Abstract
Plants, when exposed to abiotic or biotic stress, produce several pathogenesis-related proteins to counteract the effects of stress. Osmotin is one of the important pathogenesis-related proteins induced during several stress conditions. We have developed improved salt stress tolerant transgenic chilli pepper plants (Capsicum annum L. var. Aiswarya 2103) by ectopic expression of the Nicotiana tabaccum osmotin gene using Agrobacterium tumefaciens EHA105 as a vector. Four-week-old chilli pepper leaves were used as an explant and A. tumefaciens EHA105 harboring pBINASCOSM plasmid that contains osmotin gene under the control of CaMV 35S promoter and npt II as a selectable marker was used in co-cultivation. Transgene integration and expression were analyzed using molecular, immunochemical, and biochemical assays. PCR and Southern blot analysis confirmed that osmotin gene has been successfully integrated into the genome of chilli pepper plants. The osmotin gene was stably segregated and expressed in T2 generation transgenic chilli pepper plants, and it was confirmed by Western blot analysis. Biochemical assays of these putative transgenic plants revealed enhanced levels of chlorophyll, proline, glycinebetaine, APX, SOD, DHAR, MDHAR, GR, and relative water content. Yield potential of the putative transgenic chilli pepper plants was evaluated under salinity stress conditions in a green house. The putative transgenic chilli pepper plants overexpressing the osmotin gene were morphologically similar to wild-type plants and produced 3.32 kg chilli pepper fruits per plant at 300 mM NaCl concentration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinity is one of the prominent abiotic factors affecting crop yields in arid and semiarid irrigated areas. Almost three-quarters of the earth surface is covered by salt water, and thus it is not surprising that salts affect a significant proportion of the world’s land surface. Salt-affected soil contains sufficient concentrations of soluble salts, which cause toxicity to common crop plants. In agriculture, salt stress has its own negative impact on the growth and economic yield of many important crops (Maas and Hoffman 1977). Salinity imposes some potential effects on plant tissues: a water deficit that results from the relatively high solute concentrations in the soil, ion-specific stresses resulting from altered K+/Na+ ratio and Na+ and Cl− concentrations that are detrimental to plants and interference with the uptake of essential nutrients. Hence, the plant water potential should be lowered in order to maintain water uptake in salt environment. Higher accumulation of salt ion will cause toxic effect on cells, and thus must be separated from the metabolic machinery of the cells. This is achieved by compartmentation of solutes (Flowers and Yeo 1986).
Plants respond to adverse environmental conditions by expression of specific genes and synthesis of a large number of stress-related proteins, which plays a crucial role in stress adaptation (Skriver and Mundy 1990). Significant progress has been reported in the identification of genes involved in abiotic stress tolerance and their transfer to economically important crop plants for increased stress tolerance. Among the several stress-related proteins, osmotin is one of the unique proteins, which is induced in response to both abiotic and biotic stresses in plants (Parkhi et al. 2009) and highly conserved in the Solanaceae family (Zhu et al. 1995). Osmotin is a stress-responsive protein adapted to salinity and desiccation and accumulates in salt-adapted cells (Bressan et al. 1987). Osmotin is an abundant cationic 26-kDa protein that belongs to the family of PR-5 type proteins (Linthorst 1991). Osmotin proteins have peptide sequences, which are similar to the bifunctional trypsin/α-amylase inhibitor from maize, the sweet protein thaumatin, the tomato protein NP 24, the tobacco anti viral protein gp 22, potato PR protein C, tobacco, and barley thaumatin-like proteins (La Rosa et al. 1992). Osmotin protein is induced in cultured tobacco cells adapted to grow under salt stress or polyethylene glycol (PEG)-mediated water stress (Singh et al. 1985). It has been demonstrated that tobacco osmotin gene expression is activated by abscisic acid, sodium chloride, wounding, viral infection, ethylene, and cold (La Rosa et al. 1992; Zhu et al. 1996). Osmotin provides osmotolerance to plants probably by facilitating the compartmentation of solutes (Barthakur et al. 2001) or by being involved in metabolic or structural alterations during osmotic adjustment (Raghothama et al. 1993). Overexpression of osmotin induces proline accumulation and confers tolerance to osmotic stress in transgenic tobacco (Barthakur et al. 2001). Singh et al. (1987) hypothesized that the synthesis of osmotin protein could induce synthesis and accumulation of certain solutes or could be involved in metabolic or structural changes. The constitutive expression of an osmotin gene in transgenic tobacco, cotton, and strawberry showed improved tolerance to salinity and drought stress (Barthakur et al. 2001; Parkhi et al. 2009; Husaini and Abdin 2008).
Chilli pepper (Capsicum annum L.) is an important vegetable and spice crop valued for its aroma, taste, pungency, and flavor. In addition to the importance as a vegetable, chillies have also received attention recently for their potential as nutraceuticals. Although used primarily for seasoning, it is now recognized that chilli pepper has played a major nutritional role in many cultures by supplying them with a primary source of vitamin C. Compared to the other vegetable crops, chilli pepper plants are more sensitive to salinity stress. Improving the salinity stress tolerance of chilli pepper has become a top priority objective in most chilli pepper growing areas. Based on these, we attempted to transform the osmotin gene under the control of CaMV 35S promoter into chilli pepper for a potential improvement of growth under salt stress. A successful transgene integration of osmotin was analyzed using molecular, immunochemical, and biochemical analyses.
Materials and methods
Preparation of osmotin gene constructs and plant transformation
All recombinant DNA techniques were performed essentially as described by Sambrook et al. (1989), using modifications recommended by the enzyme manufacturers.
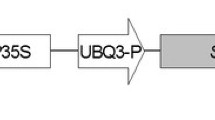
The tobacco (Nicotiana tabacum L. cv. Wisconsin 38) osmotin gene was isolated from the expression vector pRSET (Invitrogen life science technologies, India) as a 1,300-bp Pst I and Kpn I fragment and ligated to pUC18 plasmid (Genei, Bangalore, India) carrying 800-bp CaMV 35S promoter and 200-bp octopine synthase gene terminator (OSCT) and designated as pASC14. The expression cassette of CaMV 35S promoter: osmotin: octopine synthase gene terminator was released by double digestion of plasmid pASC14 with Hind III and EcoR I and subcloned into pBIN20 plasmid. The resulting recombinant binary vector was labeled as pBINASCOSM (Fig. 1). The binary vector pBINASCOSM was introduced into Agrobacterium tumefaciens EHA105 by triparental mating using the pRK2013 helper vector. The selected transgenic strain of Agrobacterium tumefaciens EHA105 was grown overnight at 28°C in minimal AB medium supplemented with the appropriate antibiotics (50 mg l−1 kanamycin and 10 mg l−1 refampicin) for the plant transformation.
Schematic representation of the binary vector pBINASCOSM used in chilli pepper transformation. The T-DNA region of the pBINASCOSM binary vector showing the assembly of osmotin expression cassette (CaMV 35S P: osmotin: OSCT) and kanamycin expression cassette (Nos P: npt II: Nos Ter). CaMV 35S P CaMV 35S promoter; OSCT Octopine synthase gene terminator; Nos P Nopaline synthase promoter; Nos Ter Nopaline synthase terminator; npt II Neomycin phosphor transferase enzyme
Transformation of chilli pepper plants
The plant transformation was carried out by using 4 weeks old chilli pepper plant leaf samples. Leaf discs of 3 mm × 3 mm were prepared from surface sterilized plant leaves and inoculated on MS basal medium with the adoxil surface of the leaf disc in contact with the medium for 24 h. After 24 h the leaf discs were co-cultivated with the Agrobacterium tumefaciens EHA105 carrying the osmotin gene construct and placed on co-cultivation media [CCM (M.S media, 30 g l−1 sucrose, 5 mg l−1 BA, 1.0 mg l−1 IAA, 1.0 mg l−1 nicotinic acid, 7 g l−1 agar)] for 72 h in dark. Transformed leaf discs were selected by transferring the leaf discs onto the selection media [SM (CCM containing 50 mg l−1 kanamycin, 200 mg l−1 cefotaxime, and 250 mg l−1 augmentin)], and incubated at 22 ± 2°C in dark for 3–4 weeks for shoot induction. The regenerated shoots were multiplied on shoot multiplication media [SMM (SM containing 4 mg l−1 AgNo3) at 22 ± 2°C under 16 h photoperiod for 2 weeks. The multiplied shoots were then transferred onto shoot elongation media [SEM (M.S media, 30 g l−1 sucrose, 3 mg l−1 BA, 1.0 mg l−1 IAA, 1.0 mg l−1 nicotinic acid, 3 mg l−1 GA3 and 7 g l−1 agar)] for 2 weeks at 22 ± 2°C under 16 h photo period and rooted on MS containing 0.5 mg l−1 IBA and 50 mg l−1 kanamycin. After 5 weeks, the well-rooted shoots were removed from the culture bottles, washed with sterile distilled water, and transferred into the pots containing a mixture of autoclaved peat and sand (1:1 ratio). The pots were covered with polythene bag for 2 weeks to maintain high humidity and then transferred to green house for further growth. One-month-old green house grown T2 generation chilli pepper plants were treated with different concentrations of NaCl (0–500 mM) for 15 days, those plants survived at high NaCl concentration, and non-transgenic plants (NT) were randomly selected for molecular, immunochemical and biochemical analysis.
Molecular analysis of transgenic chilli pepper plants
PCR amplification
For PCR analysis, DNA samples from T2 generation randomly selected putative transformants (15 plants), and non-transgenic (NT) chilli pepper leaves were isolated by the method described by Della Porta et al. (1983). The osmotin gene (1,000 bp) was amplified using primers 5′-AGCCGATTTCTAACTGGCACT-3′ and 5′-CGGTATAAGGCTTCTAAGGGC-3′. The npt II gene (732 bp) was amplified by using primers 5′-GAGGCTATTCGGCTATGACTG-3′ and 5′- ATCGGGAGAGGCGATACCGTA-3′. All polymerase chain reactions were performed using a Peltier effect thermal cycler (MJ Research, Waltham, MA). Samples containing 60 ng genomic DNA were subjected to initial denaturation at 94°C for 5 min followed by 30 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min followed by 7-min final extension at 72°C. Sixty nanogram of pBINASCOSM plasmid was used as positive control. The amplified PCR fragments were analyzed by 1.2% agarose gel electrophoresis at 100 volts.
Southern blot hybridization
Southern blot analysis was performed to verify integration and copy number of the transgene. Genomic DNA (10 μg) of transformed and non-transformed (NT) chilli pepper plants was digested with 20 U BamH I and resolved in 0.8% agarose gel. Following agarose gel electrophoresis, DNA was transferred to nylon membrane (Hybond-N+, Amersham biosciences, UK) as described by Sambrook et al. (1989). PCR-generated osmotin gene fragment (1,000 bp) was used as probe. The probe was labeled according to the manufacturer’s instructions using a non-radioactive labeling kit (ECL random prime labeling and detection system, Amersham biosciences, UK). The nylon membrane was hybridized at 42°C with osmotin probe for 20 h. The hybridized nylon membrane was washed with 5 × SSC, 0.1% SDS, and 1 × SSC, 0.1% SDS at 42°C, and exposed to X-ray film (Kodak, India) for 30 min. The film was developed and fixed using an X-ray film development kit (Kodak).
Immunochemical analysis of transgenic chilli pepper plants
Induction of osmotin protein and preparation of osmotin antibody
The osmotin gene (pRSET-osmotin) was induced in the E.Coli DH5α using 0.1 M IPTG. Purification of recombinant osmotin protein was performed by affinity chromatography using Ni–NTA column (QIAGEN, Hilden, Germany), and the purified osmotin protein was used to immune rabbit for the preparation of osmotin antibody. The dilution factor of the antiserum was calculated by dot western.
Western blot analysis
For Western blotting, extraction of total soluble proteins from transgenic and non-transgenic (NT) chilli pepper plants was carried out by homogenizing 100 mg of leaf samples with 500 μl of protein extraction buffer (0.05 M Na2HPO4, pH 7.2, 0.5 M NaCl, 1 mM EDTA, 1 mM PMSF, and 10 mM 2-mercaptoethanol). The extracted soluble proteins were resolved using 15% SDS–PAGE and transferred onto a nitrocellulose membrane (Millipore, Bangalore, India). Immunoblot analysis was carried out using rabbit antiserum against osmotin protein (1:500 dilution) as primary antibody, alkaline phosphatase conjugated goat anti rabbit IgG as secondary antibody (1:750 dilutions), and visualized by BCIP-NBT (Genei, Bangalore, India) staining.
Biochemical analysis of transgenic chilli pepper
For all biochemical assays, we used Western blotting positive and non-transgenic chilli pepper plants.
Leaf disc senescence assay
Leaf disc senescence assay was performed by incubating the leaf discs of 4 × 4 cm in different concentrations of NaCl (0, 25, 50, 100, 200, 300, 400, and 500 mM) for about 1 week at 24 ± 2°C under a 16-h photoperiod. After 1-week incubation, the total chlorophyll content of the leaf discs was estimated according to the method described by Hiscox and Israelstam (1979). Each treatment comprised three bottles in replicates of two.
Estimation of free proline, glycinebetaine, and H2O2 concentrations
Hundred milligrams of freshly harvested leaf samples was homogenized using 3% sulfosalicylic acid, and free proline concentration was estimated at 560 nm using colorimeter (Bates et al. 1973). Glycinebetaine concentration was estimated according to the method described by Cromwell and Rennie (1953). H2O2 level was determined by the method of Velikova et al. (2000).
Assay of APX and SOD activities
APX activity was estimated based on the method of Chen and Asada (1989). Five hundred milligrams of leaf tissue was homogenized with 1.5 ml of 100 mM sodium phosphate buffer (pH 7.0) containing 1 mM EDTA and 5 mM ascorbate. Fifty microliter of the extract was treated with the reaction mixture consisting of 50 mM phosphate buffer (pH 7.0), 0.5 mM ascorbate, and 0.2 mM H2O2. The enzyme activity was recorded based on the change in absorbance at 260 nm (E = 2.8 mM−1 cm−1). The results were calculated in terms of μM of ascorbate oxidized per minute. Superoxide dismutase activity was assayed by following the method of Giannopolitis and Ries (1977).
Dehydroascorbate reductase (DHAR) activity
Dehydroascorbate reductase was assayed by observing the increase in absorbance at 265 nm of a 1 ml reaction mixture containing 50 mM potassium phosphate buffer (pH 7.0), 0.1 mM EDTA, 2.5 mM reduced glutathione, 0.2 mM dehydroascorbate, and 80 μl extract (Doulis et al. 1997).
Monodehydroascorbate reductase (MDHAR) activity
Monodehydroascorbate reductase activity was determined as described previously (Miyake and Asada 1992). The decrease in absorbance at 340 nm was monitored in a 1 ml reaction mixture containing 50 mM HEPES/KOH (pH 7.6), 0.1 mM NADPH, 2.5 mM ascorbate, 80 μl extract, and 0.4 units ascorbate oxidase.
Glutathione reductase (GR) activity
Glutathione reductase activity was determined as described by Foyer and Halliwell (1976). The oxidized glutathione (GSSG)-dependent oxidation of NADPH was followed at 340 nm in a 1 ml reaction mixture containing 100 mM sodium phosphate buffer (pH 7.8), 0.5 mM GSSG, 80 μl extract, and 0.1 mM NADPH.
Malondialdehyde (MDA) assay
Malondialdehyde was assayed by homogenizing 200 mg of leaf sample in 10% TCA solution. The reaction was performed by adding 2 ml of supernatant to 3 ml of 0.6% thiobarbituric acid dissolved in 10% TCA, and the mixture was incubated at room temperature for 2 h followed by boiling at 100°C for 1 h. The OD was measured at 450, 532, and 600 nm. By using the following formula, MDA concentration was estimated.
Evaluation of relative water content and ion leakage ratio of cell membranes
Leaf sections of similar size were used to estimate relative water content (RWC) and ion leakage ratio. The estimation of RWC was carried out according to the method of Brini et al. (2007). Membrane damage was reckoned and assayed by measuring ion leakage from leaf discs according to the method of Fan et al. (1997).
Salt stress tolerant trails
T2 generation transgenic and non-transgenic chilli pepper seeds were germinated in plastic cups containing sterile peat and sand (1:1 ratio) in green house. After 15 days of germination, the plantlets were equally watered bi-weekly with different concentrations of NaCl (25–400 mM). For control, another set for each plant was maintained, which was similarly watered without NaCl (0 mM). Each treatment comprised five earthen pots in replicates of two.
Yield performance of transgenic chilli pepper plants
For yield performance, we selected high salt stress tolerant T2 generation transgenic and non-transgenic chilli pepper plants. Transgenic and non-transgenic chilli pepper plants were tested for yield under three different conditions of NaCl stress (0, 100, and 300 mM) in green house. One-month-old chilli pepper plants were equally watered bi-weekly without NaCl, with 100, and 300 mM NaCl containing water till fruit setting and ripening. The fresh ripened chilli pepper fruits were harvested, and the fresh weight was measured using weighing balance.
Statistical analysis
Statistical analyses were carried out by two-way classification of ANOVA to evaluate whether the means were significantly different, taking P < 0.05 as significance level.
Results
Identification of transgenic chilli pepper plants
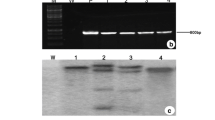
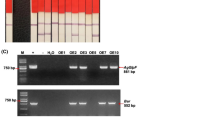
Transgenic chilli pepper plants that sustained high salt concentration were raised using Agrobacterium tumefaciens EHA105 carrying pBINASCOSM plasmid. From 350 co-cultivated leaf discs, 135 kanamycin-resistant shoots established roots and survived in different stages. Randomly selected 15 chilli pepper plants were subjected to molecular, immunochemical, and biochemical analyses. Putative transgenic plants were screened for the presence of the transgene integration by PCR and Southern hybridization. Primers for the npt II and osmotin gene were designed to amplify the DNA fragment of 732 and 1,000 bp, respectively. The presence of npt II gene was confirmed in all 15 putative transgenic chilli pepper plants and in the pBINASCOSM plasmid (Fig. 2a), whereas only 10 chilli pepper plants and pBINASCOSM showed 1,000-bp amplified product of osmotin gene (Fig. 2b). There was no amplification in the DNA of non-transgenic plants. These 10 chilli pepper plants were designated as CO1 to CO10 (Chilli osmotin 1–10) and selected for Southern blot hybridization. Southern blot hybridization showed the horseradish peroxidase (HRP)-labeled osmotin probe hybridized to BamH I-digested genomic DNA of all 10 transgenic chilli pepper plants and the pBINASCOSM plasmid carrying the osmotin gene (Fig. 2c), but there was no hybridization with the DNA of non-transgenic (NT) chilli pepper. These transgenic plants showed up to four copies of osmotin gene integration (Fig. 2c, lane 7), and the hybridization patterns were non-identical due to different transformation events. These results confirmed the presence of osmotin gene in transgenic chilli pepper plants. By Western blot analysis, a 26-kDa osmotin protein, specifically recognized by the anti-osmotin antibody, confirmed the expression of osmotin gene in six transgenic chilli pepper plants (CO1, CO2, CO4, CO5, CO8, and CO10). Remaining transgenic plants did not generate a positive signal (Fig. 2d; lanes 5, 8, 9, and 11). This may be due to the insufficient amount of protein transfer to nitrocellulose membrane or absence of gene expression or problem in the segregation of the osmotin gene. In transgenic plants, the osmotin protein accumulated about 0.081 (CO2) to 0.857 μg mg−1 (CO8) of total soluble protein. Proteins from non-transgenic (NT) plants did not react with the anti-osmotin antibody. The expression of osmotin was followed in 6 independent chilli pepper plants, and all the six plants were used for biochemical analysis to confirm salinity/salt stress tolerant capacity.
a PCR amplification of npt II gene from the genomic DNA of transformants. Lanes 1 and 19 100 bp ladder marker; Lane 2 pBINASCOSM plasmid as positive control; Lane 3 Non-transgenic (NT) chilli pepper plant DNA as negative control; Lanes 4–18 Transgenic chilli pepper plant genomic DNA samples. b PCR amplification of osmotin gene from the genomic DNA of transformants. Lanes 1 and 19 100 bp ladder marker; Lane 2 pBINASCOSM plasmid as positive control; Lane 3 Non-transgenic (NT) chilli pepper plant DNA as negative control; Lanes 4–18 Transgenic chilli pepper plant genomic DNA samples. c Analysis of transgenic chilli pepper plants by Southern blot hybridization. Lane 1 pBINASCOSM plasmid carrying osmotin gene as positive control; Lane 2 Non-transgenic (NT) chilli pepper sample as negative control; Lanes 3–12 Transgenic chilli pepper plant (CO1–CO10) DNA samples digested with BamH I enzyme and probed with osmotin specific probe; Lane 13 Molecular weight marker. d Western blotting to analyze the expression of osmotin protein. Lane 1 Non-transgenic (NT) chilli pepper sample as negative control; Lane 2 Purified osmotin protein as positive control; Lanes 3–12 Transgenic chilli pepper plant (CO1–CO10)) samples expressing osmotin gene
Salt stress tolerance
The transgenic plants over expressing the osmotin gene showed clear phenotypic difference with respect to growth and development when compared to non-transgenic plants under salt stress (Fig. 3). A noteworthy observation was the better growth performance of transgenic plants that showed near-normal phenotype with marginal growth reduction even at 300 mM NaCl, providing evidence for functional expression in plants of the transgene following introgression. In case of non-transgenic plants, the stress effect was severe even at 100 mM NaCl exhibiting prominent growth inhibition and wilted leaves. Above 100 mM NaCl, all non-transgenic chilli pepper plants became totally bleached out or drastically wilted. The transgenic chilli pepper plants behaved similar to that of non-transgenic chilli pepper plants in all aspects when grown under non stress conditions.
Phenotypic appearance of 1-month-old T2 generation transgenic and non-transgenic chilli pepper plants irrigated with NaCl containing water. a–c Transgenic chilli pepper plants irrigated with 100, 200 and 300 mM NaCl containing water, respectively; d–f Non-transgenic (NT) chilli pepper plants irrigated with 25, 50 and 100 mM NaCl containing water, respectively; g Green house grown 3 months old T2 generation transgenic chilli pepper plant irrigated with 300 mM NaCl containing water; h Green house grown T2 generation transgenic chilli pepper plant containing the green chillies
Biochemical analysis
In leaf disc senescence assay, the transgenic leaf discs retained the chlorophyll up to 300 mM NaCl concentration, whereas non-transgenic leaf discs were completely bleached above 100 mM. To confirm the visual phenotype, total chlorophyll content was estimated. The findings ascertained that transgenic plantlets have significant amount of chlorophyll than non-transgenic chilli pepper under salt stress (Fig. 4a).
Response of chilli pepper plants exposed to various concentration of NaCl for 15 days under green house conditions. a Chlorophyll concentration (mg g−1 of fw.); b Proline concentration (μg g−1 of fw.); c Glycinebetaine concentration (mg g−1 of fw.); and d Hydrogen peroxide concentration (μmol g−1 of fw.). The bars indicate mean ± standard error
There were no differences in the concentration of proline and glycinebetaine between the transgenic and non-transgenic chilli pepper when grown under optimal growth conditions. However, a more dramatic increase in proline (Fig. 4b) and glycinebetaine (Fig. 4c) concentration was observed in the transgenic chilli pepper plants when grown under salt stress conditions compared to non-transgenic chilli pepper plants.
H2O2 content was negatively correlated with changes in the activities of the antioxidant enzymes. Salt stress caused a marked increase in the level of H2O2 in non-transgenic chilli pepper with an increase in NaCl concentration. In comparison to the non-transgenic plants, the H2O2 content of the transgenic plants increased at a slow pace (Fig. 4d). This slow increase in H2O2 level might have been responsible for the much better tolerance of transgenic chilli pepper plants grown under different concentrations of NaCl.
The activities of the antioxidant enzymes such as APX and SOD were almost similar in transgenic and non-transgenic plants under optimal growth conditions. However, after 15 days of NaCl treatment, the activities of the APX and SOD were drastically reduced in non-transgenic plants, whereas transgenic plants showed marked increase in enzyme activities with increase in NaCl concentration (Fig. 5a, b).
Response of chilli pepper plants exposed to various concentration of NaCl for 15 days under green house conditions. a APX concentration (μmol mg−1 of protein min−1); b SOD activity (Units mg−1 of protein min−1); c DHAR activity (μmol min−1 mg−1); d MDHAR activity (μmol min−1 mg−1). The bars indicate mean ± standard error
There were no noticeable differences in DHAR, MDHAR, and GR activities between transgenic and non-transgenic chilli pepper plants under normal growth conditions. However, during salinity stress, transgenic plants showed increased concentrations of DHAR (Fig. 5c), MDHAR (Fig. 5d), and GR (Fig. 6a). Such type of increase was not found in non-transgenic plants.
MDA is the product of lipid peroxidation caused by reactive oxygen species (ROS) generation during oxidative stress. MDA is considered as a marker of oxidative lipid injury, and its levels were widely used as a measure of damage due to various biotic and abiotic stresses. Under normal growth conditions, both transgenic and non-transgenic plants showed similar concentration of MDA. On the other hand, non-transgenic plant showed substantially higher concentration of MDA than transgenic plants under salt stress (Fig. 6b).
There were no noticeable differences in RWC between the transgenic and non-transgenic chilli pepper plants under normal growth conditions. After 15 days of NaCl treatment, RWC of the transgenic plants was comparatively higher than that of non-transgenic plants. The percentages of RWC in transgenic and non-transgenic plants were 82 and 46%, respectively, at 100 mM NaCl concentration (Fig. 6c).
In terms of ion leakage, the transgenic and non-transgenic plants were similar under optimum growth conditions. However, under salt stress conditions, there was a considerable increase in ion leakage of non-transgenic chilli pepper than transgenic chilli pepper plants (Fig. 6d). This clearly indicates that lesser damage has been experienced by the membrane system of transgenic plants under salt stress.
The transgenic chilli pepper plants were morphologically similar to non-transgenic plants under normal and salt stress condition. At 100-mM NaCl concentration, non-transgenic plants produced about 2.2 kg of chilli pepper fruits per plant, whereas at the same concentration of NaCl, transgenic plants produced 3.3 kg of chilli pepper fruits per plant. At 300 mM concentration, transgenic chilli pepper plants produced about 3.32 kg of chilli pepper fruits per plant, and non-transgenic plants were completely perished (Table 1).
Discussion
Salinity, which has an adverse effect on the growth and the productivity of crops, has emerged into a global threat among the agricultural community by affecting approximately 20% of the globally irrigated agricultural land (Flowers and Yeo 1995; Munns 2002). Almost all major vegetable crops, including chilli pepper are susceptible to increased salinity (Shannon and Grieve 1999). Plants respond to salt stress in part by modulating gene expression, which eventually leads to the renovation of cellular homeostasis, detoxification of toxins, and recovery of growth (Xiong and Zhu 2002). Thus, one important aspect of salt-stress studies is the survey of genes in plants in response to salt stress. In the past 10 years, most of the salt stress responding genes has been identified. The identification, cloning, and expression of salinity stress tolerant-related genes in economically important plants have created an opportunity to improve crop plants against salinity stress. Osmotin protein is an important member of the PR5 group proteins that expressed during biotic and abiotic stresses. Overexpression of osmotin gene in tobacco (Barthakur et al. 2001; Sokhansanj et al. 2006), tomato (Sarad et al. 2004), strawberry (Husaini and Abdin 2008), and chilli pepper (in the present study) reported enhanced tolerance to NaCl-mediated salinity stress. In this study, the Nicotiana tabacum osmotin gene under the control of CaMV 35S promoter was successfully transferred into the chilli pepper by using Agrobacterium tumefaciens EHA105. The integration of the osmotin gene into chilli pepper genome was confirmed by PCR, Southern hybridization, and the expression of osmotin protein was ascertained by Western blotting.
Under high salt, drought, and low temperature conditions, many plants produce osmolytes such as free proline and glycinebetaine, which may function as osmoprotectants (Shi-Qing Gao et al. 2009). The Petunia and Lycium barbarum L. plants expressing Δ1 pyrroline-5-carboxylate synthetase and ATHK1 genes, respectively, accumulated more amount of free proline and survived during drought and salinity conditions (Yamada et al. 2005; Chen et al. 2009). Overexpression of CBF3/DREB1A in transgenic Arabidopsis plants produced a higher level of free proline than in wild-type plants under stress conditions (Gilmour et al. 1998). The proline content of wheat overexpressing GhDREB confers enhanced tolerance to drought, high salt, and freezing. Free proline content of osmotin-overexpressing tobacco (Barthakur et al. 2001), cotton (Parkhi et al. 2009), and strawberry (Husaini and Abdin 2008) was much more than that of non-transgenic plants. In our study, osmotin gene overexpressing in chilli pepper plants accumulated higher levels of proline than non-transgenic plants under salinity stress conditions, suggesting that overexpression of osmotin activated the expression of downstream genes involved in proline biosynthesis, which in turn, enhanced tolerance to salinity stress in transgenic chilli pepper plants. Moreover, osmotin overexpressing transgenic chilli pepper plants maintained higher level of chlorophyll than non-transgenic plants under high salt stress conditions. A possible explanation was that overexpression of osmotin activated the expression of downstream genes that prevented chlorophyll decomposition, thus ensuring normal photosynthesis, and improving tolerance to high salt stress (Zhang et al. 2004).
Reactive oxygen species (ROS) like superoxide, hydrogen peroxide, hydroxyl radicals, and singlet oxygen accumulate in higher level in plant tissues as a result of excess salt and cellular dehydration and are toxic to the cells. Under environmental stress, the balance between ROS production and the degradation activity of the antioxidant system is impaired (Dhindsa and Matowe 1981). The elevated levels of proline contributing to the detoxification of ROS and free radicals (Floyd and Nagy 1984) by forming long-lived adducts with them in response to osmotic stress have been confirmed in tobacco (Hong et al. 2000). The antioxidant enzymes include superoxide dismutase, ascorbate peroxidase, and glutathione reductase. The tomato plants over expressing Mn-SOD gene showed more amount of APX activity than non-transgenic plants under salt stress (Yueju Wang et al. 2007). SOD activity has been found to be enhanced in osmotically stressed leaf discs of rape plants compared to unstressed control plants (Aziz and Larher 1998).
The transgenic chilli pepper plants showed lower accumulation of H2O2 than the non-transgenic chilli pepper plants at different NaCl concentrations. The increased amount of NaCl increased the H2O2 content in all plants. To detoxify the accumulated H2O2, chilli pepper plants synthesized APX and SOD. The peroxidase and dismutase activities had to be largely enhanced to scavenge the bulk amount of hydrogen peroxide and sustain growth during salt stress. The transgenic chilli pepper plants showed higher concentrations of APX and SOD than the non-transgenic chilli pepper plants under salt stress. This reflects the importance of APX and SOD as a scavenging system. Benavides et al. (2000) reported the enzymes responsible for H2O2 detoxification during salt tolerance in Solanum tuberosum to be APX and CAT. However, they also suggested that APX was likely to be more important than CAT in detoxification.
In addition to APX, monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), and glutathione reductase (GR) also catalyze important steps in ascorbate–glutathione cycle (Noctor and Foyer 1998), which leads to the production of reducing agents such as ascorbate and glutathione. The increase in DHAR, MDHAR, and GR activities ensures efficient regeneration of ascorbate and glutathione, which can scavenge increased levels of H2O2 under stress conditions. Increases in the activity of DHAR and GR in stressed transgenic tubers and tobacco also demonstrated improved tolerance to various oxidative stresses (Foyer et al. 1991, 1995; Aono et al. 1995).
The content of MDA serves as an indicator of lipid peroxidation in plant cells (Shalata and Tal 1998). It was reported that under the salt stress, the SOD activity of salinity tolerant plants was higher, and the level of peroxidation of membrane lipids was much lower than that of sensitive plants (Wu et al. 2003; Shalata and Tal 1998). In our study, the MDA content of non-transgenic chilli pepper plants treated with 100 mM NaCl increased remarkably, while those of transgenic plants increased at a meager rate. This observation suggested that the degree of damage to cell membrane of transgenic plants is significantly lesser than that of non-transgenic chilli pepper plants. Therefore, the higher activities of antioxidant enzymes in transgenic chilli pepper had played their role in elimination of ROS, and thus prevented the membrane lipids from peroxidation.
Relative water content is an important and a major determinant of metabolic activity and leaf survival. It was proposed to be a better indicator of the healthy status of plant cell than water potential (Sinclair and Ludlow 1985). In the present study, transgenic chilli pepper showed higher amount of RWC than non-transgenic chilli pepper plants. It has been hypothesized that osmotin may be involved in osmotic adjustment of cells either by facilitating the accumulation or compartmentation of solutes or by regulating metabolic or structural alterations during osmotic adjustment (Singh et al. 1987).
We concluded that salt stress seriously affected the growth and development of non-transgenic chilli pepper plants; however, the cloning and expression of osmotin gene into chilli pepper enhanced salt tolerance. The presented results suggested that overexpression of osmotin in chilli pepper plays more pivotal role in preventing the accumulation of ROS and protects the cell against ROS caused by the salt stress, thereby enhancing salt tolerance in transgenic plants. This study will help to explain the differential and essential roles of osmotin in adaptive responses of plant cells to environmental stresses, especially salinity stress.
Abbreviations
- CaMV 35S:
-
Cauliflower mosaic virus 35S promoter
- APX:
-
Ascorbate peroxidase
- SOD:
-
Superoxide dismutase
- DHAR:
-
Dehydro ascorbate reductase
- MDHAR:
-
Monodehydroascorbate reductase
- GR:
-
Glutathione reductase
- MS:
-
Murashige and Skoog medium
- BA:
-
6-Benzyladenine
- IAA:
-
Indole-3-acetic acid
- GA3 :
-
Gibberellic acid
- IBA:
-
Indole-3-butyric acid
- AgNO3 :
-
Silver nitrate
References
Aono M, Saji H, Sakamoto A, Tanaka K, Kondo N (1995) Paraquat tolerance of transgenic Nicotiana tabacum with enhanced activities of glutathione reductase and superoxide dismutase. Plant Cell Physiol 36:1687–1691
Aziz A, Larher F (1998) Osmotic stress induced changes in lipid composition and peroxidation in leaf discs of Brassica napus L. J Plant Physiol 153:754–762
Barthakur S, Babu V, Bansal KC (2001) over expression of osmotin induces proline accumulation and confers tolerance to osmotic stress in transgenic tobacco. J Plant Biochem Biotechnol 10:31–37
Bates LS, Waldren RP, Teare JD (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–207
Benavides MP, Marconi PL, Gallego SM, Comba ME, Tomaro ML (2000) Relationship between antioxidant defense system and salt tolerance in Solanum tuberosum. Aust J Plant Physiol 27:273–278
Bressan RA, Singh NK, Handa AK, Mount R, Clithero J, Hasegawa PM (1987) Stability of altered gene expression in cultured plant cells adapted to salt. In: Monti L, Porceddu E (eds) Drought resistance in plants, physiological and genetic aspects. Commission of the European Communities, Brussels, pp 41–57
Brini F, Hanin M, Lumbreras V, Amara J, Khoudi H et al (2007) Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep 26:2017–2026
Chen GX, Asada K (1989) Ascorbate peroxidase in tea leaves: occurrence of two isozymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol 30:987–998
Chen Ni, Liu Yan, Liu Xin, Chai Juan, Zhang Hu, Guo Guangqiv, Liu Heng (2009) Enhanced tolerance to water deficit and salinity stress in transgenic Lycium barbarum L. plants ectopically expressing ATHK1, an Arabidopsis thaliana histidine kinase gene. Plant Mol Biol Rep 27:321–333
Cromwell BT, Rennie SD (1953) The estimation and distribution of glycinebetaine (Betaine) in Beta vulgaris L. and other plants. Biochem J 55(1):189–192
Della Porta SL, Wood J, Hicks JB (1983) A plant DNA mini preparation: verson II. Plant Mol Biol Rep 1:19–21
Dhindsa RS, Matowe W (1981) Drought tolerance in two mosses correlated with enzymatic defense against lipid peroxidation. J Exp Bot 22:79–91
Doulis AG, Debian N, Kingston-Smith AH, Foyer CH (1997) Differential localization of antioxidants in maize leaves. Plant Physiol 114:1031–1037
Fan L, Zhen S, Wang X (1997) Antisense suppression of phospholipase D retards abscisic acid and ethylene promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9:2183–2196
Flowers TJ, Yeo AR (1986) Ion relations of plants under drought and salinity. Aust J Plant Physiol 13:75–91
Flowers TJ, Yeo AR (1995) Breeding for salinity resistance in crop plants-Where next? Aust J Plant Physiol 22:875–884
Floyd RA, Nagy ZS (1984) Formation of long lived hydroxyl free radical adducts of proline and hydroxyl-proline in a Fenton reaction. Biochim Biophys Acta 790:94–97
Foyer CH, Halliwell B (1976) The presence of glutathione and glutathione reductase in chloroplasts: a proposed role in ascorbic acid metabolism. Planta 133:21–25
Foyer C, Lelandais M, Galap C, Kunert KJ (1991) Effect of elevated cytosolic glutathione reductase activity on the cellular glutathione pool and photosynthesis in leaves under normal and stress conditions. Plant Physiol 97:863–872
Foyer CH, Souriau N, Perret S, Lelandais M, Kunert KJ, Pruvost C (1995) Overexpression of glutathione reductase but not glutathione synthetase leads to increases in antioxidant capacity and resistance in photo inhibition in poplar trees. Plant Physiol 109:1047–1057
Gao S-Q, Chen M, Xia L-Q, Xiu H-J, Xu Z-S, Li L-C, Zhao C-P, Cheng X-G, Ma Y-Z (2009) A cotton (Gossypium hirsutum) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep 28:301–311
Giannopolitis CN, Ries SK (1977) Superoxide dismutases. I. Occurrence in higher planta. Plant Physiol 59:309–314
Gilmour SJ, Zarka DG, Stockinger EJ (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcription activators as an early step in cold-induced cor gene expression. Plant J 16:433–442
Hiscox JD, Israelstam GF (1979) A method for extraction of Chlorophyll from leaf tissue without maceration. Can J Bot 59:463–469
Hong ZL, Lakkineni K, Zhang ZM, Verma DPS (2000) Removal of feedback inhibition of DELTA1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol 122:1129–1136
Husaini AM, Abdin MZ (2008) Development of transgenic strawberry (Fragaria x ananassa Dutch.) plants tolerant to salt stress. Plant Sci 174:446–455
La Rosa PC, Chen Z, Nelson DE, Singh NK, Hasegawa PM, Bressan RA (1992) Osmotin gene expression is post transcriptionally regulated. Plant Physiol 100:409–415
Linthorst HJM (1991) Pathogenesis related proteins of plants. Crit Rev Plant Sci 10:123–150
Maas EV, Hoffman GJ (1977) Crop salt tolerance—current assessment. J Irrigation Drainage Div 103:115–134
Miyake C, Asada K (1992) Thylakoid-bound ascorbate peroxidase in spinach chloroplasts and photoreduction of its primary oxidation product monodehydroascorbate radicals in the thylakoids. Plant Cell Physiol 35:539–549
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Ann Rev Plant Physiol Plant Mol Biol 49:249–279
Parkhi V, Kumar V, Sunilkumar G, Campbell LAM, Singh NK, Rathore KS (2009) Expression of apoplastically secreted tobacco osmotin in cotton confers drought tolerance. Mol Breed 23:625–639
Raghothama KG, Liu D, Nelson DE, Hasegawa PM, Bressan RA (1993) Analysis of an osmotically regulated pathogenesis related osmotin gene promoter. Plant Mol Boil 23:1117–1128
Sambrook J, Fritch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring, Harbor, NY
Sarad N, Rathore M, Singh NK, Kumar N (2004) Genetically engineered tomatoes: new vista for sustainable agriculture in high altitude regions. In: Proceedings of the Fourth International Crop Science Congress Brisbane, Australia
Shalata A, Tal M (1998) The effect of salt stress on lipid peroxidation and antioxidants in the leaf of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Physiol Plant 104:169–174
Shannon MC, Grieve CM (1999) Tolerance of vegetable crops to salinity. Sci Hortic 78:5–38
Sinclair TR, Ludlow MM (1985) Who taught plants thermodynamics? The unfulfilled potential of plant water potential. Aust J Plant Physiol 12:213–217
Singh NK, Handa AK, Hasegawa PM, Bressan RA (1985) Proteins associated with adaptation of cultured tobacco cells to NaCl. Plant Physiol 79:126–137
Singh NK, Bracker CA, Hasegava PM et al (1987) Characterization of osmotin. Plant Physiol 85:529–536
Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2:503–512
Sokhansanj S, Sadat N, Niknam V (2006) Comparison of bacterial and plant genes participating in proline biosynthesis with osmotin gene, with respect to enhancing salinity tolerance of transgenic tobacco plants, Russ. J Plant Physiol 53:110–115
Velikova V, Yordanov J, Edreva A (2000) Oxidative stress and some antioxidant systems in acid rain treated bean plants. Protective role of exogenous polyamines. Plant Sci 151:59–66
Wang Y, Wisniewski M, Meilen R, Uratsu SL, Cui M, Dandekar A, Fuchigami L (2007) Ectopic expression of Mn-SOD in Lycopercicum esculentum leads to enhanced tolerance to salt and oxidative stress. J Appl Horti 9(1):3–8
Wu FB, Zhang GP, Dominy P (2003) Four barley genotypes respond differently to cadmium: lipid peroxidation and activities of antioxidant capacity. Environ Exp Bot 50:67–78
Xiong L, Zhu JK (2002) Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ 25:131–139
Yamada M, Morishita H, Urano K et al (2005) Effects of free proline accumulation in petunias under drought stress. J Exp Bot 56:1975–1981
Zhang HW, Huang ZJ, Xie BY, Chen Q, Tian X, Zhang XL, Zhang HB, Lu XY, Huang DF, Huang RF (2004) The ethylene, jasmonate, abscisic acid and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box containing genes and salt tolerance in tobacco. Planta 220:262–270
Zhu B, Chen THH, Li PH (1995) Activation of two osmotin-like protein genes by abiotic stimuli and fungal pathogen in transgenic potato plants. Plant Physiol 108:929–937
Zhu B, Chen THH, Li PH (1996) Analysis of late blight disease resistance and freezing tolerance in transgenic potato plants expressing sense and antisense genes for an osmotin-like protein. Planta 198:70–77
Acknowledgments
We thank Sri Krishnadevaraya University, Anantapur, Andhra Pradesh, India for providing financial support to carry out the present work. The authors are grateful to Prof. M.V. Rajam, Department of Genetics, Delhi University—South campus, India for the critical correction and evaluation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Subramanyam, K., Sailaja, K.V., Subramanyam, K. et al. Ectopic expression of an osmotin gene leads to enhanced salt tolerance in transgenic chilli pepper (Capsicum annum L.). Plant Cell Tiss Organ Cult 105, 181–192 (2011). https://doi.org/10.1007/s11240-010-9850-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-010-9850-1