Abstract
A protocol was developed for regeneration and Agrobacterium-mediated genetic transformation of Lesquerella fendleri. Calli were first induced from hypocotyls and cotyledons on MS plus 0.5 mg l−1 BA, 1 mg l−1 NAA and 1 mg l−1 2,4-D, then co-cultivated for 2–3 days in darkness on MS supplemented with 0.5 mg l−1 BA, 0.2 mg l−1 NAA and 100 μmol l−1As together with Agrobacterium tumefaciens strain EHA105/pCAMBIA1301 that harbored genes for uidA (GUS) and hygromycin resistance. Following co-cultivation, calli transfected by A. tumefaciens were transferred to MS with 0.5 mg l−1 BA, 0.2 mg l−1NAA, 500 mg l−1 Cef and 10 mg l−1 hygromycin and cultured for 10 days, then the hygromycin was increased to 20 mg l−1 on the same medium. After 4 weeks the resistant regenerants were transferred to MS with 0.5 mg l−1BA, 0.2 mg l−1 NAA, 500 mg l−1 Cef and 25 mg l−1 hygromycin for further selections. Transgenic plants were confirmed by polymerase chain reaction analysis, GUS histochemical assay and genomic Southern blot hybridization. With this approach, the average regeneration frequency from transfected calli was 22.70%, and the number of regenerated shoots per callus was 6–13. Overall results described in this study demonstrate that Agrobacterium-mediated transformation is a promising approach for improvement of this Lesquerella species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lesquerella is considered to be a promising new oil crop because it contains significant quantities of hydroxy fatty acid (HFA) that can be used by industry for the manufacture of resins, waxes, nylons, plastics, corrosion inhibitors, cosmetics, coatings, and lubricating grease (Thompson et al. 1989; Arquette and Brown 1993; Dierig et al. 1993). Within this genus, Lesquerella fendleri L. is a good candidate for domestication because it has the highest agronomic potential, low seed dormancy and low fruit dehiscence (Thompson and Dierig 1994; Ploschuk et al. 2003). L. fendleri seeds contain over 25% oil by weight where the primary fatty acid (55–60%) content is the C20:1 lesquerolic hydroxy fatty acid (http://www.nnfcc.co.uk/crops/info/lesquerella.pdf). Theoretically the oil and lesquerolic acid content in L. fendleri can be increased through genetic engineering. The prerequisite for a successful gene transfer is the establishment of an efficient transformation protocol. This paper reports the development of a genetic transformation protocol for this species using calli induced from cotyledons and hypocotyls as explants.
Materials and methods
Plant material and bacterial strains
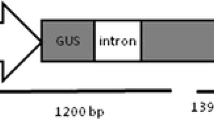
Seeds of Lesquerella fendleri L.were obtained from Prof. Dr. Sten Stymne, Swedish University of Agricultural Sciences, Alnarp, Sweden. The disarmed Agrobacterium tumefaciens strain EHA105 harboring binary vector pCAMBIA1301 (http://www.cambia.org/, Center for the Application of Molecular Biology to International Agriculture, Australia), which contains a uidA (gus) gene as the reporter gene and a hygromycin resistance gene (hygromycin phosphotransferase, Hpt) as the selectable marker, both driven by the cauliflower mosaic virus (CaMV) 35S promoter, was used for transformation studies. A. tumefaciens EHA105/pCAMBIA1301 was inoculated into modified liquid YEB medium (1 g l−1 Yeast extract, 5 g l−1 Beef extract, 5 g l−1 Peptone, 5 g l−1 Sucrose, 0.5 g l−1 MgSO4·7H2O, pH 7.2) containing 50 mg l−1 rifampicin and 50 mg l−1 kanamycin and incubated for 24 h at 28°C with reciprocal shaking (150 cycles min−1).
Callus induction and shoot regeneration
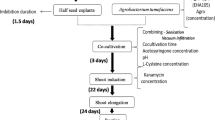
The seeds were surface sterilized for 15 min in 0.1% HgCl2, followed by three rinses in sterile distilled water. They were then germinated under aseptic conditions on MS basal medium (Murashige and Skoog 1962) with 30 g l−1 sucrose and 7 g l−1 agar (Sigma). Calli were first induced from cotyledons and hypocotyls of 7-day old seedlings on MS medium with different BA, NAA and 2,4-D combinations (Table 1), then transferred to MS medium containing 0.2 mg l−1NAA, 0.5 mg l−1 BA for regeneration. Regenerated shoots were transferred to basal MS for rooting. Above cultures were incubated at 25°C under a 16/8-h (day/night) photoperiod. Variance analyses on effects of BA, NAA and 2,4-D were carried out by running Orthogonal Design Assistant II 3.1.
Transformation and regeneration
Agrobacterium tumefaciens suspensions were pelleted by centrifugation at 2,000 rpm for 10 min and then re -suspended to an OD600 of 0.6 in liquid inoculation medium that consisted of MS medium supplemented with 3% sucrose. Each time 80 hypocotyls or cotyledons were incubated in 40 ml of A. tumefaciens inoculation medium for 5 min, then blotted on a sterile filter paper, and co-cultivated in darkness at 28°C with A. tumefaciens for 2–3 days on agar-solidified (0.7%) MS medium supplemented with 0.5 mg l−1 BA, 0.2 mg l−1 NAA and 100 μmol l−1As. Following co-cultivation, hypocotyls/cotyledons were rinsed three times with sterile distilled water containing 500 mg l−1 Cef, blotted dry on sterile filter paper, and cultured on selective regeneration medium (MS salts, 0.5 mg l−1 BA, 0.2 mg l−1 NAA) supplemented with 500 mg l−1 Cef and 10 mg l−1 hygromycin. Ten days later, hypocotyls and cotyledons were transferred to MS medium containing 0.5 mg l−1 BA, 0.2 mg l−1 NAA and 20 mg l−1 hygromycin for subsequent regeneration. Regenerated adventitious shoots under this selection pressure were transferred to the same medium with higher concentrations of hygromycin (25 mg l−1) for further selections.
Meanwhile, calli were first induced from cotyledons and hypocotyls on MS plus 0.5 mg l−1 BA, 1 mg l−1 NAA and 1 mg l−1 2,4-D, then co-cultivated with A. tumefaciens and selected as described above. After several rounds of selections at 25 mg l−1 hygromycin the regenerated shoots were transferred to MS basal medium containing 25 mg l−1 hygromycin for rooting and then moved to the soil.
PCR analysis
To check for the presence of transgenes in putative transformants, PCR analysis for the uidA gene was performed using genomic DNAs isolated from hygromycin-resistant and untransformed L. fendleri leaves as templates. Plasmid DNA of pCAMBIA 1301 was used as the positive control. The forward and reverse primers used for the uidA gene were 5′ACCCCAACCCGTGAAATCAAAAAACTC 3′ and 5′CCCGCTTCGAAACCAATGCCTAAA3′, respectively. The PCR reaction mixture (10 μl) contained 1 μl of 10 × PCR buffer, 1 μl MgCl2 of 25 mM, 0.2 μl dNTPs of 0.5 mM, 0.2 μl forward primer of 5 μM, 0.2 μl reverse primer of 5 μM, 0.1 μl Taq DNA Polymerase, 0.5 μl template, and 6.8 μl ddH2O. The enzyme and primers were from Invitrogen Company (Shanghai, China). The PCR reaction profile included 35 cycles of strand separation at 94°C for 20 s, annealing at 54°C for 25 s and extension at 72°C for 1 min. The program was extended for 8 min at 72°C. The amplification products were analyzed on 0.8% agarose-goldview (SBS Gene Company, Shanghai, China) gels.
GUS histochemical assay
A histochemical GUS assay was performed using the method of Jefferson (1987). The assay solution contained 0.5 mM potassium ferrocyanide, 0.3% (v/v) Triton X-100, and 1 mg/ml 5-bromo-4-chloro-3-indolyl-D-glucuronide (X-gluc) in 50 mM phosphate buffer, pH 7.0. Leaflets randomly collected from regenerated hygromycin-resistant and non-transformed control plants were placed in micro-centrifuge tubes containing about 0.8 ml staining solution. The tubes were incubated for 2 h at 37°C. The GUS-staining was observed after extracting chlorophyll with 70% ethanol.
Southern blot analysis
The integration of foreign genes in host genome was determined by Southern blot analysis (Holtke et al. 1995). After RNase treatment, the genomic DNA was digested with Hind III (NEB) and separated by electrophoresis (0.8% [w/v] agarose). Size-fractionated DNA was transferred to Hybond N+ membrane (Amersham Pharmacia Biotech, Piscataway, NJ) by standard Southern-blot techniques (Sambrook and Russell 2002). DIG High labeled probe was made using the PCR-amplified uidA coding sequence and with a digoxigenin (DIG)-11-dUTP using DIG High Prime DNA labeling reagents (Roche, Mannheim, Germany). Hybridization, washing, and detection were performed according to the instruction manual of the DIG High Prime DNA labeling and detection starter kit II (Roche). Hybridization signals were detected by exposing the membrane to an X-ray film at 15–25°C for 15–25 min.
Results
Callus induction and shoot regeneration
Our results indicated that calli could be induced from cotyledons and hypocotyls of L. fendleri in about 20 days after inoculation onto MS with different hormone combinations (Table 1 and Fig. 1a,b). Direct comparison indicated that BA was the most important factor for callus induction from hypocotyls and cotyledons. Variance analysis indicated that BA had significant influence on the callus induction from hypocotyls but it did not influence the callus induction from cotyledons significantly. Variance analysis also indicated that neither NAA nor 2,4-D influenced the callus induction significantly, but this conclusion needs to be confirmed by more experiments. Most calli induced from media listed in Table 1 could regenerate after transferred to the regeneration medium, but the regeneration speed varied. Some hypocotyls and cotyledons regenerated directly into adventitious shoots when the induction medium contained no 2, 4-D while calli induced on medium with 1 mg l−1 BA, 0.2 mg l−1 NAA, 2 mg l−1 2,4-D could not be regenerated. Regenerated shoots were rapidly amplified after transferred onto fresh medium containing 0.5 mg l−1 BA and 0.2 mg l−1 NAA (Fig. 1c). Roots were induced in 4 weeks when regenerated shoots were transferred to MS basal medium (Fig. 1e).
Transformation and recovery of resistant shoots
Adventitious shoots were formed from the hypocotyls and cotyledons in about one month after co-cultivation with A. tumefaciens and some of them survived after transferred to fresh medium with higher concentrations of hygromycin (25 mg l−1). However subsequent PCR gave no positive amplification.
Under the selection pressure of 20 mg l−1 hygromycin, some A. tumefaciens-transfected calli swelled and developed into adventitious shoots within 6 weeks (Fig. 1d). The average regeneration frequency from transfected calli was 22.70%, and the number of regenerated shoots per callus was 6–13. During the process of selection, subculturing explants onto fresh medium containing 25 mg l−1 hygromycin greatly reduced the number of escapes. Some of the shoots that were initially green grew yellow, leaving some continued to grow vigorously. After several rounds of selections shoots of about 3–4 cm in length were rooted on MS basal medium containing 25 mg l−1 hygromycin. All shoots subsequently formed roots on this medium in 4 weeks (Fig. 1e) and more than 80% of the rooted plantlets survived after transferred to the soil (Fig. 1f).
PCR analysis of regenerated plants for uidA gene
All hygromycin-resistant plants exhibited amplification of 1.08 kb band corresponding to the coding region of the uidA gene, indicating the presence of transgene in the transformed plants (Fig. 2).
Histochemical GUS assay
Histochemical staining revealed that leaves of putative transgenic plants were strongly positive for GUS activity (Fig. 3a), while leave samples from non-transformed plantlets did not stain blue (Fig. 3b). Since the GUS construct in EHA105/pCAMBIA1301 used in the present study contained an intron (Hajdukiewicz et al. 1994), the observed expression should not be coming from bacterial contamination.
DNA blotting analysis
Southern blot analysis confirmed the integration of the T-DNA region in the transformed plant genome (Fig. 4). Usually the transgenic plants contained one copy of uidA gene (Fig. 4, lane 4, 6, 7, 8, 10), some plants contained two copies (Fig. 4, lane 3, 5, 9) and one plant contained probably three copies of uidA gene (Fig. 4, lane11). No hybridization was detected in the non-transformed control plants (Fig. 4, lane2).
Southern hybridization analysis of non-transformed and transformed plantlets harboring uidA gene. Lane 1. pCAMBIA1301; lane 2. untransformed L. fendleri; lane 3–11. transformed L. fendleri; Genomic DNA from each line was digested with HindIII and probed with the uidA gene fragment. The bands represent the transgene insertions. The migration of DNA size markers is shown on the left in kb
Discussion
In this study we established a simple and reliable procedure for A. tumefaciens-mediated transformation of L. fendleri using calli as explants and this system should facilitate the improvement of this species. It was reported that L. fendleri had a high capacity for plant regeneration from cultured cells (Skarzhinskaya et al. 1996) and a plastid transformation protocol was established for this species (Skarzhinskaia et al. 2003). In the present study it was shown that adventitious shoots could be induced directly from hypocotyls and cotyledons when the induction media contained no 2,4-D. Considering the calli induction frequency and regeneration speed, we chose MS supplemented with 0.5 mg l−1 BA, 1 mg l−1 NAA and 1 mg l−1 2,4-D as the callus induction medium and MS supplemented with 0.5 mg l−1 BA and 0.2 mg l−1 NAA as the shoot regeneration medium in the transformation experiment.
Lesquerella fendleri calli were chosen as explants for the A. tumefaciens-mediated transformation because the regenerated shoots from transfected hypocotyls/cotyledons did not give positive PCR amplification. The calli were first induced without hygromycin, then co-cultivated with A. tumefaciens, then transferred onto medium containing hygromycin for shoot regeneration. This method is sometimes called two-step method and is often used for plant transformation (Li et al. 2006a, b). Our explanation is that calli are much looser than hypocotyls and cotyledons and the calli cells have more contact surface with A. tumefaciens during the co-cultivation, and thus makes transformation easier and decreases the possibility of escapes. According to our experience the possibility of chimeric regenerants might not be ruled out, but the percentage of un-transformed cells can be minimized by repeated selections and by increasing the selection pressure.
Abbreviations
- MS:
-
Murashige and Skoog medium
- NAA:
-
α-Napthaleneacetic acid
- BA:
-
6-Benzylaminopurine
- 2,4-D:
-
2,4-Dichlorophenoxy
- GUS:
-
β-Glucuronidase
- CaMV:
-
Cauliflower mosaic virus
- Hpt:
-
Hygromycin phosphotransferase
- X-Gluc:
-
5-Bromo-4-chloro-3-indolyl-b-d-glucoronide
- PCR:
-
Polymerase chain reaction
- As:
-
Acetosyringone
- Rif:
-
Rifampicin
- Cef:
-
Cefotaxime
References
Arquette JG, Brown JH (1993) Development of a cosmetic grade oil from Lesquerella fendleri L.seed. In: Janick J, Simon JE (eds) New crops. Wiley, New York, pp 367–371
Dierig DA, Thompson AE, Nakayama FS (1993) Lesquerella commercialization efforts in the United States. Ind Crops Prod 1:289–293
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989–994
Holtke HJ, Ankenbauer W, Muhlegger K, Rein R, Sagner G, Seibl R, Walter T (1995) The digoxigenin (DIG) system for non–radioactive labeling and detection of nucleic acids—an overview. Cell Mol Biol 41:883–905
Li D, Sun Q, Huang M, Zhang J, Bai S, Zheng L, Zhao J, Qiu D, Li L, Yang Z, You M, Liu G, Zhang Y, Zhang C, Li S (2006a) Agrobacterium–mediated genetic transformation of Elymus breviaristatus with Pseudomonas pseudoalcaligenes insecticidal protein gene. Plant Cell Tiss Organ Cult DOI: 10.1007/s11240-007-9229-0
Li DX, Zhang J, Zhao J, Zhang Y, Li L, Liu SJ, Chen F, Yang ZR (2006b) Transformation of calli of siberian wildrye grass (Elymus sibiricus L. cv. chuancao No.2) mediated by Agrobacterium. J Plant Physiol Mol Biol 32(1):45–51
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Ploschuk EL, Cerdeiras G, Windauer L, Dierig DA, Ravetta DA (2003) Development of alternative Lesquerella species in Patagonia (Argentina): potential of L. angustifolia. Ind Crops Prod 18:1–6
Sambrook J, Russell DW (2002) Molecular cloning: a laboratory manual, 3rd edn. Science Press, Beijing (in Chinese)
Skarzhinskaya M, Landgren M, Glimelius K (1996) Production of intertribal somatic hybrids between Brassica napus L. and Lesquerella fendleri L.(Gray) Wats. Theor Appl Genet 93:1242–1250
Skarjinskaia M, Svab Z, Maliga P (2003) Plastid transformation in Lesquerella fendleri L., an oilseed Brassicacea. Transgenic Res 12:115–22
Thompson AE, Dierig DA, Johnson ER (1989) Yield potential of Lesquerella fendleri L. (Gray) Wats., a new desert plant resource for hydroxy fatty acids. J Arid Environ 16:331–336
Thompson AE, Dierig DA (1994) Initial selection and breeding of Lesquerella fendleri L., a new industrial oil seed. Ind Crops Prod 2:91–106
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, W., Wang, C., Huang, BL. et al. Agrobacterium tumefaciens-mediated transformation of Lesquerella fendleri L., a potential new oil crop with rich lesquerolic acid. Plant Cell Tiss Organ Cult 92, 165–171 (2008). https://doi.org/10.1007/s11240-007-9319-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-007-9319-z