Abstract
Transgenic chilli pepper (Capsicum annuum L.) plants tolerant to salinity stress were produced by introducing the wheat Na+/H+ antiporter gene (TaNHX2) via Agrobacterium-mediated transformation. Cotyledonary explants were infected with Agrobacterium tumefaciens strain LBA4404 harboring a binary vector pBin438 that contains a wheat antiporter (TaNHX2) gene driven by the double CaMV 35S promoter and NPT II gene as a selectable marker. PCR and semiquantitative RT-PCR analysis confirmed that the TaNHX2 gene had been integrated and expressed in the T1 generation of transgenic pepper plants as compared to the non-transformed plants. Southern blot analysis further verified the integration and presence of TaNHX2 gene in the genome of chilli pepper plants. Biochemical assays of these transgenic plants revealed enhanced levels of proline, chlorophyll, superoxide dismutase, ascorbate peroxidase, relative water content, and reduced levels of hydrogen peroxide (H2O2), malondialdehyde compared to wild-type plants under salt stress conditions. The present investigation clearly showed that overexpression of the TaNHX2 gene enhanced salt stress tolerance in transgenic chilli pepper plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinity is the most prevalent abiotic stress that severely reduces the growth and yield of many important food crops worldwide to a great extent. About 20 % of world irrigated agricultural land was adversely salt affected in more than 100 countries (Flowers and Yeo 1995; Rengasamy 2006). High-salinity stress disturbs ion homeostasis in plant cells. Ion-specific stresses resulting from altered K+/Na+ ratio and Na+ and Cl− concentrations are detrimental to plants. This ion flux causes a loss of homeostasis and plants respond to this condition by removing the sodium from the cytosol, through the vacuolar Na+/H+ antiporter. A vacuole Na+/H+ antiporter, sequestrating excess Na+ into the vacuole by using the electrochemical gradient of protons generated by the vacuolar H+-translocating enzymes (H+-ATPase and the H+-PPase), has been identified as an important salt tolerance determinant (Apse et al. 1999; Gaxiola et al. 1999; Quintero et al. 2000; Blumwald 2000; Zhang and Blumwald 2001; Zhang et al. 2001; Zhu 2003; Yamaguchi and Blumwald 2005; Leidi et al. 2010). Different Na+/H+ antiporters have been identified and intensively studied in Arabidopsis, rice, beetroot, cotton, maize, salsa, clover, and tomato (Gaxiola et al. 1999; Fukuda et al. 1999; Xia et al. 2002; Wu et al. 2004; Chen et al.2007; Li et al. 2007; Tang et al. 2010; Bhaskaran and Savithramma 2011). Among the several Na+/H+ antiporter genes, the wheat TaNHX2 gene is one of the salt stress-induced gene. The over expression of the TaNHX2 gene in transgenic soybean, alfalfa, tomato, and rice showed improved tolerance to salinity (Cao et al. 2011; Zhang et al. 2012; Yarra et al. 2012; Wu et al. 2012). Bojórquez-Quintal et al. (2014) hypothesized that the expression of HKT1 transporter could induce accumulation of proline there by increases salt tolerance in Capsicum chinense Jacq. Salt tolerance is a complex mechanism controlled by many genes (Bojórquez-Quintal et al. 2014). Therefore, manipulating the expression level of certain salt stress-induced genes is necessary to improve an agronomically important crop plants.
Chilli pepper (Capsicum annum L.) is an important vegetable and indispensable spice crop of the Solanaceae family and grown for supplying its pungent fruits, which are used both ripe and green to impart pungency to the food. Green chillies are rich in vitamin A and C, minerals, and proteins while dry chillies are rich in vitamin A and D which offers hotness and pungency to the food. The demand for chillies has increased recently because of their high nutraceutical compound capsaicin. Compared to the other vegetable crops, economic yield of chilli pepper plants are more adversely affected by biotic (Egea et al. 2002; Venkataiah et al. 2003; Suzuki and Mori 2003,) and abiotic stress (Ochoa-Alejo and Ramírez-Malagón 2001; Subramanyam et al. 2011). Transgenic approaches provide a powerful tool for gene function analysis in plants. However, chilli pepper plants are still recalcitrant to current transformation technologies, limiting the genetic improvement against biotic and abiotic stress (Kothari et al. 2010). To our knowledge only one report has been published on the metabolic engineering of salt tolerance in chilli pepper using an osmotin gene (Subramanyam et al. 2011). Hence, developing salt-tolerant chilli pepper plants is essential for sustaining food production.
In the present study, transgenic chilli pepper plants were generated by overexpressing the wheat Na+/H+ antiporter gene TaNHX2 via Agrobacterium-mediated transformation, and further, possible mechanisms of salt stress tolerance for chilli pepper plants were addressed by biochemical assays.
Materials and methods
Plant material
Seeds of C. annuum (Cv G4) were obtained from regional agricultural research station (RARS), Lam, Guntur, Andhra Pradesh, India. The seeds were surface-sterilized under aseptic conditions in a laminar flow chamber by immersion in an aqueous solution of 0.1 % (w/v) mercuric chloride (HgCl2) for 3–4 min followed by 4–5 rinses with autoclaved deionized water, and then blot dried. Surface-sterilized seeds were placed on Murashige and Skoog’s basal salt solution (Murashige and Skoog 1962) supplemented with 2 % sucrose and 0.8 % (w/v) agar (Himedia, India). The medium was adjusted to pH 5.7 ± 0.1, autoclaved at 120 °C and 1.1 kPa for 20 min, and allowed to cool before the addition of surface-sterilized seeds. The seeds were germinated in a growth room environment at 25 ± 2 °C with a light regime of 16 h and irradiance of 65 μmol m−2 s−1 provided by two fluorescent lamps (Philips, India, 110 W). Cotyledons of germinated seedlings (8–10 days old) without petiole and apical parts were cut into desirable size each of 1.0 cm2 and used as explants.
Transformation vector and bacterial strain
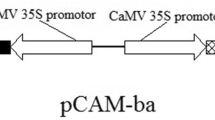
The plasmid pBin438 containing the TaNHX2 gene driven by a double Cauliflower Mosaic Virus (CaMV) 35S promoter (Yu et al. 2007) was kindly provided by Professor Shouyi Chen, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China. Agrobacterium tumefaciens strain LBA4404 harboring a binary vector pBin438-TaNHX2 was used for stable transformation of chilli pepper.
Plant transformation
Cotyledons from 10-day-old plants were used as explants for transformation. Explants were placed on pre-culture medium [PCM = MS Salts + 0.5 mg l−1 thidiazuron (TDZ) + 0.2 mg l−1 indole-3-acetic acid (IAA)] for 2 days and then infected with A. tumefaciens for 15 min at 28 °C with gentle shaking. Explants were blotted on sterile filter paper to remove excess bacterial solution before transfer to co-cultivation medium [CCM = MS salts + 0.5 mg l−1 TDZ + 0.2 mg l−1 IAA + 100 µM acetosyringone (AS)] and incubated in the dark at 25 ± 1 °C for 2 days. After co-cultivation, cotyledon explants were transferred to selective shoot induction medium (SSIM = MS Salts + 0.5 mg l−1 TDZ + 0.2 mg l−1 IAA + 80 mg l−1 kanamycin + 200 mg l−1 cefotaxime). For shoot proliferation and an initial selection, cotyledon explants were cultured for 3 weeks on regeneration medium (MS Salts + 0.5 mg l−1 TDZ + 0.2 mg l−1 IAA) containing 80 mg l−1 kanamycin and 250 mg l−1 cefotaxime. After 3 weeks of shoot proliferation, shoots (2–4 mm length) that had emerged from wounded ends of co-cultivated cotyledon explants were separated and transferred to selective shoot elongation medium (SSEM = MS Salts + 0.5 mg l−1 TDZ +0.2 mg l−1 IAA + 10 mg l−1 AgNO3 + 80 mg l−1 kanamycin + 200 mg l−1 cefotaxime). After 3–4 weeks, the elongated shoots were transferred onto shoot rooting media (SRM = ½ MS Salts + 40 mg l−1 kanamycin + 200 mg l−1 cefotaxime). The regenerated plantlets with well-developed shoots and roots were finally transferred to plastic pots filled with compost and then transferred to the greenhouse for seed production. The experiments were repeated at least three times and three replicates kept per experiment with approximately 225 explants for transformation studies. The data were statistically analyzed by analysis of variance (ANOVA, P < 0.05) followed by Duncan’s (Duncan 1955) multiple range test for mean comparison.
PCR confirmation of transgenic plants
For an initial selection of transformants, total genomic DNA was isolated from young leaves of putative transgenic and wild-type plants using a standard cetyl trimethylammonium bromide (CTAB) protocol, as described by Murray and Thompson (1980). PCR was performed by using a thermocycler (Biorad-C1000, Hercules, CA, USA). A primer set (forward 5′-ATGGGGTACCAAGTGGTGGC-3′ and reverse 5′-ATGAGAGGTAGGCCATGAGC-3′) was used to amplify a specific 800 bp of DNA sequence in transgenic plants corresponding to the TaNHX2 gene. For the detection of TaNHX2 gene, PCR conditions were set as initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 59 °C for 30 s, extension at 72 °C for 50 s, and final extension at 72 °C for 10 min. Plasmid DNA served as a positive control, and DNA from a non-transformed control plant was used as negative controls. Amplified PCR products were visualized and photographed using gel documentation system (Bio-rad; Gel docXR+) after electrophoresis on a 1 % (w/v) agarose (Seakem LE, Kolen, Germany) gel and detected by ethidium bromide staining.
Southern blot hybridization
Southern blot analysis was performed to verify the integration and copy number of the transgene. Genomic DNA (10 µg) from PCR positive and non-transformed plants was digested with HindIII. The digested DNA was separated by electrophoresis on a 0.8 % agarose gel and then blotted onto a nylon membrane (Hybond-N+, Amersham biosciences, UK). The membranes were pre-hybridized for 3 h using the membrane supplier’s protocol and hybridized at 65 °C. Following hybridization for at least 16 h, the filters were washed with a solution containing 2× SSC, 0.1 % SDS and then washed in 1× SSC, 0.1 % SDS at 65 °C. A 0.80-kb PCR product of the TaNHX2 gene labeled with digoxigenin (DIG) was used as a probe for DNA blot hybridization. After transfer to nylon membrane and hybridizing with DIG-labeled probe, the insertion copy number of the transgene was observed on autoradiography film.
Gene expression analysis by semiquantitative reverse transcription PCR
To detect the expression of transgenes by reverse transcription PCR (RT-PCR), total RNA was isolated from young leaves of T1 transgenic plants using the protocol described by Camacho-Villasana et al. (2002) and treated with DNase I (Takara, Dalian, China) to remove DNA traces. Total RNA (2 µg) was used as a template for the synthesis first-strand cDNA with M-MLV reverse transcriptase, RNase H Minus kit (Promega, Madison, WI, USA). PCR of the TaNHX2 gene was carried out according to the conditions described above. The house-keeping gene actin was used as an internal control to check the expression levels of transgenes. The primer sequences of actin were forward: 5′-CCACCACAGCCGAACGGG-3′ and reverse: 5′-ACCCGGGAACATGGTGGAACC-3′, which gives a 325-bp product with cDNA. The amplified products were electrophoresed on 2.0 % agarose gel.
Biochemical analysis of transgenic chilli pepper plants under salt stress
To examine the salt tolerance in vivo, we used a method described previously (Bhaskaran and Savithramma 2011) with slight modifications. Different transgenic lines and the wild type were planted in plastic pots (with holes at the bottom) containing Agra-Vermiculite (PULL Rhenen, 3911 TX Rhenen, the Netherlands), irrigated with 50 ml of Hoagland solution for 2 weeks in the greenhouse. The concentrations of NaCl supplementation were increased incrementally by 50 mM every 4 days for each group, to the final concentration of 50, 100, 150, and 200 mM, respectively. The stress treatment comprised of three plastic pots in two replicates. After salt treatment for 2 weeks, the plants were used for biochemical analysis.
Estimation of free proline content and H2O2 concentration
Proline content was estimated from the leaves of T1 plants after different concentrations of salt treatment independently along with the non-transformed plants by using the method of Lei et al. (2007). Hydrogen peroxide levels were determined according to the method described by Velikova et al. (2000). Five hundred milligrams of leaf tissues were homogenized in an ice bath with 5 ml 0.1 % (w/v) TCA. The homogenate was centrifuged at 12,000×g for 15 min. Fifty microliters of the extract was treated with 10 mM potassium phosphate buffer (pH 7.0) and 1 M KI. The absorbency of supernatant was read at 390 nm.
Determination of ascorbate peroxidase (APX), superoxide dismutase (SOD) activities, and malondialdehyde (MDA) concentration
After treatment with different concentrations of NaCl, APX activity was measured according to the protocol described by Chen and Asada, 1989. Leaf tissues (500 mg) were homogenized in an ice bath with 1.5 ml 10 mM sodium phosphate buffer (1 mM EDTA, 5 mM ascorbate). The homogenate (0.5 ml) was treated with the reaction mixture (50 mM phosphate buffer, pH 7.0, 0.5 mM ascorbate, 0.2 mM H2O2). The enzyme activity was calculated at absorbency of 260 nm. Superoxide dismutase (SOD, EC1.15.1.1) activity (Wang et al. 2012) and malondialdehyde (MDA) concentration were determined in transgenic and non-transformed chilli pepper plants, using the method described previously by Subramanyam et al. (2011).
Leaf disk assay
The leaf disk assay was performed using a method described previously (Bhaskaran and Savithramma 2011; Jha et al. 2013) with slight modifications to analyze transgenic plants for their salt tolerance conferred by the expression of the TaNHX2. Healthy and fully expanded chilli pepper leaves from WT and transgenic C. annuum plants (T1 generation) of similar age were detached. Leaf disks 5 mm in diameter were punched out and floated in 6 ml of half-strength Hoagland media with different concentrations of sodium chloride (0, 100, 150, and 200 mM) for 72 h. The leaf disks were kept under 16 h light/8 h dark at 25 ± 2 °C. The effects of this treatment on leaf disks were assessed by observing phenotypic changes. Leaf disks kept in half-strength Hoagland solution were taken as control.
Evaluation of relative water and chlorophyll contents
Relative water content (RWC) and chlorophyll content from non-transformed and three transgenic lines (Cap1, 3, 4) were measured after 2 weeks of salt treatment. RWC was measured by using the formula
where FW, fresh weight; DW, dry weight; TW, turgid weight (Yarra et al. 2012). The total chlorophyll content of the leaves was estimated according to the method described by Hiscox and Israelstam (1979). The experiment was repeated three times with three different transgenic lines (n = 3).
Determination of ion leakage
Ion leakage was measured based on the method of Fan et al. (1997). Leaves of transgenic and non-transgenic plants after salinity treatment were detached and washed in deionized water. Ion leakage was determined by measurement of electrolyte leaked from transgenic and non-transgenic plant leaves. Four leaves of each treatment (0, 50, 100, 150, and 200 mM NaCl) were immersed in 5–8 ml of 0.4 M mannitol at 23 °C with gentle shaking for 3 h, and the bathing solution was measured for conductivity. The total conductivity was determined after boiling the sample for 10 min. The conductivity due to leakage was expressed as the percentage of the initial conductivity versus the total conductivity.
Data analysis
The significance of the effect of the transgene under salt treatment was performed in Microsoft excel 2010 and analyzed by Student’s t test or ANOVA. All data were presented as mean ± standard deviation (SD) (n = 3).
Results
Selection and molecular confirmation of transgenic chilli pepper plants
The pBin438 vector with the TaNHX2 gene from wheat driven by a double CaMV 35S promoter (Fig. 1a) was introduced into C. annuum L. through Agrobacterium-mediated transformation (Figure S2). Ten-day-old cotyledons were pre-cultured on PCM (Table S1) for 2 days and then infected with Agrobacterium followed by 2 days of co-cultivation. When co-cultivated explants were cultured on selection medium (SSIM media, Table S1) for 3 weeks, multiple shoot buds (10–12) initiated (Fig. S1a, c, d) from the cut ends of cotyledon explants, whereas no shoot buds were observed from untransformed cotyledons (Fig. S1b). For initial elongation, the explants that survived on selection medium were transferred to MS media containing 0.5 mg l−1 TDZ + 0.2 mg l−1 IAA +10 mg l−1 AgNO3 + 80 mg l−1 kanamycin + 200 mg l−1 cefotaxime (SSEM medium, Table S1) for 2 weeks (Fig. S1e), subsequently transferred to fresh SSEM medium, and the number of shoots elongated was recorded in the next 30 days (Fig. S1f). The explants showed elongation with 5–7 shoots on average, and the shoots longer than 3 cm (Fig. S1g) were excised and subcultured on SRM medium (Table S1; Fig. S1h) for rooting. Based on the percent of co-cultivated cotyledon explants producing shoots on kanamycin medium, the average transformation efficiency of 18.17 % was achieved (Table S2). After 10–15 days on SRM medium, putative transgenic plantlets with well-developed roots were transferred to pots (Fig. S1i) and raised to maturity to obtain T1 seeds in greenhouse conditions.
Molecular confirmation of transgenic Chilli pepper (Capsicum annuum L.) plants. a Schematic map of plant expression gene construct pBIN438-TaNHX2. Expression of the TaNHX2 is driven by the double cauliflower mosaic virus 35S promoter. b PCR amplification of the TaNHX2 gene (800 bp) from the genomic DNA of T1 transgenic Lanes 1–4 (Cap-1–4) and Lane M is 1 Kb Molecular weight marker ladder; Lane W is wild-type plant (negative control), Lane P is pBIN438-TaNHX2 (positive control). c Analysis of transgenic chilli pepper plants (Lanes 1–4) by southern blot hybridization after digesting genomic DNA with HindIII and probing with digoxigenin labeled TaNHX2. d Reverse transcriptase PCR of T1 transgenic lines. Amplification was observed in three transgenic lines (Cap-1, Cap-3, and Cap-4) while there was no amplification from line 2 (Cap-2) and wild-type (W) plants; Actin gene was used as an internal control
Molecular analysis for the detection of genomic integration of the transgene (TaNHX2) was carried out on T1 transgenic plants. The PCR analysis of fragment of 800 bp with TaNHX2 gene primers was amplified from genomic DNA of four (Cap1–4) kanamycin-resistant transgenic lines, whereas corresponding bands were not detected in the non-transformed control plants with both the primers (Fig. 1b). Stable integration of the TaNHX2 gene into the genomic DNA was further confirmed by Southern blot analysis of transformed C. annuum plants. Among the four transgenic lines tested, two lines (Fig. 1c) showed single integration, and the other lines showed multiple integration. No probe hybridization was observed in the control plants (Fig. 1c). The expression of TaNHX2 in T1 transgenic plants was confirmed by RT-PCR analysis; a 800-bp amplified fragment, specifically amplified by the gene specific primers, confirmed the expression of TaNHX2 in three (Cap-1, Cap-3, and Cap-4) of the four Southern-positive T1 plants (Fig. 1d), whereas no expression was found in transgenic (Cap-2) and wild-type plants (Fig. 1d).
Overexpression of TaNHX2 in chilli pepper enhanced tolerance to the salt stress
To investigate the growth of TaNHX2-overexpressing lines under salt stress in soil, chilli pepper plants were irrigated with different concentration of Hoagland solution containing 0, 50, 100, 150, and 200 mM NaCl for 15 days. As shown in Fig. 2, under normal conditions, there was no obvious morphological differences between the wild-type and transgenic plants. However, during the treatment with increasing concentration of NaCl, the growth of wild-type plants was inhibited, and most of the plants were severely stunted at above 100 mM of NaCl (Fig. 2d), whereas the transgenic chilli pepper plants grew well and showed a normal phenotype at different concentrations of NaCl (Fig. 2a–c). These results indicate that the growth of chilli pepper plants overexpressing TaNHX2 under high salt concentrations showed increased tolerance than that of the wild-type plants.
Phenotypic differences of 2 months-old greenhouse grown wild-type and T1 transgenic plants expressing the TaNHX2 antiporter under different concentrations of salt stress. a–c T1 transgenic chilli pepper plants (Cap-1, Cap-3 and Cap-4) irrigated with different concentrations of NaCl. d Wild-type Chilli pepper plants irrigated with different concentrations of NaCl
Overexpressing TaNHX2 induces antioxidant systems and proline accumulation
To evaluate the oxidative stress induced by high salt concentrations, We performed physiological and biochemical assessments to determine the chlorophyll content, leaf relative water content, proline, H2O2 accumulation, and the activities of SOD, APX, and MDA contents in three transgenic (Cap-1, Cap-3, Cap-4) and wild-type plants (WT) under different salt stress conditions (0, 50, 100, 150, and 200 mM NaCl) (Fig. 3). We also performed leaf disk incubation assay in transgenic (Cap-3) and WT lines. The damage caused by stress was reflected in the degree of bleaching observed in the leaf disks after 72 h. Deterioration of the WT plant leaf disks was visually observed and correlated with a substantial loss of chlorophyll in comparison with transgenic line (Cap-3) (data not shown).
Biochemical analysis of wild-type and T1 transgenic plants grown under normal conditions and different salt treatments. a Proline content (µg g−1 of fw.). b Hydrogen peroxide concentration (µmol g−1 of fw.). c Malondialdehyde concentration (nmol g−1 of fw.). d Superoxide dismutase activity (Units mg−1 of protein min−1). e APX activity (µmol mg−1 of protein min−1). f Relative water content. g Chlorophyll content. h Ion leakage (% of conductivity). The graph represents mean ± SD value and asterisks indicate significant differences compared with WT values (*P < 0.05)
Proline is an important osmoprotectant that protects cells from damage under salt stress. As shown in Fig. 3a, the proline concentration was increased in transgenic lines when compared to wild-type chilli pepper plants. In presence of 150 and 200 mM salt concentration, proline content was significantly higher in TaNHX2-overexpressing lines (Cap-3 and Cap-4) than wild-type control plants (Fig. 3a).
Free radical formation and membrane damage levels were analyzed by observing the H2O2 and MDA content in transgenic and wild-type plants under different concentrations of NaCl. Salt stress caused a significant increase in the level of H2O2 in wild-type plants with an increase in NaCl concentration. In comparison with the wild-type plants, all transgenic lines exhibited a slow pace increase in H2O2 levels (Fig. 3b). MDA is considered as a marker of oxidative lipid injury, and its levels were widely used as a measure of damage due to various biotic and abiotic stresses. Based on Fig. 3c, under normal conditions, TaNHX2-overexpressing transgenic lines did not differ significantly from the wild-type plants. After salt treatment for 2 weeks, the MDA concentrations in TaNHX2-overexpressing lines were significantly decreased than wild-type plants (Fig. 3c). In comparison with the transgenic plants, the MDA concentrations were increased in wild-type plants, thereby increasing oxidative damage to membranes and reducing the resistance to salt stress. Our data suggested that TaNHX2 overexpression in chilli pepper plants reduces oxidative lipid injury and increases the better tolerance of transgenic chilli pepper plants grown under high salt concentrations.
The activities of the antioxidant enzymes APX and SOD were similar in transgenic and non-transgenic plants under optimal growth conditions. When subjected to salt stress, all transgenic lines exhibited significantly increased SOD and APX activities compared to the wild-type plants (Fig. 3d, e). As shown in Fig. 3f, under normal conditions, the RWC of all transgenic lines did not change significantly compared to the wild-type plants. However, when subjected to saline conditions, all transgenic lines exhibited an increased RWC. The percentage of RWC in the wild-type and transgenic plants were 37.4 and 80.2 %, respectively, at different stress conditions (Fig. 3f). We also estimated leaf chlorophyll content of both transgenic (Cap-1, Cap-3, Cap-4) and the wild-type plants under different salt stress conditions (50, 100, 150, 200 mM NaCl). The transgenic plants retained significant amount of chlorophyll content compared to the wild-type plants under salt stress (Fig. 3g).
Analysis of ion leakage
The ion content of wild-type and transgenic chilli pepper plants were analyzed. When plants were grown under normal growth conditions, the transgenic chilli pepper plants were morphologically similar to wild-type plants. However, after salt treatment the transgenic plants showed lower levels of ion leakage than that of wild-type plants (Fig. 3h). These data suggested that lesser damage has been occurred in the membrane system of transgenic chilli pepper plants expressing the TaNHX2 gene under salt stress.
Discussion
Progressive changing of global climatic conditions could lead to serious losses in agricultural productivity, as well as adversely affect biodiversity and global food security (Wheeler and von Braun 2013; Gill et al. 2014). Crop plants frequently encounter a variety of abiotic stresses among which salinity stress is one of the major cause of crop failure worldwide (Gill et al. 2014).On a world scale, no toxic substance restricts plant growth more than does salt. Almost three quarters of the earth surface is covered by salt water. Plants have created complex mechanisms, which help them in adapting to adverse environmental conditions. One such mechanism in higher plants is the sequestration of Na+ in the central vacuole by making use of putative Na+ (or) K+/H+ antiporters (Apse et al. 1999; Gaxiola et al. 1999).
Genetic engineering of pepper (Capsicum annuum L.) for abiotic stress tolerance is crucial for improving agricultural productivity is dependent upon the development of efficient and reliable regeneration and transformation protocols. Significant progress has been reported in the chilli pepper regeneration and transformation. However, regeneration of chilli pepper is highly genotypic dependent, and efficiency of Agrobacterium-mediated transformation has been limited in this system. (Wang et al. 1991; Ye et al. 1993; Christopher and Rajam 1997; Jayashankar et al. 1997; Subhash and Christopher 1997; Manoharan et al. 1998; Li et al. 2003; Lee et al. 2004). In this work, we addressed the use of a synthetic cytokinin (TDZ) which induced multiple shoots after 30 days of culture, but they did not elongate further. Previous studies reported the influence of silver nitrate (AgNO3) on multiple shoot production and elongation (Valera-Montero and Ochoa-Alejo 1992; Li et al. 2003). In the present study we added AgNO3 to the SSEM for multiple shoot production and elongation (Fig. S1e, f). For root induction transgenic chilli pepper plants were transferred to hormone-free media and we observed a better rooting on half-strength MS medium containing 40 mg l−1 kanamycin and 200 mg l−1 cefotaxime (Table S1; Fig. S1h). These results were consistent with the studies by Yarra et al. (2012). The highest transformation efficiency of 18.17 % was achieved by using this protocol (Table S2).
Overexpression of TaNHX2 gene was previously described in different plant species, including soybean, alfalfa, tomato and rice (Cao et al. 2011; Zhang et al. 2012; Yarra et al. 2012; Wu et al. 2012), However, very little is known about the role of TaNHX2 in salt stress tolerance.
In the present investigation, we generated transgenic TaNHX2 overexpressing chilli pepper plants (T1) that were sustained at high salt concentration. PCR and Southern blot analysis confirmed that TaNHX2 gene has been integrated and expressed in the T1 transgenic chilli pepper plants. RT-PCR revealed that three of the four transformed chilli pepper lines (Cap-1, Cap-3, and Cap-4) were expressing the TaNHX2, but the line 2 (Cap-2) did not show detectable levels of transcripts. This may be due to the low level or no expression of TaNHX2. Significant studies were reported with an inverse relation between transgene copy number and transgene expression level (Koprek et al. 2001; Kohli et al. 2003). In this study we used different TaNHX2-overexpressing lines (single and multiple insertion copies) for biochemical analyses The transgenic lines (Cap-4) expressing TaNHX2 accumulated a markedly superior level of proline, chlorophyll, SOD, ascorbate peroxide (APX), RWC in their leaves under severe saline conditions (200 mM NaCl) as compared with the multiple gene transgenic lines.
In this study, we showed that expression of the TaNHX2 gene in transgenic chilli pepper plants enhanced salinity stress tolerance. Biochemical assays of these transgenic plants revealed enhanced levels of proline, chlorophyll, RWC, SOD, and APX activities and reduced levels of H2O2, MDA contents, indicating that the TaNHX2 gene is playing an important role in the salt stress tolerance mechanism in chilli pepper plants.
Accumulation of osmolytes is the most common and important responses of plants, subjected to salt stress (Zhou et al. 2014). Among the different compatible solutes, proline has been proposed to act as an important compatible osmolyte and osmoprotective compound, and its accumulation has been reported in plants exposed to high salinity (Zhang et al. 2001; Subramanyam et al. 2011; Zhou et al. 2014). In the present investigation, the transgenic chilli pepper plants expressing TaNHX2 accumulated more proline than non-transgenic wild-type plants (Fig. 3a). Under high (150 and 200 mM) salt concentration, proline content was significantly higher in TaNHX2-overexpressing lines (Cap-3 and Cap-4) than wild-type control plants (Fig. 3a) suggesting that the expression of TaNHX2 might activate the key enzymes of the proline biosynthetic pathway (Bursy et al. 2007), whereas the increase in RWC and chlorophyll content in transgenic lines were higher than that of non-transgenic plants (Fig. 3f, g). Therefore, it has been explained that the TaNHX2 gene expression in transgenic chilli pepper plants improved the osmotic adjustment of cells by maintaining RWC and chlorophyll content to restore the growth of plants in response to damage caused by salinity stress. The results of the present study are in concurrence where the TaNHX2 gene expression in transgenic tomato (Yarra et al. 2012).
In plants, during salinity stress, the formation of reactive oxygen species (ROS) increases as by-products of various metabolic pathways (Mishra et al. 2011; Nounjan et al. 2012). To overcome salt-mediated oxidative stress, plants detoxify ROS by up-regulating antioxidative enzymes like SOD and APX. (Türkan and Demiral 2009; Dong et al. 2010; Mishra et al. 2011).
H2O2 is an important ROS, which causes apoptosis in plants (Houot et al. 2001). In the present study, the non-transgenic chilli pepper plants showed more concentration of H2O2 than transgenic chilli pepper plants at different NaCl concentrations (Fig. 3b). However, in comparison with the wild-type plants, salt stress caused a slow pace increase in H2O2 in transgenic chilli pepper plants (Fig. 3b). This slow increase in H2O2 level probably is responsible for the much better tolerance of transgenic chilli pepper plants grown under high salt concentration. The activities of SOD, APX, and the concentration of MDA are typically used to measure oxidative damage to membranes in response to salt stress (Mittova et al. 2004). In our study at the biochemical level, under different concentrations of NaCl the activities of SOD and APX were increased significantly (Fig. 3d, e), Under normal conditions, no TaNHX2-overexpressing transgenic lines showed a significantly altered MDA concentration, whereas under salt stress, TaNHX2-overexpressing transgenic lines showed increased levels of MDA but lesser than that of wild-type plants (Fig. 3c). According to these experimental data, the decrease in the concentrations of MDA at higher salt concentrations in TaNHX2-overexpressing transgenic lines is due to the high activity of SOD and APX. The obtained results were in agreement with earlier reports (Subramanyam et al. 2011; Zhou et al. 2014). The results suggest that TaNHX2 has been integrated in the genome of transgenic chilli pepper plants, thereby decreases oxidative damage to membranes and increase the resistance to salt stress.
In conclusion, the chilli pepper plants carrying the TaNHX2 gene showed elevated levels of proline and induced antioxidant enzyme systems, resulting in tolerance of higher concentration of NaCl. The obtained results pave the way to understand the role of the TaNHX2 gene under salt stress condition.
References
Apse M, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Bhaskaran S, Savithramma DL (2011) Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J Exp Bot 62:5561–5570
Blumwald E (2000) Sodium transport and salt tolerance in plants. Curr Opin Cell Biol 12:431–434
Bojórquez-Quintal E, Velarde-Buendía A, Ku-González Á, Carillo-Pech M, Ortega-Camacho D, Echevarría-Machado I, Igor P, Martínez-Estévez M (2014) Mechanisms of salt tolerance in habanero pepper plants (Capsicum chinense Jacq.): proline accumulation, ions dynamics and sodium root-shoot partition and compartmentation. Front Plant Sci 5(605):10–3389
Bursy J, Pierik AJ, Pica N, Bremer E (2007) Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J Biol Chem 282:31147–31155
Camacho-Villasana YM, Ochoa-Alejo N, Walling L, Bray EA (2002) An improved method for isolating RNA from dehydrated and nondehydrated chili pepper (Capsicum annuum L.) plant tissues. Plant Mol Biol Rep 20:407–414
Cao D, Hou W, Liu W, Yao WW, Wu C, Liu X, Han T (2011) Overexpression of TaNHX2 enhances salt tolerance of ‘composite ‘and whole transgenic soybean plants. Plant Cell Tissue Organ Cult 107:541–552
Chen GX, Asada K (1989) Ascorbate peroxidase in tea leaves: occurrence of two isozymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol 30(7):987–998
Chen M, Chen Q, Niu X, Zhang R, Lin H, Xu C, Wang X, Wang G, Chen J (2007) Expression of OsNHX1 gene in maize confers salt tolerance and promotes plant growth in the field. Plant Soil Environ 53(11):490
Christopher T, Rajam MV (1997) In vitro plant regeneration and Agrobacterium mediated transformation in red pepper. In Vitro 33:73A
Dong J, Wan G, Liang Z (2010) Accumulation of salicylic acid-induced phenolic compounds and raised activities of secondary metabolic and antioxidative enzymes in Salvia miltiorrhiza cell culture. J Biotechnol 148:99–104
Duncan DB (1955) Multiple range and multiple F tests. Biometrics 11:1–42
Egea C, Dickinson MJ, Candela M, Candela ME (2002) β-1,3-glucanase isoenzyme and genes in resistant and susceptible pepper (C. annuum) cultivars infected with Phytophthora capsici. Physiol Plant 107:312–318
Fan L, Zheng S, Wang X (1997) Antisense suppression of phospholipase D alpha retards abscisic acid-and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9:2183–2196
Flowers TJ, Yeo AR (1995) Breeding for salinity resistance in crop plants: where next? Aust J Plant Physiol 22:875–884
Fukuda A, Nakamura A, Tanaka Y (1999) Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim Biophys Acta 1446:149–155
Gaxiola RA, Rao R, Sherman A, Grisafi F, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNHX1 and AVP1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96:1480–1485
Gill SS, Ritu G, Renu T, Narendra T (2014) Genetic engineering of crops: a ray of hope for enhanced food security. Plant Signal Behav 9:e28545
Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Houot V, Etienne P, Petitot AS, Barbier S, Blein JP, Suty L (2001) Hydrogen peroxide induces programmed cell death features in cultured tobacco BY-2 cells, in a dose-dependent manner. J Exp Bot 52:1721–1730
Jayashankar S, Bagga S, Phillips GC (1997) Sweet pepper (Capsicum annuum) transformation using Agrobacterium rhizogenes. HortScience 32:454
Jha B, Mishra A, Jha A, Joshi M (2013) Developing transgenic Jatropha using the SbNHX1 gene from an extreme halophyte for cultivation in saline wasteland. PLoS ONE 8(8):e71136
Kohli A, Twyman RM, Abranches R, Wegel E, Stoger E, Christou P (2003) Transgene integration, organization and interaction in plants. Plant Mol Biol 52:247–258
Koprek T, Rangel S, Mcelroy D, Louwerse JD, Williams-carrier RE, Lemaux PG (2001) Transposon-mediated single-copy gene delivery leads to increased transgene expression in barley. Plant Physiol 125:1354–1362
Kothari SL, Joshi A, Kachhwaha S, Ochoa-Alejo N (2010) Chilli peppers—a review on tissue culture and transgenesis. Biotechnol Adv 28:35–48
Lee YH, Kim HS, Kim JY, Jung M, Park YS, Lee JS et al (2004) A new selection method for pepper transformation: callus-mediated shoot formation. Plant Cell Rep 23:50–58
Lei Y, Korpelainen H, Li C (2007) Physiological and biochemical responses to high Mn concentrations in two contrasting Populus cathayana populations. Chemosphere 68:686–694
Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, Cubero B, Fernández JA, Bressan RA, Hasegawa PM, Quintero FJ, Pardo JM (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J 61:495–506
Li D, Zhao K, Xie B, Zhang B, Luo K (2003) Establishment of a highly efficient transformation system for pepper (Capsicum annuum L.). Plant Cell Rep 21:785–788
Li J, Jiang G, Huang P, Ma J, Zhang F (2007) Overexpression of the Na+/H+ antiporter gene from Suaeda salsa confers cold and salt tolerance to transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult 90:41–48
Manoharan M, Sree Vidya CS, Lakshmi Sita G (1998) Agrobacterium-mediated genetic transformation in hot chilli (Capsicum annuum L. var. Pusa jwala). Plant Sci 131:77–83
Mishra P, Bhoomika K, Dubey RS (2011) Differential responses of antioxidative defense system to prolonged salinity stress in salt-tolerant and salt-sensitive Indica rice (Oryza sativa L.) seedlings. Protoplasma 250(1):3–19
Mittova V, Guy M, Tal M, Volokita M (2004) Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. J Exp Bot 55:1105–1113
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nounjan N, Nghia PT, Theerakulpisut P (2012) Exogenous proline and trehalose promote recovery of rice seedlings from salt-stress and differentially modulate antioxidant enzymes and expression of related genes. J Plant Physiol 169:596–604
Ochoa-Alejo N, Ramírez-Malagón R (2001) In vitro chilli pepper biotechnology. In Vitro Cell Dev Biol Plant 37:701–729
Quintero FJ, Blatt MR, Pardo JM (2000) Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett 471:224–228
Rengasamy P (2006) World salinization with emphasis on Australia. J Exp Bot 57:1017–1023
Subhash K, Christopher T (1997) Organogenesis and transformation in Capsicum baccatum. In Vitro 33:54A
Subramanyam K, Sailaja K V, Subramanyam K, Rao DM, Lakshmidevi K (2011) Ectopic expression of an osmotin gene leads to enhanced salt tolerance in transgenic chilli pepper (Capsicum annum L.). Plant Cell Tiss Org Cult 105(2):181-192
Suzuki K, Mori M (2003) Carotenoid composition of new cultivar of Capsicum annuum during maturation and its high capsanthin content. J Jpn Soc Food Sci Technol 50:324–326
Tang R, Li C, Xu K, Du Y, Xia T (2010) Isolation, functional characterization, and expression pattern of a vacuolar Na(+)/H(+) antiporter gene TrNHX1 from Trifolium repens L. Plant Mol Biol Rep 28:102–111
Türkan I, Demiral T (2009) Recent developments in understanding salinity tolerance. Environ Exp Bot 67(1):2–9
Valera-Montero L, Ochoa-Alejo N (1992) A novel approach for chili pepper (Capsicum annuum L.) plant regeneration: shoot induction in rooted hypocotyls. Plant Sci 84:215–219
Velikova V, Yordanov J, Edreva A (2000) Oxidative stress and some antioxidant systems in acid rain treated bean plants. Protective role of exogenous polyamines. Plant Sci 151:59–66
Venkataiah P, Christopher T, Subhash K (2003) Thiadiazuron induced high frequency adventitious shoot formation and plant regeneration in Capsicum annuum L. J Plant Biotechnol 5:245–250
Wang W, Yang M, Pan N, Chen ZH (1991) Plant regeneration and transformation of sweet pepper (Capsicum frutescens). Acta Bot Sin 33:780–786 (in Chinese)
Wang N, Hua H, EgrinyaEneji A, Li Z, Duan L, Tian X (2012) Genotypic variations in photosynthetic and physiological adjustment to potassium deficiency in cotton (Gossypium hirsutum L.). J Photochem Photobiol, B 110:1–8
Wheeler T, von Braun J (2013) Climate change impacts on global food security. Science 341:508–513
Wu CA, Yang GD, Meng QW, Zheng CC (2004) The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol 45:600–607
Wu LM, Chen W, Zhao Y, Feng SG, Ying QC, Liu JJ, Wang HZ (2012) Salt tolerance enhancement of transgenic rice with Na+/H+ antiporter gene driven by root specific promoter PmPgPR10. Chin J Rice Sci 26(6):643–650
Xia T, Apse MP, Aharon GS, Blumwald E (2002) Identification and characterization of a NaCl-inducible vacuolar Na+/H+ antiporter in Beta vulgaris. Physiol Plant 116:206–212
Yamaguchi T, Blumwald E (2005) Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci 10:615–620
Yarra R, He SJ, Abbagani S, Ma B, Bulle M, Zhang WK (2012) Overexpression of a wheat Na+/H+ antiporter gene (TaNHX2) enhances tolerance to salt stress in transgenic tomato plants (Solanum lycopersicum L.). Plant Cell Tissue Organ Cult 111(1):49–57
Ye Z, Li H, Zhang J, Jing Y (1993) Genetic transformation and plant regeneration in pepper. Acta Bot Sin 35:88–93 (in Chinese)
Yu JN, Huang J, Wang ZN, Zhang JS, Chen SY (2007) An Na+/H+ antiporter gene from wheat plays an important role in stress tolerance. J Biosci 32:1153–1161
Zhang H, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19:765–768
Zhang HX, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98:12832–12836
Zhang YM, Liu ZH, Wen ZY, Zhang HM, Yang F, Guo XL (2012) The vacuolar Na+–H+ antiport gene TaNHX2 confers salt tolerance on transgenic alfalfa (Medicago sativa). Funct Plant Biol 39(8):708–716
Zhou J, Jingjing W, Yufang B, Like W, Luozhong T, Xiang Yu, Misato O, Taku D, Qiang Z (2014) Overexpression of PtSOS2 enhances salt tolerance in transgenic poplars. Plant Mol Biol Rep 32:185–197
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Acknowledgments
This work was supported by University Grants Commission (UGC) under One-Time Research Grant scheme to SA, and the author MB is greatly thankful to Council of Scientific and Industrial Research (CSIR), India, for financial support in the form of CSIR-SRF Fellowship for his Doctoral studies. The authors thank Professor Shouyi Chen and Prof. Jinsong Zhang, Center for Genome Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China, for providing the plasmid pBin438-TaNHX2 and Dr. Wanke Zhang for technical assistance. The authors also thank the anonymous reviewers for their valuable comments and suggestions that had improved this manuscript greatly.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bulle, M., Yarra, R. & Abbagani, S. Enhanced salinity stress tolerance in transgenic chilli pepper (Capsicum annuum L.) plants overexpressing the wheat antiporter (TaNHX2) gene. Mol Breeding 36, 36 (2016). https://doi.org/10.1007/s11032-016-0451-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0451-5