Abstract
Genetic transformation of an elite white poplar genotype (Populus alba L., cv. ‘Villafranca’) was performed with MAT vectors carrying the ipt and rol genes from Agrobacterium spp. as morphological markers. The effects associated with the use of different gene promoters and distinct in vitro regeneration protocols were evaluated. Poplar plantlets showing abnormal ipt and rol phenotypes were produced only in the presence of exogenous growth regulators. The occurrence of abnormal ipt and rol phenotypes allowed the visual selection of transformants. The ipt-type MAT vector pEXM2 was used to monitor the activity of the yeast site-specific recombination R/RS system in the transformed white poplar cells. Results from these experiments demonstrated that recombinase-mediated excision events occurred during the early stages of in vitro culture, thus causing the direct production of ipt marker-free transgenic plants with normal phenotype at an estimated frequency of 36.4%. Beside this unexpected finding, transgenic ipt-shooty plants were obtained at a frequency of 63.6% and normal shoots were subsequently recovered after a prolonged period of in vitro culture. Although the transformation efficiency observed in this study, using both ipt and nptII genes as selection markers, was similar to that previously reported with standard vectors carrying only the nptII gene, the easy identification of ipt transformants, the early recombinase-mediated excision events and finally the relatively short time period required to produce ipt marker-free transgenic plants support for the choice of MAT vectors as a reliable strategy for the future production of marker-free GM poplars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Crop improvement based on genetic transformation has led to the production of genetically modified (GM) plants expressing genes of interest associated with antibiotic or herbicide resistance genes, used as selection markers. The latter, necessary for the isolation of transgenic plants, are no longer needed in mature plants. The persistence of resistance genes in the GM plants is undesirable (Hohn et al. 2001; Goldstein et al. 2005).
Different approaches for the removal of selectable marker genes from GM plants have been described and sexual crosses have been traditionally used to segregate out the marker gene from the gene of interest (Hohn et al. 2001). In the MAT (Multi-Auto-Transformation) system, developed by Ebinuma and coworkers (Ebinuma et al. 1997; Sugita et al. 1999; Ebinuma and Komamine 2001), the ipt and rol oncogenes from Agrobacterium tumefaciens and Agrobacterium rhizogenes, respectively, are used as morphological markers due to their ability to induce abnormal phenotypes. The oncogenes are then removed from GM plants, using the yeast site-specific recombination system R/RS (Araki et al. 1987) and consequently marker-free transgenic plants can be easily regenerated. To date, the MAT vectors have been utilized to transform a limited number of annual species, such as Nicotiana tabacum, Antirrhinum majus, Oryza sativa and Nierembergia caerulea (Sugita et al. 1999; Cui et al. 2000, 2001; Sugita et al. 2000; Ebinuma and Komamine 2001; Endo et al. 2002; Khan et al. 2006). Among perennial plants, only the hybrid aspen (Populus sieboldii × P. grandidentata) and sweet orange (Citrus sinensis L.) have been tested with the ipt-type MAT system (Ebinuma and Komamine 2001; Matsunaga et al. 2002; Ballester et al. 2006). For all these systems, transformation/regeneration protocols were carried out in the absence of both selective chemical agents and hormones and resulted in the production of ipt or rol marker-free transgenic plants, which still harboured the nptII, hpt, uidA or GFP sequences located on the T-DNA region, outside the excised cassette. Except for rice, in all other species engineered with the MAT vector system, transformants were firstly selected based on the presence of the ipt-shooty or rol phenotype and, subsequently, only the phenotypically normal transgenic plants emerging from abnormal shoots were isolated (two-step transformation). However, the two-step procedure is laborious since it requires several in vitro subcultures and a prolonged period for the production of marker-free transgenic plants. For this reason, any further improvement of the methodology, saving time and work efforts, is strongly recommended. In this context, the direct regeneration of transgenic plants lacking their morphological marker, ipt or rol, as a consequence of early excision events (single-step transformation) could represent a reliable strategy for a more effective use of MAT vectors. Using an ipt-type MAT vector, the direct regeneration of ipt marker-free transgenic rice plants with normal phenotype from scutellum tissues was described by Endo et al. (2002). This single-step transformation method represents a useful transformation approach for those plant species that rely on somatic embryogenesis for in vitro regeneration. In the case of rice cells, it has been suggested that the overproduction of cytokinin associated with the overexpression of ipt gene causes the induction of embryogenic tissues and that, following excision, a change in the cytokinin level could determine the direct regeneration of shoots from embryogenic tissues (Endo et al. 2002).
Among woody species, white poplar (Populus alba L.) is considered a good biomass producer (about 9 oven dry t ha−1 year−1), due to its remarkable resprouting ability after coppicing (Confalonieri et al. 2000). GM white poplar plants from the elite clonal cultivar ‘Villafranca’, expressing selectable marker genes (Balestrazzi et al. 2000) and relevant traits such as herbicide tolerance (Confalonieri et al. 2000), insect pest resistance (Delledonne et al. 2001; Balestrazzi et al. 2006), production of antioxidant compounds (Giorcelli et al. 2004), phytoremediation (Zelasco et al. 2004) and biomass production (Zelasco et al. 2006) have been obtained.
In this study we evaluate the efficacy of the MAT vector system to transform the white poplar (Populus alba L.) elite clonal cultivar ‘Villafranca’. Both ipt and rol genes were used as morphological markers for the visual selection of transformants. We also investigated the effects of different gene promoters and regeneration protocols on the efficiency of white poplar transformation. Moreover, we tested the effectiveness of the yeast R/RS system in removing the morphological marker ipt gene from the transgenic white poplars, using an ipt-type MAT vector. Finally, an efficient short-term and cost-effective transformation/regeneration procedure producing ipt marker-free transgenic plants is reported. Our results clearly demonstrate that MAT vectors can be considered a useful tool for the production of marker free transgenic plants in those agronomically relevant species which can be reproduced only by vegetative propagation
Materials and methods
Plant material, plasmids and A. tumefaciens strain
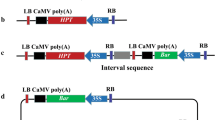
The clonal cultivar ‘Villafranca’ (Populus alba L.) was obtained by controlled crossing in 1954 at the Poplar Research Institute (Casale Monferrato, Italy) and registered for commercial use in Italy and Hungary. Aseptic shoot cultures of ‘Villafranca’ were maintained and propagated in vitro as previously described (Confalonieri et al. 2000). All the constructs used in this work, prepared and supplied by the Nippon Papers Industries (Tokyo, Japan), are derivatives of the pBI121 binary vector (Ebinuma and Komamine 2001). A schematic representation of these constructs is described in Fig. 1. The binary control vectors pIPT5, pIPT10 and pIPT20 contain the ipt gene associated with the 35SCaMV, native ipt and rbcS3B promoters, respectively (Fig. 1A). Plasmid pROL20 is a rol-type vector carrying the rolABC genes and their native promoters (Fig. 1A). The excision monitor control vector pEXM2 contains a ‘hit and run’ cassette, composed of the native ipt gene and the R gene, flanked by two recognition sites (RS) (Fig. 1B). In this construct the coding region of the reporter gene uidA and the associated 35SCaMV promoter are separated by one of the recognition sites. The recombinase-mediated excision of the ‘hit and run’ cassette leads to the inactivation of the uidA gene. In this model system, the nptII marker gene has been inserted into the multiple cloning site located outside the ‘hit and run’ cassette (Fig. 1B). All the above described constructs were transferred to the A. tumefaciens strain EHA105, a non oncogenic derivative of strain A281 which harbours the hypervirulent helper Ti plasmid pTiBo542 (Hood et al. 1993).
Schematic representation of the binary control plasmids pIPT5, pIPT10, pIPT20 and pROL20 (A) and the ipt-type MAT vector pEXM2 (B) RB, right border; LB, left border; 35S-p, cauliflower mosaic virus 35S promoter; nptII, neomycin phosphotransferase-coding sequence; T, nopaline synthase gene terminator; p, promoter; uidA, β-glucuronidase-coding sequence; RS, recognition site; R, recombinase-coding sequence; ipt, isopentenyl transferase gene; ipt-p and ipt-T, promoter and terminator regions of the ipt gene; rolA, rolB and rolC, Agrobacterium rhizogenes genes from Ri plasmid with their associated promoter and terminator regions; RbcS, ribulose-1,5-biphosphate carboxylase small subunit; EXM, excision monitor. Gene-specific oligonucleotide primers are indicated by bars. The black arrow indicates the region of the pBI121 vector in which the different cassettes represented as 35S-p-ipt-T, ipt-p-ipt-ipt-T, rbcS-p-ipt-T and RolA-RolB-RolC have been inserted to produce the pIPT5, pIPT10, pIPT20 and pROL20 constructs. Transcription orientation for each cassette of the pEXM2 vector is indicated by grey arrows. The position of the nptII hybridization probe is also indicated
A. tumefaciens-mediated transformation and in vitro selection/regeneration
Agrobacterium tumefaciens-mediated transformation and in vitro shoot regeneration of Populus alba L. cv. ‘Villafranca’ was performed using two different protocols. The first procedure was described by Confalonieri et al. (2000). After co-cultivation and a callus induction step, shoot regeneration was achieved in the presence of the cytokinin analog thidiazuron (TDZ) and benzyl-aminopurine (BAP). In the second procedure, internodal explants were co-cultivated as described (Confalonieri et al. 2000) but placed and subcultured on a modified (3/4 macrosalts) MS medium (Murashige and Skoog 1962) without growth regulators. Three replications of 50 internodal stem explants each were used for all the tested vectors. Transformation efficiency was defined as the frequency of stem explants which produced kanamycin-resistant (KmR) calli or shoots. Data were arcsin√x transformed before statistical analysis.
Molecular analyses
Plant DNA for PCR analysis was extracted as described by Rogers and Bendich (1988) and subsequently purified using the GFXTM PCR DNA and Gel Band Purification kit (Amersham Biosciences). PCR was carried out as described by Confalonieri et al. (1994) in a Perkin-Elmer Thermo-Cycler 480 and amplification products were analysed on 1.4% (w/v) agarose gels (Duchefa Biochemicals). The following conditions were used to amplify the 599 bp fragment region of nptII gene: 35 cycles at 94°C, 2 min, 65°C, 1 min, 72°C, 1 min. The gene-specific oligonucleotide primers were K-1 (5′-AGGCTATTCGGCTATGACTGG-3′) and K-2 (5′-GCGGTCCGCCACACCCAGCCG-3′), respectively. Oligonucleotide primers IPT-1 (5′-CTTGCACAGGAAAGACGTCG-3′) and IPT-2 (5′-AATGAAGACAGGTGTGACGC-3′) were used to amplify the 800 bp fragment corresponding to the ipt gene with the following conditions: 35 cycles at 94°C, 50 s, 56°C, 50 s, 72°C, 1 min. Oligonucleotide primers REC-1 (5′-TCTGGGACGCCTCGGGAACTTC-3′) and REC-2 (5′-TGTCCCAGAGACTAAGACTGGTAC-3′) were used to detect the 666 bp fragment corresponding to the coding region of the R gene at the following conditions: 35 cycles at 94°C, 50 s, 68°C, 50 s, 72°C, 1 min. Primers RS1-A (5′-CAGCGCATCGCCTTCTATCG-3′) and RS1-B (5′-GATGTCTTTCCTGTGCAAG-3′) were used to amplify the 1.08 kb fragment spanning 42 bp of the nptII region, 255 bp of the Nos terminator, the RS site (58 bp) and 736 bp of the native ipt promoter. Primers RS2-A (5′-GTGGTCCCAAAGATGGACC-3′) and RS2-B (5′-TGCATCGGCGAACTGATC-3′) were then used to amplify the 422 bp fragment spanning 188 bp of the 35SCaMV region, the RS site (58 bp) and 176 bp of the uidA gene. In each case amplification was performed at the following conditions: 94°C, 50 s, 58°C, 50 s, 72°C, 1 min (RS1-A and RS1-B), 94°C, 50 s, 56°C, 50 s, 72°C, 50 s (RS2-A and RS2-B). When the R/RS system is activated, the absence of these amplification products is expected while, in the presence of the ‘hit and run’ cassette, both the 1.08 kb and the 422 bp fragments are produced. For the detection of the rolC gene sequence, primers ROL-1 (5′-AAATGCGAAGTAGGCGCTCCG-3′) and ROL-2 (5′-TACGTCGACTGCCCGACGATGATG-3′) were used and amplification of the predicted 0.2 kb fragment was obtained at 94°C, 50 s, 60°C, 50 s, 72°C 1 min. Oligonucleotide primers uidA-1 (5′-TATGCGGGCAACGTCTGG-3′) and uidA-2 (5′-TCACGCGGCTATCAGCTCTT-3′) were designed to amplify the 1.0 kb fragment from the uidA coding region at the following conditions: 94°C, 50 s, 54°C, 50 s, 72°C 2 min. For Southern blot hybridization analysis, plant genomic DNAs (10 μg) were digested with the indicated restriction enzymes (M-Medical S.r.l.), separated on 0.4% (w/v) agarose gels and transferred to a nylon membrane (HybondTM N+, Amersham Biosciences) according to manufacturer’s instructions. The nptII fragment (599 bp) was used as probe and labelled with [α-32P]-dCTP using the Hexa Label PlusTM DNA labelling kit (M-Medical S.r.l.). Filters were hybridized as described by Sambrook et al. (1989).
Results
Role of ipt and rol oncogenes as morphological markers for the selection of transformation events in Populus alba L. cv. ‘Villafranca’.
Transformation experiments were carried out to evaluate the role of the ipt and rolABC oncogenes as morphological markers in P. alba L. cv. ‘Villafranca’. The effects of ipt gene expression controlled by different promoters and regeneration protocols were also investigated. KmR calli were obtained only in the presence of the cytokinin analogs TDZ and BAP, using the procedure described by Confalonieri et al. (2000). The frequency of stem internodes that produced KmR calli was 2%, 8% and 14%, respectively for the pIPT5, pIPT10 and pIPT20 constructs (data not shown). As expected, the frequency of stem internodes from the untransformed control line that produced calli was 100%. After several subcultures, a reduced number of KmR calli developed shoots with either normal or abnormal phenotype and the plant transformation efficiency ranged from 1 to 5% (data not shown). Analysis of variance revealed no significant differences (P = 0.54) in plant transformation efficiency among different constructs. All the ipt-shooty lines showed a reduced apical dominance, did not root on a hormone-free medium and new abnormal shoots and calli were continuously produced at contact sites with the agar medium (Fig. 2A). Only the abnormal ipt-shooty phenotype was observed among plants regenerated following transformation with the pIPT5 and pIPT10 constructs. Using the pIPT20 construct, both KmR plants with a typical ipt-shooty phenotype and KmR plants with normal phenotype were regenerated. The phenotypically normal shoots grew and rooted on kanamycin-containing medium lacking growth regulators. The frequency of stem explants producing phenotypically normal plants was 1% while the pIPT20 ipt-shooty lines were regenerated at a higher frequency (5%). Stem internodal explants co-cultivated with EHA105 carrying pROL20 produced KmR plants with normal and rol phenotype with a similar frequency (2–3%). The presence of hairy roots is a typical feature of the rol phenotype (Fig. 2B). In this study we demonstrated that the ipt and rolABC genes can be used as visually selectable markers for the transformation of white poplar, and produced abnormal phenotypes clearly distinguishable from the untransformed control plant lines (Fig. 2C).
Phenotypes deriving from the overexpression of the ipt and rolABC oncogenes in P. alba cv. ‘Villafranca’. (A) Poplar shoot regenerated following transformation by A. tumefaciens EHA105 carrying pIPT20 and showing a typical ipt-shooty phenotype with absence of both apical dominance and rooting ability and callus production at the contact sites with the agar medium (indicated by arrow). Bar: 0.5 cm. (B) Poplar plantlet regenerated following transformation by EHA105 carrying pROL20 and showing a typical rol phenotype. Root proliferation is evident. Hairy roots are indicated by arrow. Bar: 0.5 cm. (C) Non transformed poplar plantlet with normal phenotype. Bar: 1 cm
Molecular analysis of the ipt and rol poplar lines
In order to confirm the presence of transgene sequences, four different putative transgenic lines, regenerated following co-cultivation with EHA105 carrying pIPT20, and two lines obtained following co-cultivation with EHA105 carrying pROL20, were analysed by PCR. Results from these experiments are summarized in Fig. 3. The predicted nptII fragment (599 bp) was amplified in all the tested pIPT20 lines (Fig. 3A, lanes 3–6), except for the non transformed control line (Fig. 3A, lane 2). Oligonucleotide primers IPT-1 and IPT-2 revealed the presence of the predicted ipt fragment (800 bp) (Fig. 3B, lanes 3–6). No amplified products were evidenced using DNA extracted from the non transformed plant (Fig. 3B, lane 2). The positive controls obtained using pIPT20 plasmid as template showed the expected amplification products (Fig. 3A, lane 1 and Fig. 3B, lane 1, respectively). In the case of pROL20, the presence of the nptII sequence was demonstrated in both the tested lines (Fig. 3C, lanes 3 and 4) while the same fragment was not amplified in the non transformed control line (Fig. 3C, lane 2). The rolC sequence was also detected in the same poplar lines (Fig. 3D, lanes 3 and 4) while no amplification products were observed in the non transformed control line (Fig. 3D, lane 2). The positive controls obtained using pROL20 as template are shown in Fig. 3C (lane 1) and Fig. 3D (lane 1), respectively. The same transgenic lines were further analysed by Southern hybridization to confirm the stable integration of the nptII sequence into the poplar genome (Fig. 3E). Restriction analysis of poplar DNA performed with HindIII was expected to produce a DNA fragment (2.0 kb) spanning NosP-nptII-NosT region of the T-DNA and a variable portion corresponding to the flanking plant chromosomal region. Hence each hybridization band should represent a single copy of the transgene sequence. As expected, no hybridization signal was evident in the non transformed control line (Fig. 3E, lane 1). The nptII probe recognized a single fragment of approximately 10.0 kb in one of the tested pIPT20 lines (Fig. 3E, lane 2). Single hybridization signals ranging between 7.0 and 8.0 kb were evident in the other poplar lines (Fig. 3E, lanes 3–5). An hybridization signal of approximately 9.0 kb was observed for one of the pROL20 lines (Fig. 3E, lane 6). A smaller band of approximately 3.0 kb was observed for the second pROL20 line (Fig. 3E, lane 7).
Molecular analyses of the pIPT20 and pROL20 lines. (A) Detection of nptII transgene sequence in the pIPT20 lines. Control plasmid (pIPT20) and non transformed line are shown (lane 1 and 2). PCR analysis of DNAs extracted from four putative transgenic lines of P. alba cv. ‘Villafranca’ regenerated following co-cultivation with EHA105 carrying pIPT20 (lanes 3–6). (B) Detection of ipt transgene sequence in the pIPT20 lines. Control plasmid (pIPT20) and non transformed line are shown (lane 1 and 2). PCR analysis of the putative transgenic pIPT20 poplar lines (lanes 3–6). (C) Detection of nptII transgene sequence in the pROL20 lines. Control plasmid (pROL20) and non transformed line are shown (lane 1 and 2). PCR analysis of two putative transgenic lines obtained following co-cultivation with EHA105 carrying pROL20 (lanes 3 and 4). (D) Detection of the rolC transgene sequence in the pROL20 lines. Control plasmid (pROL20) and non transformed line are shown (lane 1 and 2). PCR analysis of the putative transgenic pROL20 poplar lines (lanes 3 and 4). M, Gene RulerTM 100 bp DNA Ladder (M-Medical S.r.l.). (E) Southern blot analysis of the pIPT20 and pROL20 lines. Lane 1, non transformed control line; lanes 2–5, pIPT20; lanes 6 and 7, pROL20. DNAs were digested with HindIII and probed using the [α-32P]-dCTP-labelled 599 bp nptII fragment
Activation of the site-specific recombination system R/RS in white poplar cells transformed with the pEXM2 vector
To evaluate the efficacy in white poplar of the R/RS system for site-specific recombination associated with the expression of the ipt gene as morphological marker, a transformation experiment was performed with EHA105 carrying pEXM2. Eighteen independent pEXM2 KmR callus lines were recovered. Approximately three-four months after co-cultivation, both normal and abnormal shoots were regenerated. Eleven KmR plant lines, corresponding to a transformation frequency of 7.3%, continued to grow on kanamycin-containing medium lacking growth regulators. Four of them (2.6%) were characterized by the presence of shoots with normal phenotype and rooting (single-step transformation) while the remaining seven (4.6%) exhibited the typical ipt-shooty phenotype. The ipt-shooty lines were, as expected, unable to form roots (Fig. 4A). Four months later three ipt-shooty lines were able to develop normal shoots. The occurrence of new shoots with normal morphology from an ipt-shooty line is represented in Fig. 4B. The other ipt-shooty lines continued showing their abnormal phenotype.
(A) Three different pEXM2 lines showing the typical ipt-shooty phenotype. The presence of callus tissue at the contact sites with the agar medium is indicated by arrows. Bar: 1 cm. (B) New normal shoots elongating from an ipt-shooty plantlet are indicated by arrows. Bar: 0.5 cm. (C) Molecular analysis of genomic DNA from three different ipt-shooty lines regenerated following transformation of P. alba cv. ‘Villafranca’ with EHA105 carrying pEXM2. The nptII transgene sequence was evidenced by PCR analysis using K-1 and K-2 oligonucleotide primers (nptII, lanes 5–7). The ipt transgene sequence was revealed using IPT-1 and IPT-2 oligonucleotide primers (ipt, lanes 8–10). (D) The sequence corresponding to the coding region of the R gene from yeast was evidenced using the oligonucleotide primers REC-1 and REC-2 (lanes 3–5). (E) The presence of the “hit and run” cassette was demonstrated using oligonucleotide primers RS1-A and RS1-B (RS1) and oligonucleotide primers RS2-A and RS2-B (RS2) (lanes 5–7 and 8–10, respectively). All the oligonucleotide sets were used with the pEXM2 plasmid (positive control, C–E, lane 1) and with the non transformed line (C–E, lane 2). M, Gene RulerTM 100 bp DNA Ladder (M-Medical S.r.l.). (F) Southern blot analysis of the three different ipt-shooty lines regenerated following transformation of P. alba cv. ‘Villafranca’ with EHA105 carrying pEXM2. Lane 1, non transformed control line; lanes 2–4, ipt-shooty. DNAs were digested with HindIII/EcoRI and probed using the [α-32P]-dCTP-labelled 599 bp nptII fragment
Molecular analysis of the pEXM2 lines
All the ipt-shooty and the phenotypically normal pEXM2 lines were tested by PCR. Results from this analysis are summarized in Figs. 4 and 5, respectively. When PCR analysis was performed on five selected ipt-shooty pEXM2 lines, the predicted nptII and ipt fragments were amplified. Results from molecular analyses performed on three representative ipt-shooty pEXM2 lines are shown in Fig. 4C, D, E and F. The presence of both nptII and ipt fragments was evidenced in the pEXM2 lines (Fig. 4C, nptII, lanes 5–7; ipt, lanes 8–10). No amplification products were obtained with the non transformed control (Fig. 4C, lanes 3 and 4). The pEXM2 plasmid was used as positive control (Fig. 4C, lanes 1 and 2). The same pEXM2 lines were tested for the presence of the R gene sequence encoding the yeast recombinase. As shown in Fig. 4D (lanes 3–5), PCR performed on genomic DNAs with gene-specific oligonucleotide primers allowed amplification of the 666 bp fragment corresponding to the R gene coding region. Positive and negative controls are also reported (Fig. 4D, lanes 1 and 2, respectively). In addition, the two different amplification products corresponding to the flanking regions of the RS sites were detected using specific oligonucleotide sets (Fig. 4E, RS1, lanes 5–7; RS2, lanes 8–10). Positive and negative controls are visible in Fig. 4E (lanes 1–4). Southern hybridization was carried out on the same ipt-shooty pEXM2 lines (Fig. 4F). In this case DNA samples underwent the double digestion HindIII/EcoRI since the use of the single HindIII digestion revealed identical hybridization patterns for all the tested lines (not shown). As expected no hybridization signal was evident in the non transformed control line (Fig. 4F, lane 1). The nptII probe recognized three different DNA fragments ranging between 8.0 and 20 kb in one of the tested pEXM2 lines (Fig. 4F, lanes 2 and 3). Two hybridization signals of approximately 9.0 and 18.0 kb were evident in the second poplar line (Fig. 4F, lane 3). An hybridization band of approximately 8.0 kb was detected in the third line (Fig. 4F, lane 4). Similarly, all the tested pEXM2 lines with normal shoots were found to contain the predicted 599 bp nptII fragment (Fig. 5B, nptII, lanes 3–6). The expected nptII fragment was not amplified in the non transformed control line (Fig. 5B, nptII, lane 1). A positive control (pEXM2 vector) was also tested (Fig. 5B, nptII, lane 2). The 800 bp ipt fragment was not amplified in the same lines, thus indicating the excision of the ‘hit and run’ cassette and the negative results obtained using the RS2-A and RS2-B oligonucleotide primer sets seem to further confirm that the excision had occurred (data not shown). When PCR was carried out using oligonucleotide primers designed on the uidA coding sequence, the expected 1.0 kb fragment located outside the ‘hit and run’ cassette was detected (Fig. 5D, lanes 3–6). Positive and negative controls are also shown (Fig. 5B nptII, lanes 1 and 2; uidA, lanes 1 and 2). Results from PCR analysis confirmed that all the phenotypically normal pEXM2 lines regenerated in this transformation experiment were ipt marker-free transgenic poplar plants deriving from the early excision of the ‘hit and run’ cassette and the consequent removal of the morphological marker ipt gene.
Production of ipt marker-free transgenic poplar plants by ‘single-step’ transformation. (A) Phenotypically normal poplar plantlet resulting from the early excision of the ipt gene caused by the 35SCaMV-driven recombinase activity. Bar: 1 cm. (B) Molecular analysis of genomic DNA from phenotypically normal shoots regenerated following transformation of P. alba cv. ‘Villafranca’ with EHA105 carrying pEXM2. The nptII transgene sequence was evidenced by PCR analysis using K-1 and K-2 oligonucleotide primers (nptII, lanes 3–6). The uidA sequence was detected using the gene-specific oligonucleotides uidA-1 and uidA-2 (uidA, lanes 3–6). All the oligonucleotide primer sets were used with the pEXM2 plasmid (positive control, lane 1) and with the non transformed control line (lane 2). M, Gene RulerTM 100 bp DNA Ladder (M-Medical S.r.l.). (C) Southern blot analysis of the four different pEXM2 lines with normal phenotype (lanes 2–5). Non transformed control line (lane 1). DNAs were digested with HindIII and probed using the [α-32P]-dCTP-labelled 599 bp nptII fragment
The presence and stable integration of the nptII sequence was subsequently confirmed by Southern blot hybridization analysis using the gene-specific probe (Fig. 5C). HindIII digestion of genomic DNAs from the non transformed control line and the four marker-free transgenic pEXM2 lines was carried out. As expected, no hybridization bands were found in the genomic DNA from the non transformed control. Hybridization fragments of approximately 15–18 kb were detected in two different lines (Fig. 5C, lanes 2 and 3) while a stronger signal of approximately 6.0 kb was detected in the other two lines (Fig. 5C, lane 4 and 5).
Discussion
The efficacy of the morphological marker genes ipt and rolABC from Agrobacterium spp. and the activity of the yeast site-specific recombination R/RS system carried by an ipt-type MAT vector were evaluated using an elite Populus alba genotype. The typical morphological changes (lack of apical dominance and rooting ability) induced by the overexpression of the ipt oncogene were observed in the transgenic white poplar lines obtained following transformation with the pIPT5, pIPT10 and pIPT20 constructs. These constructs were already tested in hybrid aspen by Ebinuma and Komamine (2001). These authors reported that the use of pIPT20 construct resulted into regeneration of transgenic shoots at the highest frequency. Differently from this report, no statistically significant differences in transformation efficiency were observed in our experimental conditions. The ipt oncogene from A. tumefaciens is considered a regeneration-promoting factor due to its ability to stimulate plant regeneration from explants by the organogenic pathway (Zuo et al. 2002). Overexpression of the ipt gene produces high cytokinin levels and consequently the direct formation of shoots from the organogenic cells. In our experimental conditions, the co-cultivated poplar internodal explants regenerated shoots only in the presence of growth regulators. These results suggest that the amount of cytokinins produced by the activity of the bacterial isopentenyltransferase could not be sufficient for stimulating the organogenesis pathway in the white poplar cells. Another possible explanation might be related to the response of ‘Villafranca’ to genetic transformation with bacterial oncogenes. Balestrazzi and coworkers (2000) reported the occurrence of tumors when the same genotype was inoculated with A281 and 82.139 wild type A. tumefaciens strains. When excised tumors were cultivated in the absence of growth regulators, shoots with normal phenotype were regenerated. Based on this evidence, it might be hypothesized that additional functions, provided by other oncogene sequences contained in the T-DNA, such as the iaaH and iaaM genes involved in auxin biosynthesis, may be required to promote shoot differentiation in ‘Villafranca’. Von Schwartzemberg et al. (1994) reported on poplar transformation with the ipt gene, using the clone 717-1B4 (Populus tremula × P. alba) with the aim to provide a model system for the study of cytokinin metabolism. Although the poplar genotype and the callus induction protocol differed from our experimental conditions, similarly, regeneration of ipt-shooty plant lines was achieved in the presence of the cytokinin TDZ.
The rolABC genes from the Ri T-DNA of A. rhizogenes have been also used as morphological markers due to their ability to induce hairy roots. The rol genes have been already proved to modify the architecture of transgenic poplars as reported by Nilsson et al. (1996). Transgenic aspen engineered with the CaMV35S-rolC construct and showing both reduced size and small leaves were described by Fladung et al. (1997). The expression of the rolABC coding sequences in transgenic aspen was associated with reduced apical dominance and breaking of axillary shoot buds (Tzfira et al. 1998). Moreover, since the expression of rolC in aspen produced light-green coloured leaves, this phenotypic feature was used to test in forest trees the recombination events mediated by the transposable element Ac from maize (Fladung et al. 1997).
The rol-type MAT vector system has been tested in N. tabacum (Ebinuma and Komamine 2001) and A. majus (Cui et al. 2000, 2001). These studies proved that the rolABC genes can be used as a selection marker for Agrobacterium-mediated transformation. The rolABC genes, introduced for the first time into white poplar genotype using the pROL20 construct, were able to induce the typical abnormal phenotype usually associated with their expression in planta (Fig. 2B). Plantlets were characterized by the presence of abundant hairy roots on stems. Moreover, transgenic plants with bushy phenotype were obtained (data not shown). Similar observations were already reported by Nilsson et al. (1996) who transformed P. tremula × P. tremuloides with a different construct carrying only the rolC sequence. Although kanamycin was used in our experiments as selection agent during callus induction and regeneration phases, the morphological changes induced in white poplar by ipt and rolABC oncogenes proved the feasibility of the MAT vector system as a tool for the gene transfer and the visual detection of transformants.
The efficacy of the yeast site-specific recombination R/RS system in removing the marker gene ipt in white poplar cells was evaluated using the control vector pEXM2. When the pEXM2 construct was used to transform the ‘Villafranca’ cultivar, the frequency of co-cultivated explants that produced KmR plants, was 7.2%. This value was similar to the average transformation efficiency (7–11%) already reported for ‘Villafranca’ (Balestrazzi et al. 2000; Delledonne et al. 2001), achieved using only the nptII gene as selectable marker. Endo et al. (2002) has shown that the effectiveness of the nptII gene as selection marker was enhanced when it was co-transformed with the ipt gene. Recent work from Ballester et al. (2006) showed that ipt overexpression in Citrus sinensis greatly enhanced transformation efficiency, although they used a different transformation MAT vector. Our results did not confirm these findings. Among the eleven pEXM2 plant lines, a fraction corresponding to 63.6% showed the expected ipt-shooty phenotype. The latter was evident at approximately three-four months from the beginning of the experiment, while an additional four-month period was necessary to see the appearance of normal shoots from the ipt-shooty poplar plantlets (Fig. 4B). Our data demonstrated that the yeast recombinase system R/RS was active in removal of ipt marker gene from transgenic poplar cells and produced the intended results. Similar observations have been recently reported by Khan et al. (2006) and Ballester et al. (2006) who produced ipt marker-free transgenic Nierembergia caerulea and Citrus sinensis plants, respectively. More interestingly, the use of the pEXM2 construct generated KmR lines with normal phenotype (36.4%) which were obtained only three-four months after the beginning of the transformation experiment. The production of ipt marker-free transgenic plants without the recovery of ipt-shooty intermediates (single-step transformation) was already reported in rice by Endo et al. (2002), using a different ipt-type MAT vector. Activation of the 35SCaMV promoter during the early developmental stage has been documented in plant tissues (Ohta et al. 1990; Vancanneyt et al. 1990) and this might explain the results obtained in the white poplar system. In this study we have used a pEXM2 control vector in which the nptII gene is located outside the ‘hit and run’ cassette and the regeneration of ipt marker-free shoots was achieved by means of exogenous cytokinins in the presence of kanamycin as selective agent. These experimental conditions could not represent the best way to generate marker-free transgenic poplars, however they resulted into useful information concerning the effectiveness of the in vitro procedure required when testing MAT vectors.
In conclusion, our study illustrates the feasibility of ipt and rolABC genes as efficient positive morphological markers for the visual selection of transformants in an elite white poplar genotype. Furthermore, this is the first demonstration of the efficacy of the R/RS system in promoting efficiently the early excision of the ipt marker gene in a woody plant species, with the consequent rapid production of ipt marker-free transgenic plants. The occurrence of single-step transformation events is also described, suggesting for possible future improvements in the production of marker-free GM poplars. However, further experimental work will be necessary to acquire detailed information on the temporal mechanism involved in the precocious activation of yeast recombinase and to assess the practicality and efficiency of the single-step phenomenon.
Abbreviations
- BAP:
-
Benzyl-aminopurine
- NAA:
-
α-Naphthaleneacetic acid
- TDZ:
-
Thidiazuron
References
Araki H, Jearnpipatkula A, Tatsumi H, Sakurai T, Ushino K, Muta T, Oshima Y (1987) Molecular and functional organization of yeast plasmid pSR1. J Mol Biol 182:191–203
Balestrazzi A, Carbonera D, Confalonieri M (2000) Agrobacterium tumefaciens-mediated transformation of elite white poplar (Populus alba L.) and regeneration of transgenic plants. J Genet Breed 54:263–267
Balestrazzi A, Allegro G., Confalonieri M (2006) Genetically modified trees expressing genes for insect pest resistance. In: Fladung M, Ewald D (eds) Tree transgenesis: recent developments. Springer-Verlag Berlin Heidelberg, pp 253–273
Ballester A, Cervera M, Pena L (2006) Efficient production of transgenic citrus plants using isopentenyl transferase positive selection and removal of the marker gene by site-specific recombination. Plant Cell Rep, DOI 10.1007/s00299-006-0197-3
Confalonieri M, Balestrazzi A, Bisoffi S (1994) Genetic transformation of Populus nigra by Agrobacterium tumefaciens. Plant Cell Rep 13:256–261
Confalonieri M, Belenghi B, Balestrazzi A, Negri S, Facciotto G, Schenone G, Delledonne M (2000) Transformation of elite white poplar (Populus alba L.) cv. ‘Villafranca’ and evaluation of herbicide resistance. Plant Cell Rep 19:978–982
Cui M, Takayanagi K, Kamada H, Nishimura S, Handa T (2000) Transformation of Antirrhinum majus L. by a rol-type multi-auto-transformation (MAT) vector system. Plant Sci 159:273–280
Cui M, Takayanagi K, Kamada H, Nishimura S, Handa T (2001) Efficient shoot regeneration from hairy roots of Antirrhinum majus L. transformed by the rol-type MAT vector system. Plant Cell Rep 20:55–59
Delledonne M, Allegro G, Belenghi B, Balestrazzi A, Picco F, Levine A, Zelasco S, Calligari P, Confalonieri M (2001) Transformation of white poplar (Populus alba L.) with a novel Arabidopsis thaliana cysteine proteinase inhibitor and analysis of insect pest resistance. Mol Breed 7:35–42
Ebinuma H, Sugita K, Matsunaga E, Yamakado M (1997) Selection of marker-free transgenic plants using the isopenthenyl transferase gene. Proc Natl Acad Sci USA 94:2117–2121
Ebinuma H, Komamine A (2001) MAT (Multi-Auto-Transformation) vector system. The oncogenes of Agrobacterium as positive markers for regeneration and selection of marker-free transgenic plants. In Vitro Cell Dev Biol-Plant 37:103–113
Endo S, Sugita K, Sakai M, Tanaka H, Ebinuma H (2002) Single-step transformation for generating marker-free transgenic rice using the ipt-type MAT vector system. Plant J 30:115–122
Fladung M, Kumar S, Ahuja MR (1997) Genetic transformation of Populus genotypes with different chimeric gene constructs: transformation efficiency and molecular analysis. Trans Res 6:111–121
Giorcelli A, Sparvoli F, Mattivi F, Balestrazzi A, Tava A, Vrhovsek U, Calligari P, Bollini R, Confalonieri M (2004) Expression of the stilbene synthase (StSy) gene from grapevine in transgenic white poplar results in high accumulation of the antioxidant compounds resveratrol glucosides. Trans Res 13:203–214
Goldstein DA, Tinland B, Gilbertson LA, Staub JM, Bannon GA, Goodman RE, Mc Coy RL, Silvanovich A (2005). Human safety and genetically modified plants: a review of antibiotic resistance markers and future transformation selection technologies. J Appl Microbiol 99:7–23
Hohn B, Levy A, Putcha H (2001) Elimination of selection markers from transgenic plants. Curr Opin Biotech 12:139–143
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacteria helper plasmids for gene transfer to plants. Trans Res 2:208–218
Khan RS, Chin DP, Nakamura I, Mii M (2006) Production of marker-free transgenic Nierembergia caerulea using MAT vector system, Plant Cell Rep, DOI 10.1007/s00299-006-0125-6
Matsunaga E, Sugita K, Ebinuma H (2002) Asexual production of selectable-marker free transgenic woody plants, vegetatively propagated species. Mol Breed 10:95–106
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:73–97
Nilsson O, Moritz T, Sundberg B, Sandberg G, Olsson O (1996) Expression of the Agrobacterium rhizogenes rolC gene in a deciduous forest tree alters growth and development and leads to stem fasciation. Plant Physiol 112:493–502
Otha S, Mita S, Hattori T, Nakamura K (1990) Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 3:805–813
Rogers SO, Bendich AJ (1988) Extraction of DNA from plant tissues. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual, A6. Kluwer, Dordrecht, pp 1–10
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Sugita K, Matsunaga E, Ebinuma H (1999) Effective selection system for generating marker-free transgenic plants independent of sexual crossing. Plant Cell Rep 18:941–947
Sugita K, Matsunaga E, Kasahara T, Ebinuma M. (2000) Transgene stacking in plants in the absence of sexual crossing. Mol Breed 6:529–536
Tzfira T, Zuker A, Altman A (1998) Forest-tree biotechnology: genetic transformation and its application to future forests. Trends Biotech 16:439–446
Vancanneyet G, Schmidt R, O’Connor-Sanchez A, Willmitzer L, Rocha-Sosa M (1990) Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet 220:245–250
Von Schwartzenberg K, Doumas P, Jouanin L, Pilate G (1994) Enhancement of the endogenous cytokinin concentration in poplar by transformation with Agrobacterium T-DNA gene ipt. Tree Physiol 14:27–35
Zelasco S, Fogher C, Calligari P, Savazzini F, Bisoffi S, Pietra S, Bolchi A, Scaramelli L, Ottonello S, Confalonieri M (2004) Expression of an artificial metallothionein gene in transgenic white poplar and evaluation of cadmium tolerance. Proceedings of SIFV—SIGA Joint Congress. SIFV XLIII Annual Congress—SIGA XLVIII Annual Congress. Lecce, 15–18 September 2004, p. 223
Zelasco S, Reggi S, Calligari P, Balestrazzi A, Bongiorni C, Quattrini E, Delia G, Bisoffi S, Fogher C, Confalonieri M (2006) Expression of the Vitreoscilla hemoglobin (VHb)-encoding gene in transgenic white poplar: plant growth and biomass production, biochemical characterization and cell survival under submergence, oxidative and nitrosative stress conditions. Mol Breed 17:201–216
Zuo J, Niu Q-W, Ikeda Y, Chua N-H (2002) Marker-free transformation: increasing transformation frequency by the use of regeneration-promoting genes. Curr Opin Biotech 13:173–180
Acknowledgments
This research was supported by a grant from ‘Ministero dell’Università e della Ricerca Scientifica’ (COFIN 2003) and by a grant from Fondazione Branca-Bussolera. S. Z. and V. R. were awarded by a Research Fellowship from Fondazione Branca-Bussolera, Mairano di Casteggio (Italy). We would like to thank Tiziano Collot for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zelasco, S., Ressegotti, V., Confalonieri, M. et al. Evaluation of MAT-vector system in white poplar (Populus alba L.) and production of ipt marker-free transgenic plants by ‘single-step transformation’. Plant Cell Tiss Organ Cult 91, 61–72 (2007). https://doi.org/10.1007/s11240-007-9278-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-007-9278-4