduction is essentially required (Lawton et al. 1990; Verlinden et al. 2002).
During the past decades, progress in understanding the molecular mechanism of ethylene perception and signal transduction has been made through screening for Arabidopsis mutants defective in the ethylene triple response phenotype, and a framework has been established in this model plant. Ethylene is perceived by a family of membrane-associated receptors, which act as negative regulators of ethylene response, along with the downstream Raf-like serine/threonine kinase called CONSTITUTIVE TRIPLE RESPONSE1 (CTR1). In the absence of ethylene, the active receptors interact with CTR1 to repress the downstream response. While binding ethylene, the receptors are inactive, resulting in the inactivation of CTR1 and the downstream repression is relieved. Then the ethylene signal is transmitted through ETHYLENE-INSENSITIVE2 (EIN2) into the nucleus to active the EIN3/EIN3-like (EIL) transcription factors, which trigger transcription of downstream target genes, such as ETHYLENE RESPONSE FACTOR1 (ERF1), ultimately inducing diverse ethylene responses (Guo and Ecker 2004; Chen et al. 2005; Lin et al. 2009).
The transcription factor EIN3, located at the most downstream position of the ethylene signaling pathway, has received more attention than any other components (Chen et al. 2005). EIN3 functions as a positive regulator of ethylene response (Chao et al. 1997) and a potential integration point for cross-talk with other signals (Yanagisawa et al. 2003; Chen et al. 2005; Zhu et al. 2011). This protein belongs to a small family, of which six members (EIN3 and EIL1 to EIL5) have been identified in Arabidopsis. These members share common features for nuclear-localized transcription factors whereas only EIN3 and its most closely related EIL1 have been conclusively demonstrated to function in the ethylene signaling pathway (Chao et al. 1997; Alonso et al. 2003; Guo and Ecker 2004). To date, the EIN3/EIL gene family has also been well documented in many other plant species including tobacco (Kosugi and Ohashi 2000; Rieu et al. 2003), tomato (Tieman et al. 2001; Yokotani et al. 2003), carnation (Waki et al. 2001; Iordachescu and Verlinden 2005), rice (Mao et al. 2006), banana (Mbeguie et al. 2008), apple (Tacken et al. 2010), and Oncidium (Chen et al. 2011). These EIN3/EIL genes are almost ubiquitously expressed throughout the plants, despite slightly different patterns and levels of their expression. For example, four LeEILs are expressed in all tomato tissues examined, with LeEIL2 transcript somewhat higher than those of LeEIL1 and LeEIL3, and LeEIL4 mRNA accumulation increases during fruit ripening, whereas the three others are relatively unchanged (Tieman et al. 2001; Yokotani et al. 2003). In Arabidopsis, the regulation of EIN3 at the post-transcriptional level has been reported to be crucial in ethylene signaling. EIN3 mRNA level is not affected by ethylene, however, EIN3 protein is stabilized and accumulates in response to ethylene, while it is rapidly degraded through the 26S proteasome pathway in the absence of ethylene (Guo and Ecker 2003; Yanagisawa et al. 2003). By contrast, Arabidopsis EIL1 shows significant differences in expression upon ethylene treatment (De Paepe et al. 2004; Chen et al. 2005), and EILs in other species such as carnation (Waki et al. 2001; Iordachescu and Verlinden 2005), banana (Mbeguie et al. 2008), and Oncidium (Chen et al. 2011) have also been found to be regulated at the transcriptional level by ethylene and developmental cues.
Tree peony (Paeonia suffruticosa), a traditional ornamental plant in China, is popular for its attractive flowers around the world. Cut flower of tree peony exhibits short vase life, which causes a major problem in its shipping and handling. Previous studies revealed that cut flower opening and senescence of most tree peony cultivars, for example, ‘Luoyang Hong’, was associated with the increase of ethylene production (Jia et al. 2008), which was largely derived from the petals (Jia 2010). Moreover, exogenous ethylene was able to influence the postharvest process of the cultivar ‘Luoyang Hong’ as well as the endogenous ethylene production, further confirming the important role of ethylene in regulation of tree peony cut flower opening and senescence (Zhou et al. 2009). Ethylene biosynthesis, perception and signal transduction have become the molecular biology research hotspots in order to extend the vase life and improve the quality of this ornamental flower (Zhou et al. 2008, 2010; Gao et al. 2011). To gain a better understanding of ethylene signaling in regulating tree peony cut flower development at the transcriptional level, we isolated three full-length cDNAs of EIN3 homologous genes from petals of tree peony, and investigated their expression patterns in different tissues and during cut flower opening and senescence, as well as the influences of exogenous ethylene and 1-methycyclopropene (1-MCP), an ethylene action inhibitor, on the expression of these genes. The possible roles for PsEILs in regulation of tree peony cut flower opening and senescence are discussed.
Materials and methods
Plant materials
Tree peony (Paeonia suffruticosa ‘Luoyang Hong’) was grown in the nursery of Luoyang Tuqiao Peony and Young Plants Co., Ltd., Luoyang City, Henan Province, China. Flowers were harvested at stage 1, and different tissues including roots, stems, leaves, sepals, petals, stamens, and pistils were collected from intact plants with fully opened flowers. Flower opening stages were described previously in Guo et al. (2004): stage 1, soft bud; stage 2, pre-opening; stage 3, initial opening; stage 4, half opening; stage 5, full opening; stage 6, wilting. After transportation to the laboratory, different tissue samples were frozen in liquid nitrogen and stored at −80 °C for further use. Meanwhile, flowers were re-cut to 25 cm in stem length with the uppermost leaves left and rehydrated for 1 h. Then those flowers each were placed in glass flasks filled with 100 mL distilled water and kept for observation in a room at 20–23 °C, 50–60 % relative humidity, and a 12-h photoperiod with an illumination of ~40 μM m−2 s−1. Petal samples were detached from flowers at each stage during vase life, and frozen in liquid nitrogen before storage at −80 °C for later total RNA extraction.
Ethylene and 1-MCP treatments
To investigate the regulatory effect of ethylene on the expression of tree peony EIL genes, exogenous ethylene and 1-MCP treatments were imposed on cut flower as follows: after rehydration, flowers with their bases in distilled water were sealed in a 100 L glass chamber with 10 μL L−1 ethylene or 1 μL L−1 1-MCP for 6 h at room temperature. Control flowers were incubated in another chamber with air. After treatment, flowers were held in glass flasks with distilled water under the same aforementioned environmental conditions. Petal samples were collected at 0, 12, and 24 h after each treatment, immediately frozen and stored until extraction of total RNA.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from different tissues of tree peony according to the method described by Chang et al. (1993), and then digested with RNase-free DNase I (Takara, Japan) to remove the residual genomic DNA. The first-stand cDNA was synthesized from 1 μg of DNA-free RNA using a PrimeScript® RT reagent Kit (Takara, Japan).
Isolation of tree peony EIL cDNAs and sequence analysis
PsEIL partial cDNA fragments were isolated by reverse transcription-polymerase chain reaction (RT-PCR) using total RNA extracted from senescent tree peony petals and nested degenerate primers (EIL-OF and EIL-OR as outer primers, EIL-IF and EIL-IR as inner primers). These primer sequences were designed based on highly conserved domains among plant EIN3/EIL proteins, ELERRMW and IRKLVRQS (for outer primers) or ARRKKMS and PQRRFPL (for inner primers) (Fig. 1). The PCR program was carried out with a template denaturation at 94 °C for 5 min, followed by 30 cycles of denaturing at 94 °C for 30 s, annealing at 46 °C for 30 s, and elongating at 72 °C for 1 min, then ended with the elongation at 72 °C for 7 min. The remaining 3′ and 5′ ends of each PsEIL cDNA were isolated by rapid amplification of cDNA ends (RACE) method as follows: the 3′ end cDNA fragment was amplified according to the manufacturer’s instruction with a SMART™ RACE cDNA Amplification Kit (Clontech, Japan) by two successive 3′ RACE-PCR, using adaptors and specific primers 3′-EIL1-G1, -G2 (for PsEIL1), 3′-EIL2-G1, -G2 (for PsEIL2) or 3′-EIL3-G1, -G2 (for PsEIL3), which were designed based on each PsEIL partial cDNA sequence isolated above; the 5′ end cDNA fragment was extended using a 5′-Full RACE Kit (Takara, Japan) with nested specific primers 5′-EIL1-G1, -G2 (for PsEIL1), 5′-EIL2-G1, -G2 (for PsEIL2) or 5′-EIL3-G1, -G2 (for PsEIL3). Once three partial fragments of each cDNA had been aligned together, full-length cDNA for each PsEIL gene was generated through PCR programmed at 94 °C for 5 min, then followed by 32 cycles of 30 s at 94 °C, 30 s at 60 °C, 3 min at 72 °C, and a final elongation for 7 min at 72 °C. The details of primers used in amplifications are described in Table 1. All amplified products were cloned into the pMD18-T vector (Takara, Japan), and then putative positive clones were identified by PCR with RV-M and M13-47 sequencing primers before they were sequenced by Sunbiotech Co., Ltd. (Beijing, China).
Multiple amino acid sequence alignments of PsEILs and EIN3 (Arabidopsis thaliana, AF004216). Identities and similarities among amino acid sequences are colored in black and gray, respectively. Box regions designate the acidic region (AD), proline-rich region (PR), and five small clusters of basic amino acids (BDI-V). The putative nuclear localization signal sites are indicated by solid lines. Asterisk represents the lysine residue critical for the function of EIN3. The polyasparagine repeats present in the C-terminus of Arabidopsis EIN3 but not in that of PsEILs are marked by dots. Arrows refer to the primer amino acid sequences for nested RT-PCR
Comparisons of the nucleotide and predicted amino acid sequences were performed using the BLAST program, while the open reading frame (ORF) was identified by ORF Finder at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). For multiple sequence alignments, the deduced full-length amino acid sequences of PsEILs and Arabidopsis EIN3 were analyzed using DNAMAN (version 5.2) with default parameters, and the phylogenetic analysis was carried out based on Neighbor-Joining (NJ) model with 1000 bootstrap replications by MEGA (version 4.0).
Real-time RT-PCR analysis
For real-time RT-PCR, each primer set was designed based on the 3′ end cDNA sequence of the corresponding gene with Primer Premier 5 (Table 2 for primer sets). Gene specificity of all primer sets was tested with the following procedure. First, Primer-BLAST searches against GenBank databases were performed for primers to confirm that none of them matched with other sequences. Then primers were examined by melting peaks and dissociation curves to check that only a single product existed for each primer set. For evaluation of each primer set efficiency, a standard curve was produced using serial dilutions of a cDNA mixture from different tissues of tree peony, and efficiency values (ranging from 96 to 101 %) were automatically calculated by Bio-Rad CFX manager (version 2.0) (data not shown).
Real-time RT-PCR was performed on a Miniopticon Real-Time PCR instrument (Bio-Rad, USA) using a 20 μL reaction mixture comprised of 10 μL of SYBR® Premix Ex Taq™ (TaKaRa, Japan), 0.4 μL of each primer (10 μM), 2 μL of 10-fold diluted cDNA, and 7.2 μL of PCR-grade water. The PCR program was initiated at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 57 °C for 30 s, and 72 °C for 30 s, and completed with a melting curve analysis in every reaction. Real-time RT-PCR was repeated three times for each sample, with a no template reaction as a negative control, and the PCR products were analyzed on 2.0 % agarose gels stained with ethidium bromide to ensure their size in accordance with expectation. The transcript abundance of each PsEIL was normalized to that of the reference gene, Paeonia suffruticosa ubiquitin (PsUB), and relative transcript level was presented as mean with standard errors.
Results
Isolation and sequence analysis of EIL cDNAs from tree peony
With degenerate primers designed based on the conserved EIN3 domains, PCR products of about 350 bp in length were obtained from senescent petals of tree peony and subsequent sequencing revealed three distinct fragments. Based on the sequences of these three partial cDNA fragments, full-length cDNAs of the corresponding genes were isolated by the combination of RT-PCR and RACE-PCR and designated as PsEIL1 (GenBank accession number JQ771469), PsEIL2 (GenBank accession number JQ771470), and PsEIL3 (GenBank accession number JQ771471). PsEIL1 was 2,400 bp in full length consisting of a 196-bp 5′-untranslated region (UTR), a 1,833-bp ORF encoding 610 amino acids, and a 371-bp 3′-UTR. PsEIL2 was 2,399 bp long composed of a 261-bp 5′-UTR, a 1,824-bp ORF encoding 607 amino acids, and a 314-bp 3′-UTR. The cDNA of PsEIL3 was 2,917 bp comprising 213-bp 5′-UTR, a 1,959-bp ORF encoding 652 amino acids, and a 745-bp 3′-UTR. Deduced polypeptides encoded by PsEIL1, PsEIL2, and PsEIL3 had predicted molecular masses of 68, 69, and 73 kDa, respectively. The overall nucleotide sequence identity between the PsEILs ranged from 40 to 47 %, while the ORFs were 47–58 % identical at the nucleotide level and 35–51 % at the amino acid level. In addition, PsEIL3 shared 98 % nucleotide identity with a previously cloned cDNA fragment, Ps-EIN3-1 (EU526839), which encodes a partial tree peony EIN3 homologue of only 180 amino acid residues (Zhou et al. 2010). Multiple sequence alignment analysis revealed that three PsEILs share predicted amino acid sequence similarity with the Arabidopsis EIN3 protein, especially in the conserved N-terminal halves (Fig. 1). All the PsEILs possess an acidic region, a proline-rich region, and five small clusters of basic amino acids in the N-terminus, and have putative sites for nuclear localization signal, as well as the lysine residue (234 for PsEIL1, 244 for PsEIL2, and 249 for PsEIL3) which is critical for the function of EIN3 (Chao et al. 1997). The polyasparagine repeats present in the C-terminus of Arabidopsis EIN3 are absent in any of the PsEILs (Fig. 1).
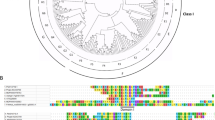
Phylogenetic analysis of multiple EIN3 homologues with complete amino acid sequences revealed that these proteins could be classified in two clusters (Fig. 2). Cluster I containing a majority of EIN3/EIL family members is separated into two subclusters. Both PsEIL2 and PsEIL3 are grouped into the first subcluster, which consists of AtEIN3 and AtEIL1, together with other homologues from various dicotyledonous species. Within this subcluster, PsEIL2 is the closest to NtEIL1 and NtEIL2, while PsEIL3 is the closest to MdEIL2 and CmEIL2. The second subcluster mainly includes EIN3 homologues from monocotyledons. PsEIL1 belongs to the same cluster (cluster II) as AtEIL2 and AtEIL3, but it proves to be more closely related to AtEIL3 than to AtEIL2.
Phylogenetic analysis of EIN3 homologues from Paeonia suffruticosa (PsEIL1, JQ771469; PsEIL2, JQ771470; and PsEIL3, JQ771471), Arabidopsis thaliana (AtEIN3, AF004216; AtEIL1, AF004213; AtEIL2, AF004214; AtEIL3, AF004215; AtEIL4, NM_121050; and AtEIL5, NM_125909), Nicotiana tabacum (NtEIL1, AY248903; NtEIL2, AY248904; NtEIL3, AY248905; NtEIL4, AY248906; and NtEIL5, AY248907), Lycopersicon esculentum (LeEIL1, AF328784; LeEIL2, AF328785; LeEIL3, AF328786; and LeEIL4, AB108840), Dianthus caryophyllus (DC-EIL1/2, AY728191; DC-EIL3, AY728192; and DC-EIL4, AY728193), Vigna radiata (VR-EIL1, AF467784; VR-EIL2, AF467783), Malus x domestica (MdEIL1, GU732484; MdEIL2, GU732485; and MdEIL3, GU732486), Cucumis melo (CmEIL1, AB063191; CmEIL2, AB063192), Actinidia deliciosa (AdEIL1, EU170633; AdEIL2, EU887511; AdEIL3, EU887512; and AdEIL4, EU887513), Musa acuminata (MA-EIL1, DQ682615; MA-EIL2, AB266318; MA-EIL3, AB266319; MA-EIL4, AB266320; and MAEIN3, AB266321), Oncidium (OgEIL1, HQ585983; OgEIL2, HQ585984), Oryza sativa (OsEIL1, AB074971; OsEIL2, AB074972; OsEIL3, AK065979; OsEIL4, AP005816; OsEIL5, AP005116; and OsEIL6, AL606636), and Zea mays (ZmEIL1, EU973374). PsEILs and the EIN3/EILs of Arabidopsis thaliana are highlighted by triangles and gray boxes, respectively. The scale below means 0.05 amino acid substitutions per site
Tissue specificity and expression patterns of PsEIL genes during cut flower opening and senescence
Real-time RT-PCR was performed to investigate the tissue specificity and expression patterns of the three PsEILs during cut flower postharvest development. Results showed that the transcripts of PsEILs were found in all tree peony tissues examined, but with different accumulation patterns and levels based on the tissue and the opening stage.
None of the three PsEIL genes were flower specific. PsEIL1 was constitutively expressed in all tissues except somewhat more abundance in roots. The closely related PsEIL2 and PsEIL3 genes exhibited different expression patterns, with PsEIL2 transcript higher in roots and lower in flower tissues, and PsEIL3 high mRNA accumulation in petals and low abundance in stamens and pistils compared with the vegetative tissues. Expression levels of these PsEIL genes were different from each other in various tissues, and the most prominent feature was that PsEIL3 transcript accumulated considerably higher in sepals, leaves, and stems, but especially in petals, compared with the other two PsEIL genes (Fig. 3).
Relative transcript levels of three PsEILs in different tissues. Real-time RT-PCR was performed to analyze the relative transcript levels of three PsEILs in roots, stems, leaves, sepals, petals, stamens, and pistils from intact tree peony with fully opened flowers. Data from real-time RT-PCR were normalized to PsUB mRNA levels, and presented as mean with standard errors (S.E.) of three replications
Throughout cut flower postharvest development, PsEIL1 transcript in petals accumulated at a relatively low level during the early opening stages, and reached the maximum abundance when cut flower became senescent (stage 6), whereas PsEIL2 and PsEIL3 in petals kept increasing gradually during the opening process until reaching peak mRNA accumulation in fully opened flowers (stage 5), and subsequently started to drop with wilting. Although the expression patterns for the two genes of PsEIL2 and PsEIL3 were similar, PsEIL3 exhibited approximately tenfold higher mRNA accumulation than PsEIL2 (Fig. 4).
a Phenotypes of tree peony cut flower at the following stages of postharvest development: soft bud (stage 1), pre-opening (stage 2), initial opening (stage 3), half opening (stage 4), full opening (stage 5), and wilting (stage 6). b Relative transcript levels of three PsEILs in petals throughout tree peony cut flower postharvest development. Real-time RT-PCR was performed to analyze the relative transcript levels of three PsEILs in petals at different flower opening stages. Data from real-time RT-PCR were normalized to PsUB mRNA levels, and presented as mean with standard errors (S.E.) of three replications
Effect of ethylene and 1-MCP treatments on PsEIL gene expression
To investigate whether the expression of the PsEILs was modulated by ethylene, transcript levels of these genes in petals after exogenous ethylene and 1-MCP treatments were determined. As shown in Fig. 5, when compared with the control, the transcript level of PsEIL1 showed no significant alteration after treatments with exogenous ethylene and 1-MCP. By contrast, as PsEIL2 and PsEIL3 transcripts were seemingly unchanged in control petals, they were both immediately enhanced to 1.5-fold at the end of ethylene treatment (0 h), and remained significantly induced at 12 and 24 h thereafter. The mRNA abundance of PsEIL3 was substantially suppressed after 1-MCP treatment, with an approximately fourfold decline from 0 to 12 h, and afterwards a 2.5-fold decrease. However, PsEIL2 transcript level was not significantly reduced by 1-MCP at any time examined.
Effect of exogenous ethylene and 1-MCP treatments on transcript levels of three PsEILs in petals. Real-time RT-PCR was performed to analyze the relative transcript levels of three PsEILs in petals at 0, 12, and 24 h after different treatments, including C (cut flower incubated with air), E (10 μL L−1 ethylene for 6 h), and M (1 μL L−1 1-MCP for 6 h). Data from real-time RT-PCR were normalized to PsUB mRNA levels, and presented as mean with standard errors (S.E.) of three replications (*P < 0.05 for ethylene or 1-MCP treatment against control)
Discussion
Ethylene plays an important role in regulating postharvest opening and senescence of cut flower, and ethylene signal transduction is a critical part in this regulation. In the present work, three full-length cDNAs for transcription factor EIN3 homologous genes, PsEIL1, PsEIL2, and PsEIL3, were isolated and characterized from tree peony. Deduced polypeptides of all the PsEILs show high sequence similarity to the Arabidopsis EIN3 protein, with many highly conserved domains in the N-terminus (Fig. 1). These regions are also well conserved in other EIN3/EIL proteins from different plant species (Chao et al. 1997; Lee and Kim 2003; Chen et al. 2011; Zou et al. 2011). In addition, PsEILs do not have the polyasparagine repeats near the C-terminus. Similar results have been found in predicted peptide sequences of Arabidopsis EIL1 and tomato LeEILs which complement the Arabidopsis ein3-1 mutant (Chao et al. 1997; Tieman et al. 2001). These structure conservations suggest that PsEILs are functional EIL genes (Chao et al. 1997). A previously cloned cDNA fragment from tree peony, Ps-EIN3-1(Zhou et al. 2010), is very similar to the corresponding region of PsEIL3. But the Ps-EIN3-1 fragment encodes a short amino acid sequence, which is less than 30 % of the full-length sequence of PsEIL3 and located in the highly conserved N-terminus. Furthermore, the nucleotide sequences of PsEIL3 and Ps-EIN3-1 are not identical, with 7 nucleotide differences within the conserved coding region, where 8 nucleotide differences exist between NtEIL3 and NtEIL4 from tobacco (Rieu et al. 2003). Thus, we could not rule out the possibility that PsEIL3 and Ps-EIN3-1 may be different EIL cDNAs in tree peony.
The PsEILs were widely expressed in various tree peony tissues with different expression patterns and levels, such data are in accordance with those from other species (Waki et al. 2001; Rieu et al. 2003; Yokotani et al. 2003; Yin et al. 2010; Chen et al. 2011) and match multiple functions of ethylene depending on tissue types, including regulating root hair formation, stem elongation, flower senescence, and leaf abscission (Abeles et al. 1992; Bleecker and Kende 2000). Within the PsEIL gene family, PsEIL1 exhibited a relatively constant expression level in all tissues and PsEIL2 mRNA level was higher in roots and lower in flower tissues. Similar expression patterns have also been observed in kiwifruit EIL gene family (Yin et al. 2008; Yin et al. 2010). Compared with the above two PsEIL genes, PsEIL3 was predominantly expressed in sepals, leaves, and stems, but especially in petals, where PsEIL3 showed the highest transcript abundance. Considering the result of a previous study that the ethylene production of the intact flower is primarily derived from the petals in tree peony ‘Luoyang Hong’ (Jia 2010), we propose that the expression of three PsEIL genes, especially PsEIL3, could contribute to the response of tree peony to ethylene.
During cut flower opening and senescence, three PsEIL transcripts in petals presented clear up-regulation, although the patterns and levels of their expression were different. Since EIN3 acts as a positive regulator in the ethylene signaling pathway (Chao et al. 1997), it would be expected that the increase in mRNA levels of PsEILs and, in turn, proteins would enhance tissue sensitivity to ethylene, which is consistent with the fact that tree peony gradually becomes more and more ethylene sensitive with flower opening (Liu 2009). It is worth noting that the production of endogenous ethylene, which influences flower diameter and postharvest opening process, increases during cut tree peony ‘Luoyang Hong’ opening and reaches a peak at full opening stage, then decreases (Jia et al. 2008). These data make it possible to put forward a hypothesis that the up-regulation of PsEILs during vase life might be associated with the increase of endogenous ethylene. In an attempt to demonstrate the regulatory effect of ethylene on PsEILs at the transcriptional level, we determined the transcript abundances of PsEILs in petals after exogenous ethylene and 1-MCP treatments.
The PsEIL1 transcript, which was relatively low at the early opening stages and then accumulated with the maximum at senescence, was not significantly affected by either ethylene or 1-MCP treatment. It is likely that the accumulation in the transcript level of PsEIL1 is due to some senescence signals rather than ethylene, as in the case of melon CmEILs (Huang et al. 2010). The pattern of PsEIL1 transcript unaffected by exogenous ethylene here is similar to those of EIN3/EILs found in Arabidopsis (Chao et al. 1997), tomato (Tieman et al. 2001), tobacco (Rieu et al. 2003), mung been (Lee and Kim 2003), rose (Ma et al. 2006), and kiwifruit (Yin et al. 2008), suggesting that their regulation may be controlled by ethylene at the post-transcriptional level. Indeed, it has been reported that ethylene induces Arabidopsis EIN3/EIL1 protein accumulation by promoting EBF1/EBF2 proteasomal degradation (Guo and Ecker 2003; Yanagisawa et al. 2003; An et al. 2010). However, phylogenetic analysis showed that PsEIL1 is located in a cluster distant to that of the two other PsEILs but together with AtEIL3, a member of the EIN3 family that might participate in regulating sulfur acquisition and metabolism rather than ethylene response (Maruyama-Nakashita et al. 2006). Thus, further study on the regulation of PsEIL1 at the protein level would be important to confirm the involvement of PsEIL1 in the ethylene signaling pathway.
Apart from the post-transcriptional regulation, previous studies have shown that transcriptional regulation of EIN3/EIL genes also played an important role in ethylene-mediated plant development. In carnation, three DC-EIL genes were regulated at the transcriptional level, in particular, one gene (DC-EIL3) was induced throughout flower development and after ethylene treatment, which was considered a possible role in regulation of flower senescence (Iordachescu and Verlinden 2005). In banana, MA-EIL2 mRNA abundance was markedly increased by ethylene and ripening, which showed a positive correlation with fruit quality development (Mbeguie et al. 2008). A recent study has reported that the increase in transcript levels of OgEIL1 and OgEIL2 in Oncidium may be attributed to ethylene (Chen et al. 2011). Similar to these findings, in the present work, PsEIL2 and PsEIL3 transcripts increased gradually during cut flower opening and reached peak abundance at full opening stage before decreasing, coincident with the climacteric phase of ethylene production (Jia et al. 2008). In addition, PsEIL2 and PsEIL3 transcripts were significantly increased following exogenous application of ethylene even at the early opening stages, but only PsEIL3 was substantially inhibited by 1-MCP treatment. Previous studies revealed that exogenous ethylene treatment effectively enhanced the ethylene production before initial opening of the flowers, and accelerated postharvest opening and senescence of tree peony ‘Luoyang Hong’ cut flower, on the other hand, 1-MCP suppressed ethylene production and prolonged the vase life (Zhou et al. 2009), indicating that the pattern of PsEIL3 mRNA accumulation was positively correlated with that of ethylene production and the postharvest process of cut flower after different treatments. Taken together, these results suggest that consistent with the high phylogenetic proximity of PsEIL3 with DC-EIL3 (Fig. 2), the transcriptional regulation of PsEIL3 is critical in ethylene signaling and possibly involved in ethylene-medicated flower opening and senescence.
In conclusion, we have isolated and characterized three EIN3 homologous genes (PsEIL1, PsEIL2, and PsEIL3) from tree peony. These PsEIL genes were differentially expressed in various tissues and during cut flower opening and senescence, indicating they are spatiotemporally regulated. PsEIL1 is likely associated with petal senescence, and possibly subject to post-transcriptional regulation by ethylene; PsEIL2 and PsEIL3 are regulated at the transcriptional level and at least PsEIL3, which was more abundant in petals and under positive feedback regulation by ethylene and 1-MCP, may play an essential role in ethylene-mediated postharvest development of tree peony cut flower. Further studies should let us gain insight into the accumulation of PsEIL proteins in relationship with the ethylene response and the process of cut flower opening and senescence. Our work should reveal targets for RNAi inhibition of critical PsEILs in ethylene signal transduction and extend the vase life of tree peony cut flower.
Abbreviations
- CTR:
-
CONSTITUTIVE TRIPLE RESPONSE
- EIN:
-
ETHYLENE-INSENSITIVE
- EIL:
-
EIN3-like
- ERF:
-
ETHYLENE RESPONSE FACTOR
- 1-MCP:
-
1-methycyclopropene
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- RACE:
-
Rapid amplification of cDNA ends
- ORF:
-
Open reading frame
- NCBI:
-
National Center for Biotechnology Information
- UTR:
-
Untranslated region
References
Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in plant biology. Academic Press, San Diego
Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100:2992–2997
An FY, Zhao QO, Ji YS, Li WY, Jiang ZQ, Yu XC, Zhang C, Han Y, He WR, Liu YD, Zhang SQ, Ecker JR, Guo HW (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-Box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22:2384–2401
Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16:1–18
Borochov A, Woodson WR (1989) Physiology and biochemistry of flower petal senescence. Hortic Rev 11:15–43
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89:1133–1144
Chen YF, Etheridge N, Schaller GE (2005) Ethylene signal transduction. Ann Bot 95:901–915
Chen SY, Tsai HC, Raghu R, Do YY, Huang PL (2011) cDNA cloning and functional characterization of ETHYLENE INSENSITIVE 3 orthologs from Oncidium Gower Ramsey involved in flower cutting and pollinia cap dislodgement. Plant Physiol Biochem 49:1209–1219
De Paepe A, Vuylsteke M, Van Hummelen P, Zabeau M, Van Der Straeten D (2004) Transcriptional profiling by cDNA-AFLP and microarray analysis reveals novel insights into the early response to ethylene in Arabidopsis. Plant J 39:537–559
Gao J, Jia PY, Wang YJ, Zhang C, Wang WR, Dong L (2011) Effects of ethylene and 1-MCP treatments on the expressions of CTR genes of tree peony. Acta Bot Boreal-Occident Sin 31:19–26
Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF (EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115:667–677
Guo H, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7:40–49
Guo WW, Dong L, Wang LY, Chen RX, Liu AQ (2004) The postharvest characteristics and water balance of some cultivars of tree-peony cut flowers. Scientia Silvae Sinicae 40:89–93
Huang SZ, Sawaki T, Takahashi A, Mizuno S, Takezawa K, Matsumura A, Yokotsuka M, Hirasawa Y, Sonoda M, Nakagawa H, Sato T (2010) Melon EIN3-like transcription factors (CmEIL1 and CmEIL2) are positive regulators of an ethylene- and ripening-induced 1-aminocyclopropane-1-carboxylic acid oxidase gene (CM-ACO1). Plant Sci 178:251–257
Iordachescu M, Verlinden S (2005) Transcriptional regulation of three EIN3-like genes of carnation (Dianthus caryophyllus L. cv. Improved White Sim) during flower development and upon wounding, pollination, and ethylene exposure. J Exp Bot 56:2011–2018
Jia PY (2010) Isolation and expression analysis of key genes related to ethylene signal transduction pathway in tree peony cut flowers. Dissertation, Beijing Forestry University
Jia PY, Zhou L, Guo WW, Wang LY, Dong L (2008) Postharvest behavior and endogenous ethylene pattern of Paeonia suffruticosa cut flowers. Acta Hortic 768:445–450
Kosugi S, Ohashi Y (2000) Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res 28:960–967
Lavee S, Parnes A, Avidan N (2010) Involvement of ethylene in growth induction of stationary tobacco pith tissue in vitro. Plant Cell Tissue Organ Cult 103:123–129
Lawton KA, Huang B, Goldsbrough PB, Woodson WR (1989) Molecular cloning and characterization of senescence-related genes from carnation flower petals. Plant Physiol 90:690–696
Lawton KA, Raghothama KG, Goldsbrough PB, Woodson WR (1990) Regulation of senescence-related gene expression in carnation flower petals by ethylene. Plant Physiol 93:1370–1375
Lee JH, Kim WT (2003) Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiol 132:1475–1488
Lin ZF, Zhong SL, Grierson D (2009) Recent advances in ethylene research. J Exp Bot 60:3311–3336
Liu YY (2009) The responses of ‘Luoyang Hong’ tree peony cut flowers at different opening stages to ethylene and 1-MCP. Dissertation, Beijing Forestry University
Lu JR, Vahala J, Pappinen A (2011) Involvement of ethylene in somatic embryogenesis in scots pine (Pinus sylvestris L.). Plant Cell Tissue Organ Cult 107:25–33
Ma N, Tan H, Xue JH, Li YQ, Gao JP (2006) Transcriptional regulation of ethylene receptor and CTR genes involved in ethylene-induced flower opening in cut rose (Rosa hybrida) cv. Samantha. J Exp Bot 57:2763–2773
Mao C, Wang S, Jia Q, Wu P (2006) OsEIL1, a rice homolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component. Plant Mol Biol 61:141–152
Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18:3235–3251
Mbeguie D, Hubert O, Fils-Lycaon B, Chillet M, Baurens FC (2008) EIN3-like gene expression during fruit ripening of Cavendish banana (Musa acuminata cv. Grande naine). Physiol Plant 133:435–448
Reid MS, Evans RY, Dodge LL, Mor Y (1989) Ethylene and silver thiosulphate influence opening of cut rose flowers. J Am Soc Hort Sci 114:436–440
Reid MS, Wu MJ (1992) Ethylene and flower senescence. Plant Growth Regul 11:37–43
Rieu I, Mariani C, Weterings K (2003) Expression analysis of five tobacco EIN3 family members in relation to tissue-specific ethylene responses. J Exp Bot 54:2239–2244
Shibuya K (2012) Molecular mechanisms of petal senescence in ornamental plants. J Jpn Soc Hort Sci 81:140–149
Tacken E, Ireland H, Gunaseelan K, Karunairetnam S, Wang D, Schultz K, Bowen J, Atkinson RG, Johnston JW, Putterill J, Hellens RP, Schaffer RJ (2010) The role of ethylene and cold temperature in the regulation of the apple POLYGALACTURONASE1 gene and fruit softening. Plant Physiol 153:294–305
Tieman DM, Ciardi JA, Taylor MG, Klee HJ (2001) Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26:47–58
Verlinden S, Boatright J, Woodson WR (2002) Changes in ethylene responsiveness of senescence-related genes during carnation flower development. Physiol Plantarum 116:503–511
Waki K, Shibuya K, Yoshioka T, Hashiba T, Satoh S (2001) Cloning of a cDNA encoding EIN3-like protein (DC-EIL1) and decrease in its mRNA level during senescence in carnation flower tissues. J Exp Bot 52:377–379
Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425:521–525
Yin XR, Chen KS, Allan AC, Wu RM, Zhang B, Lallu N, Ferguson IB (2008) Ethylene-induced modulation of genes associated with the ethylene signalling pathway in ripening kiwifruit. J Exp Bot 59:2097–2108
Yin XR, Allan AC, Chen KS, Ferguson IB (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153:1280–1292
Yokotani N, Tamura S, Nakano R, Inaba A, Kubo Y (2003) Characterization of a novel tomato EIN3-like gene (LeEIL4). J Exp Bot 54:2775–2776
Zhou L, Dong L (2008) Cloning and sequence analysis of 1-aminocyclopropane-1-carboxylic acid oxidase gene cDNA from tree peony. Acta Hortic Sin 35:891–894
Zhou L, Jia PY, Liu J, Wang WR, Huo ZP, Dong L (2009) Effect of ethylene on cut flowers of tree peony ‘Luoyang Hong’ opening and senescence process and endogenous ethylene biosynthesis. Acta Hortic Sin 36:239–244
Zhou L, Dong L, Jia PY, Wang WR, Wang LY (2010) Expression of ethylene receptor and transcription factor genes, and ethylene response during flower opening in tree peony (Paeonia suffruticosa). Plant Growth Regul 62:171–179
Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim JM, To TK, Li W, Zhang X, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo H (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108:12539–12544
Zou AL, Zhang WJ, Pan QY, Zhu SM, Yin JJ, Tian RN, Gu HW, Wang XM, Qi JL, Yang YH (2011) Cloning, characterization, and expression of LeEIL-1, an Arabidopsis EIN3 homolog, in Lithospermum erythrorhizon. Plant Cell Tissue Organ Cult 106:71–79
Acknowledgments
The authors thank Assistant Professor Mengmeng Gu (Texas A&M University, USA) and the two anonymous reviewers for critically revising the manuscript. This research was supported by the National Natural Science Foundation of China (No. 30972030).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, Y., Zhang, C., Jia, P. et al. Isolation and expression analysis of three EIN3-like genes in tree peony (Paeonia suffruticosa). Plant Cell Tiss Organ Cult 112, 181–190 (2013). https://doi.org/10.1007/s11240-012-0220-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-012-0220-z