Abstract
During a parasitological survey, Myxidium-like spores were identified in the gall bladders of marine fishes from Australian waters. This paper describes four novel species of Ellipsomyxa Køie, 2003, three novel species of Myxidium Bütschli, 1882 and six novel species of Zschokkella Auerbach, 1910 from teleosts from Australian waters using a combination of morphological, biological and molecular characters. Phylogenetic analyses showed a monophyletic relationship of all Ellipsomyxa spp. sequences with Sigmomyxa sphaerica (Thélohan, 1895) and Myxidium queenslandicus Gunter & Adlard, 2008 as sister species to the clade. The validity of genus Sigmomyxa Karlsbakk & Køie, 2012 is discussed. In phylogenetic analyses, the novel species of Myxidium fell within the ‘marine’ clade of Fiala (2006). However, the novel species of Zschokkella fell within the ‘freshwater’ clade of Fiala (2006) and formed a distinct clade with all other sequences of Zschokkella spp. from the gall bladder of marine fish and a sequence of a species of Myxobolus Bütschli 1882, also from the gall bladder of a marine fish. This is the second distinct marine lineage to emerge within the freshwater clade.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Myxidium Bütschli, 1882 was originally proposed to house Myxidium lieberkuehni Bütschli, 1882 taken from the urinary tract of the freshwater fish Esox lucius Linnaeus (see Bütschli, 1882). The currently accepted generic diagnosis, as defined by Lom & Dyková (2006), characterises species with “spores generally fusiform, straight or slightly crescent or even sigmoid, with more or less pointed ends. Shell valves smooth or with ridges, suture line bisects the spore. Two polar capsules, mostly pyriform, lie one at each end of the spore. Capsular foramina are situated in the sutural plane, at or near the end of the spore and open as a rule in opposite directions. The binucleate sporoplasm lies between the capsules. The species are typically coelozoic (rarely histozoic), forming small or large trophozoites, mono-, di- or polysporic, the latter with pansporoblast formation”. From the current diagnosis, several characters including fusiform spores stand out as unique to the genus. Until recently all species displaying sigmoid spores were also assigned to Myxidium. However, Karlsbakk & Køie (2012) recently established a new genus, Sigmomyxa Karlsbakk & Køie, 2012, characterised by spores that are spindle-shaped in valvular view and sigmoid in sutural view. Species of Sigmomyxa can be distinguished from Myxidium spp. by the ellipsoidal outline of spore valves and thin-walled protrusions associated with the polar capsule (PC) tips. Although no longer a characteristic unique to Myxidium, polar capsules situated one at each end of the spore that open as a rule in opposite directions are, when combined with spore shape, useful characters for generic placement.
All other characters used to place species within Myxidium are ambiguous at best. Mostly pyriform PCs, PCs opening at or near the end of the spore, suture line bisecting the spore and spores with more or less pointed ends, are all poorly defined characters and are open to individual interpretation. In particular, PC shape becomes confusing when PCs are neither exactly pyriform nor exactly spherical. Similarly, a suture line that bisects the spore may be straight, curved or sinuous (as described for other genera). Also, the degree of point at the ends of spores is difficult to define given the variability in spore shape. The remaining characters [i.e. two PCs, one located at each end of the spore, smooth or ridged spore valves, binucleate sporoplasm, typically coelozoic (rarely histozoic), forming small or large trophozoites, mono-, di- or polysporic, the latter with pansporoblast formation], are not generically informative as all are diagnostic of other genera assigned to the family Myxidiidae Thélohan, 1892 (see Lom & Dyková, 2006; Prunescu et al., 2007; Hartigan et al., 2012; Karlsbakk & Køie, 2012).
Genus Zschokkella Auerbach, 1910 was originally proposed to house Zschokkella hildae Auerbach, 1910 identified from the urinary bladder of three species of Gadiformes on the basis of certain morphological differences to those displayed by species of Myxidium (see Auerbach, 1910). Auerbach (1910) defined genus Zschokkella as having “spores from the side with some semicircular edges extended, at both ends a large circular polar capsule not open to the outer edges as Myxidium, but asymmetrically to the flat side. Shell thick, bivalve, the seam line runs in an arc towards the spore” (translated from German). Based on what was known at the time and the diagnostic tools available, the proposal of genus Zschokkella was a logical and valid process.
Today, genus Zschokkella is defined by “spores ellipsoidal in sutural view and slightly bent or semicircular in valvular view, with rounded or bluntly pointed ends. Shell valves smooth or with ridges. The suture is straight, curved or sinuous. Polar capsules almost spherical, open slightly subterminally and both to one side; the sporoplasm is binucleate. Trophozoites disporic to polysporic, the latter with pansporoblast formation” (Lom & Dyková, 2006). Again, several characters from the current generic diagnosis stand out as unique. Within the Myxidiidae, semicircular spores are distinct to species of Zschokkella. Although the character of PCs opening to one side is vague, if we consider the original description (PCs open asymmetrically to the flat side of the spore) then this character becomes clearly defined. As described above, the remaining diagnostic characters are either equivocal or could apply equally to other genera within the Myxidiidae.
Today, the concept of Myxidium has grown to encompass over 220 species mainly from marine and freshwater fishes, but also from reptiles, birds and one species of hyperparasite (Lom & Dyková, 2006; Eiras et al., 2011; Freeman & Shinn, 2011). Zschokkella has grown to encompass over 65 species, again, mainly from marine and freshwater fishes but also from amphibians and reptiles (Lom & Dyková, 2006). Currently there are 28 small sub-unit ribosomal DNA (SSU rDNA) sequences for species of Myxidium and 18 for Zschokkella available on GenBank, including the type-species of both genera, M. lieberkuehni and Z. hildae. Of these, 12 are replicates taken from different hosts and 12 are of undescribed species. Phylogenetic analyses based on SSU rDNA sequences separate species of Myxidium and Zschokkella into marine and freshwater groups where they form clades including species belonging to other genera (Fiala, 2006). In phylogenetic analyses, polyphyletic distribution of species is observed for both genera (Fiala, 2006; Bartošová et al., 2009; Fiala & Bartošová, 2010). Molecular analyses of SSU rDNA data show little congruency with relatedness of currently assigned myxosporean species derived from either morphological or biological factors such as host or geographic location (Fiala, 2006; Fiala & Bartošová, 2010). However, some evidence exists that tissue tropism may be an informative character to reflect phylogenetic relationships (Fiala, 2006; Fiala & Bartošová, 2010; Heiniger & Adlard, 2012; Heiniger et al., 2013; Kristmundsson & Freeman, 2013).
The confusion in taxonomic identification of species of Myxidium and Zschokkella is well documented (Diamant et al., 1994; Palenzuela et al., 2002; Lom & Dyková, 2006). It is obvious from the available literature that no clarity exists in the characters originally used to distinguish between genera to the point where new species have been assigned almost arbitrarily based on superficial morphology (see Diamant et al., 1994; Chen & Ma, 1998; Gong et al., 2003). Or indeed, more recently naming an organism based on the priority of a genus (with the assumption of future synonymy) (Diamant et al., 1994); the genetic relationship to an existing named species, regardless of the validity of this named species (Yemmen et al., 2013); or the genetic and morphological distinction from the genus sensu stricto (Karlsbakk & Køie, 2012). The clear lack of definitive boundaries between Myxidium and Zschokkella has led to inconsistencies in assigning species to either genus with several species identified as either Myxidium or Zschokkella displaying characters of both genera (Schulman, 1966; Hine, 1975; Chen & Ma, 1998; Gong et al., 2003).
The inclusion of molecular data in species descriptions and subsequent phylogenetic studies has highlighted some congruence with biological or morphological characters between closely related species. These studies have given some clarity to the systematics of small groups of species and resulted in the resurrection or establishment of new genera. Hartigan et al. (2012) proposed the re-establishment of Cystodiscus Lutz, 1889 for species infecting amphibians previously assigned to Myxidium based on a distinct genetic monophyletic lineage combined with ultrastructural and morphological data. Similarly, Prunescu et al. (2007) proposed the genus Soricimyxum Prunescu, Prunescu, Pucek & Lom, 2007 to house the only myxosporean described to date from a terrestrial mammal, based on the assumed difference in biology and mode of transmission. Palenzuela et al. (2002) proposed the genus Enteromyxum Palenzuela, Redondo & Álvarez-Pellitero, 2002 to house their new species and transferred Myxidium leei Diamant, Lom & Dyková, 1994 based on phylogenetic, morphological and biological (histozoic parasites in the epithelia of the digestive tract of marine fishes) characters.
Several revisions of the taxonomic placement of species and genera in the Myxidiidae have also resulted in the transfer of species to other genera and families. Based on close phylogenetic relationships and morphological similarities, Heiniger et al. (2011) re-established the family Coccomyxidae Léger & Hesse, 1907 for Coccomyxa Léger & Hesse, 1907 (previously assigned to family Myxidiidae), Auerbachia Meglitsch, 1968 and Globospora Lom, Noble & Laird, 1975. Køie (2003) proposed the genus Ellipsomyxa Køie, 2003 for their new species based on morphological differences and tentatively placed the genus in family Ceratomyxidae Doflein, 1899. Since its proposal, two new species have been described and Zschokkella mugilis Sitja-Bobadilla & Alvarez-Pellitero, 1993 and Leptotheca fusiformis Davis, 1917 have been transferred to the genus (Køie & Karlsbakk, 2009; Gunter & Adlard, 2010; Whipps & Font, 2013). In phylogenetic analyses, species of Ellipsomyxa are more closely related to species of Myxidium than their purported confamilial, Ceratomyxa, leading authors to speculate on the placement within Ceratomyxidae (see Gunter et al., 2009; Køie & Karlsbakk, 2009). Furthermore, species of Ellipsomyxa are more morphologically similar to species of Myxidium and Zschokkella than Ceratomyxa but remain distinguished by the position of the suture line and size and shape of their PCs (Køie, 2003; Køie & Karlsbakk, 2009).
This paper examines the taxonomic identity of ‘Myxidium/Zschokkella’-like isolates identified in teleosts from Australian waters. Based on morphological, molecular and biological characters we describe four novel species of Ellipsomyxa, three novel species of Myxidium and six novel species of Zschokkella.
Materials and methods
Host and parasite collection
Teleosts were collected using localised sprays of clove oil anaesthetic, spear, micro spear and line fishing off Heron Island (23°26′S, 151°54′E) and Lizard Island (14°40′S, 145°27′E), Great Barrier Reef, Queensland, Moreton Bay (27°11′S, 153°15′E), Queensland, and off Point Cloates (22°40′S, 113°41′E), Ningaloo Reef, Western Australia. Fish were euthanized by neural pithing. Currently valid host names conform with those from the California Academy of Sciences Catalogue of Fishes (http://research.calacademy.org/ichthyology/catalog/fishcatmain.asp). The gall bladder of each fish was examined for the presence of myxosporean infections using the preparation methods described by Heiniger et al. (2008). Infected gall bladders were preserved in 100% ethanol for DNA analysis and frozen in saline for morphological characterisation.
Morphological analysis of spores
Measurements of spores followed the guidelines of Lom & Arthur (1989). Images of thirty spores were taken with an Olympus BH2 microscope at ×400 or ×1000 magnification using a Nikon Digital Sight DS-LI digital camera (Nikon Corporation, Japan). Measurements were taken from microphotographs using the measuring tool in the Nikon NIS Elements software (Nikon Corporation, Japan) calibrated against a stage micrometer. Mean measurements and their standard deviations were calculated for each spore dimension, allowing characterisation of each isolate. All measurements are given in micrometres. Type-specimens were deposited in the Queensland Museum (QM) or Western Australian Museum (WAM), depending on the origin of material.
Morphological comparisons referred to in the Results section under ‘Remarks’ for each species were undertaken to discriminate those species which most closely resembled the new species being described. These species were selected from a morphometric dataset that included all Myxidium, Zschokkella and Ellipsomyxa spp.
SSU rDNA analysis
DNA was extracted from 600 μl of infected bile preserved in ethanol. The sample was first pelleted at 15,700×g for 10 min and the ethanol supernatant removed. DNA was extracted from the pellet as per the recommended protocol accompanying the QIAgen DNeasy Kit (QIAGEN Inc., Valencia, California). Small subunit ribosomal DNA (SSU rDNA) was amplified by PCR using the primers MyxospecF (5′-TTC TGC CGT ATC AAC TWG TTG-3′; Fiala, 2006) and 18R (5′-CTA CGG AAA CCT TGT TAC G-3′; Whipps et al., 2003). An additional primer, Myxid1R (5′-CGA TCA GAT ACC GTC CTA GTT C-3′), was designed for this study based on conserved regions of already acquired sequence data using AlleleID version 7.72 (Apte & Singh, 2007). PCR reactions and purification were performed as described by Heiniger et al. (2008), with the following exceptions: annealing temperature for each primer pair as follows: MyxospecF-18R at 51°C and MyxospecF-Myxid1R at 58°C, and extension and final extension temperatures performed at 72°C in all PCR reactions. Purified DNA was sent to Australian Genome Research Facility, The University of Queensland, Australia, for sequence determination using the same primers as used for the initial amplification.
Phylogenetic analyses
SSU rDNA sequences from the taxa described in this study were edited using Geneious Pro version 5.6.2 (Drummond et al., 2010). All sequences generated in this study were lodged in GenBank. Initial phylogenetic analyses of SSU rDNA included 131 myxosporean sequences, all with corresponding formal descriptions or morphological and photographic data available, for comparative purposes. This dataset was aligned using Muscle version 3.7 (Edgar, 2004) using the clustal W algorithm (Thompson et al., 1994) with UPGMB parameters for all iterations on the CIPRES portal (Miller et al., 2010). Maximum likelihood analysis (ML) was conducted using the RAxML algorithm (Stamatakis et al., 2008) on the CIPRES portal with the gamma rate model of heterogeneity and maximum likelihood search estimating the proportion of invariable sites parameters. Nodal support was inferred based on 1,000 bootstrap replicates. This analysis indicated that the new species reported here showed a close genetic relationship with species of Myxidium, Zschokkella, Ellipsomyxa, Sinuolinea Davis, 1917, Sigmomyxa, Coccomyxa, Auerbachia and Myxobolus. Therefore, for succinctness only the new species and closest related myxosporean SSU rDNA sequences, as determined by BLAST analyses and ML analysis described above, were used in phylogenetic analyses presented here (total of 109 sequences). Buddenbrockia plumatellae Schroder, 1910 and Tetracapsuloides bryosalmonae (Canning, Curry, Feist, Longshaw & Okamura, 1999) were used as outgroup taxa in all SSU rDNA analyses. Alignment of the SSU rDNA dataset was produced as described above. Ambiguous characters were removed from all alignments using the program GBlocks (Castresana, 2000) with less stringent parameters (56, 56, 8, 5 and “with half”). The resulting alignment was exported as fasta and nexus files, edited by eye and trimmed using MacClade version 4.08 (Maddison & Maddison, 2005). This produced an alignment of 1,238 bases for SSU rDNA. This alignment was used to conduct all phylogenetic analyses.
Maximum likelihood analyses were performed as described above. Bayesian analyses were conducted using MrBayes version 3.1.2 (Ronquist & Huelsenbeck, 2003). The software jModelTest version 2.1.3 (Posada, 2008) was used to estimate the best substitution model for the SSU rDNA datasets. Bayesian inference analysis was conducted using the TVM+I+G model predicted as the best estimator by the Akaike Information Criterion (AIC) in jModelTest. Bayesian inference analysis was run over 10,000,000 generations (ngen=10,000,000) with 2 runs each containing 4 simultaneous Markov Chain Monte Carlo (MCMC) chains (nchains=4) and every 1,000th tree saved (samplefreq=1,000). Bayesian analyses used the following parameters: nst=6, rates=invgamma, ngammacat=4, the MCMC parameters were left at the default settings, and the priors parameters of the combined dataset were set to ratepr=fixed. Samples of substitution model parameters, and tree and branch lengths were summarised using the parameters ‘sump burnin = 3,000’ and ‘sumt burnin = 3,000’. These ‘burnin’ parameters were chosen because the log likelihood scores ‘plateaued’ well before 3,000,000 replicates in the Bayesian inference analysis as determined using Tracer v1.5 (Drummond & Rambaut, 2007).
Bayesian, neighbour-joining and maximum parsimony analyses was also performed on two separate datasets. One dataset contained all SSU rDNA sequences of the novel Ellipsomyxa and Myxidium and the closest related sequences, and one containing all SSU rDNA sequences of the novel Zschokkella and the closest related sequences. Bayesian analyses were performed as described above. Neighbour-joining and maximum parsimony analyses were conducted on these smaller datasets to examine relatedness of the novel species and closest related (as determined by Bayesian, ML, and BLAST analyses). The datasets were aligned, edited and trimmed as described above. Ambiguous characters were removed from all alignments using the program GBlocks (Castresana, 2000) with less stringent parameters (8, 8, 8, 5, and “with half” for the Ellipsomyxa/Myxidium dataset and 16, 16, 8, 5 and “with half” for the Zschokkella dataset). This produced alignments of 1,287 and 1,470 bases for the Ellipsomyxa/Myxidium and Zschokkella datasets, respectively. Neighbour-joining and maximum parsimony analyses were conducted using the default parameters to construct trees using PAUP* 4.0b10 (Swofford, 2002). Parsimony analyses employed a heuristic search with 50 repetitions of random sequence addition and tree bisection and reconnection branch swapping. All characters were treated as unordered, Ts/Tv ratio was set to 1:2 and gaps were treated as missing data. The strength of resultant relationships of neighbour-joining and maximum parsimony was tested by bootstrap analyses with 10,000 replicates.
Pairwise differences were analysed using Geneious Pro version 5.6.2 to determine total nucleotide distance and percentage differences. Pairwise difference analyses were performed using three datasets containing the new species from this study and the most closely related sequences as determined by BLAST analyses. These datasets was aligned, edited and trimmed as described above. The resulting alignments were 1,343, 1,344 and 1,510 bases for datasets containing the new species of Ellipsomyxa, Myxidium and Zschokkella, respectively.
Results
Between 2005 and 2012, 3,015 individual teleosts were examined for the presence of myxosporean infections in the gall bladder from four localities, off Heron Island on the southern Great Barrier Reef, off Lizard Island on the northern Great Barrier Reef, Moreton Bay, Queensland and Ningaloo Reef, off Point Cloates, Western Australia. Teleosts from 68 families belonging to 17 orders were sampled. One hundred and fifty isolates (defined as unique host/parasite combinations) of ‘Myxidium/Zschokkella’-like spores were identified from eight orders and 27 families of fishes. Here we provide formal description for 83 isolates referable to 13 novel species of myxozoans. Mixed infections with species of Ceratomyxa were observed in 100% of apogonid hosts. Co-infections with isolates of congeners or species of other genera were not observed.
-
Class Myxosporea Bütschli, 1881
-
Order Bivalvulida Shulman, 1959
-
Family Ceratomyxidae Doflein, 1899
-
Genus Ellipsomyxa Køie, 2003
Diagnosis
Ellipsoidal thin-walled myxospores. Suture line straight, central, perpendicular, or forming an acute angle to thickness (longitudinal) axis or sinuous. In apical or polar view, two equal-sized spherical to pyriform polar capsules occur some distance from each other and from rounded spore ends. Polar capsules discharge at opposite sides some distance from spore ends (Køie, 2003).
Ellipsomyxa manilensis n. sp.
Type-host: Arothron manilensis (Marion de Procé) (Tetraodontidae).
Type-locality: Lagoon, off Lizard Island (14°40′S, 145°27′E), Great Barrier Reef, Queensland, Australia.
Site of infection: Gall bladder.
Prevalence: 1 of 2 fishes (50%).
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465649). DNA voucher (QM G465650).
Representative sequence: GenBank accession number KF179048.
Etymology: The species epithet manilensis is in reference to the type-host species, A. manilensis.
Description (Figs. 1A, 2)
[Mature spore measurements (n = 10) are shown in Table 1.] Mature spores ovoid. Small valvular protrusions associated with the tips of the polar capsules present. Suture line thin, slightly curved. Spore valves smooth. Polar capsules equal, subspherical to pyriform, opening terminally in opposite directions. Polar filament with 3–4 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
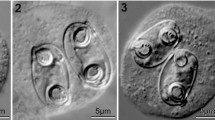
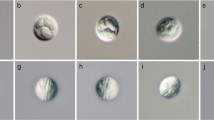
Phase contrast photomicrographs of spores of the novel species of Ellipsomyxa, Myxidium and Zschokkella. A, E. manilensis n. sp.; B–C, E. arothroni n. sp. ex Arothron hispidus (B) and Arothron stellatus (C); D, E. nigropunctatis n. sp.; E–J, E. apogoni n. sp. ex Apogon doederleini (LI) (E), A. doederleini (HI) (F), Ostorhinchus cyanosoma (LI) (G), O. cyanosoma (LI) (H); O. cookii (HI) (I), and O. aureus (NR) (J); K–L, M. scomberomori n. sp.; M–N, M. maxi n. sp.; O, M. milleri n. sp.; P–S, Z. cardinalis n. sp. ex Cheilodipterus quinquelineatus (HI) (P), C. quinquelineatus (HI) (Q), C. quinquelineatus (LI) (R), and Nectamia fusca (HI) (S); T, Z. ohlalae n. sp.; U–V, Z. compressis n. sp.; W–X, Z. jaimeae n. sp.; Y–Z, Z. bicarinatis n. sp.; 1–2, Z. balistoidi n. sp. Abbreviations: HI, Heron Island; LI, Lizard Island; NR, Ningaloo Reef. Scale-bar: 10 μm
Illustrations of mature spores of the novel Ellipsomyxa, Myxidium and Zschokkella spp. in valvular (a) and sutural (b) view. 1, E. manilensis n. sp.; 2, E. arothroni n. sp.; 3, E. nigropunctatis n. sp. in sutural view; 4, E. apogoni n. sp.; 5, M. scomberomori n. sp.; 6, M. maxi n. sp.; 7, M. milleri n. sp.; 8, Z. cardinalis n. sp.; 9, Z. ohlalae n. sp.; 10, Z. compressis n. sp.; 11, Z. jaimeae n. sp. in valvular view; 12, Z. bicarinatis n. sp.; 13, Z. balistoidi n. sp. Scale-bar: 10 μm
Remarks
There are four other described Ellipsomyxa spp. from marine fishes. Ellipsomyxa manilensis n. sp. is distinctly larger in all dimensions than all other species of Ellipsomyxa. Furthermore, E. manilensis n. sp. has fewer polar filament coils (3–4) than E. gobii Køie, 2003 (6–7), E. syngnathi Køie & Karlsbakk, 2009 (5–6), E. mugilis (Sitja-Bobadilla & Alvarez-Pellitero, 1993) (5) and E. adlardi Whipps & Font, 2012 (5–6). Ellipsomyxa manilensis n. sp. has smaller polar capsules with a greater number of polar filament coils than E. arothroni n. sp. Ellipsomyxa nigropunctatis n. sp. has shorter polar capsules than E. manilensis n. sp.
A single SSU rDNA sequence of 1,229 bases was generated for E. manilensis n. sp. (GenBank accession number KF179048). The sequence differs from the aligned sequences of other Ellipsomyxa spp. by 10–31 nucleotides (Table 2). Ellipsomyxa manilensis n. sp. is genetically most similar to E. arothroni n. sp. with 98.8% sequence identity.
Ellipsomyxa arothroni n. sp.
Type-host: Arothron hispidus (Linnaeus) (Tetraodontidae).
Other host: Arothron stellatus (Anonymous) (Tetraodontidae).
Type-locality: Lagoon, off Lizard Island (14°40′S, 145°27′E), Great Barrier Reef, Queensland, Australia.
Other localities: Ningaloo Reef (A. hispidus); off Lizard Island (A. stellatus).
Site of infection: Gall bladder.
Prevalence: Ex A. hispidus off Lizard Island: 5 of 5 fishes (100%); at Ningaloo Reef: 4 of 4 fishes (100%); ex A. stellatus off Lizard Island: 3 of 3 fishes (100%).
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465651–G465652). DNA voucher (G465653- G465655).
Representative sequence: GenBank accession number KF179049.
Etymology: The species epithet arothroni is in reference to the type-host genus, Arothron.
Description (Figs. 1B–C, 2)
[Mature spore measurements (n = 80) are shown in Table 1.] Mature spores ovoid. Small valvular protrusions associated with the tips of the polar capsules present. Suture line thin, straight, occasionally slightly curved. Spore valves smooth. Polar capsules equal, pyriform, opening sub-terminally, in opposite directions. Polar capsules situated at anterior end of the spore in valvular view. Polar filament with 4–6 coils, perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Ellipsomyxa arothroni n. sp. is distinctly larger than E. gobii (9.4–13.8 × 11.2–17.7 vs 8–9 × 10.8–12 μm) and has pyriform polar capsules rather than spherical as observed for E. gobii. Ellipsomyxa mugilis is also distinguished from E. arothroni n. sp. by its subspherical polar capsules and distinctly smaller spores (5.5–9 × 10–13.5 vs 9.4–13.8 × 11.2–17.7 μm). Similarly, E. syngnathi has distinctly smaller spores (7.2–8.6 × 9–10.8 vs 9.4–13.8 × 11.2–17.7 μm) and polar capsules (2.7–3.2 × 3.2–4.1 vs 3.1–5.4 × 4.1–7.9 μm) than E. arothroni n. sp. Ellipsomyxa arothroni n. sp. has wider spores (9.4–13.8 vs 7.1–8.8 μm) and larger polar capsules (3.1–5.4 × 4.1–7.9 vs 3.3–4.1 × 3.9–4.9 μm) than E. adlardi. Ellipsomyxa arothroni n. sp. has larger polar capsules than E. nigropunctatis n. sp. Although the spore measurements of E. arothroni n. sp. and E. nigropunctatis n. sp. overlap, they are greater for E. arothroni n. sp. in all instances.
Identical SSU rDNA sequences of 1,311 bases were generated from two fish species, A. hispidus and A. stellatus, for E. arothroni n. sp. (five isolates from individual fish with 100% sequence identity) (GenBank accession number KF179049). These sequences differ from the aligned sequences of other Ellipsomyxa spp. by 3–62 nucleotides (Table 2). Ellipsomyxa arothroni n. sp. is genetically most similar to E. nigropunctatis n. sp. with 99.8% maximum sequence identity.
Ellipsomyxa nigropunctatis n. sp.
Type-host: Arothron nigropunctatus (Bloch & Schneider) (Tetraodontidae).
Type-locality: Lagoon, off Lizard Island (14°40′S, 145°27′E), Great Barrier Reef, Queensland, Australia.
Other locality: Ningaloo Reef.
Site of infection: Gall bladder.
Prevalence: 1 of 2 fishes (50%) off Lizard Island; 0 of 1 fish at Ningaloo Reef.
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465656). DNA voucher (QM G465657).
Representative sequence: GenBank accession number KF179050.
Etymology: The species epithet nigropunctatis is in reference to the type-host species, A. nigropunctatus.
Description (Figs. 1D, 2)
[Mature spore measurements (n = 30) are shown in Table 1.] Mature spores ovoid. Small valvular protrusions associated with the tips of the polar capsules present. Longitudinal suture line thin, occasionally slightly curved. Spore valves smooth. Polar capsules equal, pyriform, opening sub-terminally, in opposite directions. Polar capsules situated at anterior (or posterior) end of the spore in valvular view. Polar filament with 5 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Ellipsomyxa nigropunctatis n. sp. is distinguished from E. gobii and E. mugilis by its pyriform polar capsules and larger spores. Ellipsomyxa nigropunctatis n. sp. has larger spores than E. syngnathi (8–12.9 × 11.9–16.3 vs 7.2–8.6 × 9–10.8 μm). Ellipsomyxa nigropunctatis n. sp. has wider spores (8–12.9 vs 7.1–8.8 μm) and larger polar capsules (2.8–4.6 × 3.5–5.7 vs 3.3–4.1 × 3.9–4.9 μm) than E. adlardi. Ellipsomyxa nigropunctatis n. sp. has smaller polar capsules than E. arothroni n. sp. Although the spore measurements of E. arothroni n. sp. and E. nigropunctatis n. sp. overlap they are greater for E. arothroni n. sp. in all instances.
A single SSU rDNA sequence of 1,564 bases was generated for E. nigropunctatis n. sp. (GenBank accession number KF179050). This sequence differs from the aligned sequences of other Ellipsomyxa species by 2–62 nucleotides (Table 2). Ellipsomyxa nigropunctatis n. sp. is genetically most similar to E. arothroni n. sp. with 99.8% maximum sequence identity.
Ellipsomyxa apogoni n. sp.
Type-host: Apogon doederleini Jordan & Snyder (Apogonidae).
Type-locality: Lagoon, off Heron Island (23°26′S, 151°54′E), Great Barrier Reef, Queensland, Australia.
Other locality: Off Lizard Island.
Site of infection: Gall bladder.
Prevalence: 10 of 14 fishes (71%) off Heron Island; 1 of 2 fishes (50%) off Lizard Island.
Other hosts and localities: Apogonidae - Ostorhinchus aureus (Lacepède), 6 of 14 fishes (43%) at Ningaloo Reef; Ostorhinchus cookii (Macleay), 4 of 35 fishes (11%) off Heron Island, 0 of 8 fishes off Lizard Island, 0 of 2 fishes at Ningaloo Reef; Ostorhinchus cyanosoma (Bleeker), 3 of 4 fishes (75%) off Lizard Island, 0 of 23 fishes at Ningaloo Reef.
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465658–G465659); Voucher - Giemsa-stained, air-dried spores (QM G465660–G465665).
Representative sequence: GenBank accession number KF179051.
Etymology: The species epithet apogoni is in reference to the type-host family, Apogonidae.
Description (Figs. 1E–J, 2)
[Mature spore measurements (n = 233) are shown in Table 1.] Mature spores ellipsoidal in sutural view and ovoid in valvular view. Suture line distinct, straight, bisecting the spore in sutural view. Small valvular protrusions associated with the tips of the polar capsules present in sutural view. Spore valves with 5–9 longitudinal striations. Polar capsules equal, pyriform, opening sub-terminally in opposite directions and on opposite sides of the suture line. Polar capsules lie along longest axis of the spore in valvular view and diagonally opposed in sutural view. Polar filament with 2–4 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Ellipsomyxa apogoni n. sp. is distinguished from all species of Ellipsomyxa, including the new species described here, by the presence of spore striations (5–9 per spore valve) and fewer polar filament coils (2–4). Ellipsomyxa gobii and E. mugilis are further distinguished by their spherical to subspherical polar capsules. Ellipsomyxa adlardi is further distinguished by its longer spores (11.3–14.4 vs 8.8–12.1 μm) and larger polar capsules (3.3–4.1 × 3.9–4.9 vs 1.7–3.4 × 2.4–4.8 μm). The spore dimensions of E. syngnathi and E. apogoni n. sp. overlap but the ranges are greater for E. apogoni n. sp. in all instances.
Identical SSU rDNA sequences of 667 bases were generated from A. doederleini, O. aureus, O. cookii and O. cyanosoma for E. apogoni n. sp. (four isolates from individual hosts with 100% sequence identity) (GenBank accession number KF179051). Due to mixed infections with Ceratomyxa species, only the primer combination of MyxospecF/Myxid1R could successfully amplify clean, uncontaminated SSU rDNA sequences. The sequence differs from the aligned sequences of other Ellipsomyxa species by 2–31 nucleotides (Table 2). Ellipsomyxa apogoni n. sp. is genetically most similar to E. nigropunctatis n. sp. with 99.7% maximum sequence identity.
-
Family Myxidiidae Thélohan, 1892
-
Genus Myxidium Bütschli 1882
Diagnosis
Spores generally fusiform, straight or slightly crescent or even sigmoid, with more or less pointed ends. Shell valves smooth or with ridges, suture line bisects the spore. Two polar capsules, mostly pyriform, lie one at each end of the spore. Capsular foramina are situated in the sutural plane, at or near the end of the spore and open as a rule in opposite directions. The binucleate sporoplasm lies between the capsules (Lom & Dyková, 2006).
Myxidium scomberomori n. sp.
Type-host: Scomberomorus semifasciatus (Macleay) (Scombridae).
Type-locality: Moreton Bay (27°11′S, 153°15′E), Queensland, Australia.
Site of infection: Gall bladder.
Prevalence: 1 of 2 fishes (50%).
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465666). DNA voucher (QM G465667).
Representative sequence: GenBank accession number KF179052.
Etymology: The species epithet scomberomori is in reference to the type-host genus, Scomberomorus.
Description (Figs. 1K–L, 2)
[Mature spore measurements (n = 30) are shown in Table 1.] Mature spores fusiform with prominent acuminated tips giving sigmoid or S-shaped appearance in sutural view. One spore valve is more flattened than the other. Spores appear to twist on longitudinal axis giving reniform appearance in sutural view. Longitudinal suture line thin. Spore valves smooth. Polar capsules equal, elongate pyriform, opening sub-terminally in opposite directions. Polar capsules situated closer to flat spore valve. Polar filament with 5 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Myxidium scomberomori n. sp. is morphologically similar to Myxidium laticurvum Kabata, 1962 (syn. M. trachinorum Canning, Curry, Anderson & Okamura, 1999; see Karlsbakk, 2001). Myxidium scomberomori n. sp. has narrower spores (5.3–8.0 μm, average 6.9 μm) than M. laticurvum (average 8.8 ± 0.32 μm, based on fresh spores reported by Canning et al., 1999). The polar capsules of M. laticurvum overlap in valvular view, a feature not present in M. scomberomori n. sp. Myxidium scomberomori n. sp. is distinguished from all other species of Myxidium by the unusual shape of its spores, reniform in sutural view.
A single SSU rDNA sequence of 1,641 bases was generated for M. scomberomori n. sp. (GenBank accession number KF179052). The sequence differs from the aligned sequences of other bivalvulidan species by 45–94 nucleotides (Table 3). Myxidium scomberomori n. sp. is genetically most similar to M. laticurvum (JN033229) with 96.2% sequence identity.
Myxidium maxi n. sp.
Type-host: Istiblennius meleagris (Valenciennes) (Bleniidae).
Type-locality: Lagoon, off Heron Island (23°26′S, 151°54′E), Great Barrier Reef, Queensland, Australia.
Site of infection: Gall bladder.
Prevalence: 3 of 5 fishes (60%).
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465668–G465670).
Representative sequences: GenBank accession numbers KF179054 and KF179055.
Etymology: The species epithet maxi is in honour of Mr Max Heiniger in recognition of his support and encouragement of this work.
Description (Figs. 1M–N, 2)
[Mature spore measurements (n = 30) are shown in Table 1.] Main body of mature spores ellipsoidal, but valvular protrusions associated with the tips of the polar capsules give spores a sigmoid or S-shaped appearance in sutural view and spindle-shaped appearance in valvular view. Longitudinal suture line thin. Spore valves smooth. Polar capsules equal, pyriform, opening terminally in opposite directions and on opposite sides of the suture line. Polar capsules lie along longest axis of the spore in valvular view and diagonally opposed in sutural view. Usually difficult to view both polar capsules clearly on the same focal plane. Polar filament with 5–6 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Myxidium maxi n. sp. is morphologically similar to M. adriaticum Lubat, Radujkovic, Marques & Bouix, 1989, M. contortum Fantham, 1930, M. maamouni Abdel-Baki, 2009, M. myxocephali Fantham, Porter & Richardson, 1940, M. tsudae Fujita, 1923 and M. incurvatum Thélohan, 1892. Furthermore, M. incurvatum and M. contortum are reported from the gall bladders of species of Blenniidae. Myxidium incurvatum is smaller in overall size (4–5 × 8–9 vs 7.1–8.9 × 11.6–15.5 μm) than M. maxi n. sp. and can be further distinguished genetically with 103 nucleotides difference. Myxidium tsudae has larger spores (8–9 × 16–19 vs 7.1–8.9 × 11.6–15.5 μm) than M. maxi n. sp. and can be further distinguished by the presence of striations on the spore valves. Myxidium maamouni has larger polar capsules (3–5 × 6–8 vs 2.5–3.5 × 4.2–6.5 μm) with more polar filament coils (9–12 vs 5–6 coils) than M. maxi n. sp. Myxidium contortum has longer spores (21–24 vs 11.6–15.5 μm) and polar capsules (6–8 vs 4.2–6.5 μm) than M. maxi n. sp. Myxidium maxi n. sp. has shorter spores than M. myxocephali (11.6–15.5 vs 13.3–17.5 μm). The spores of M. adriaticum are similar in size to M. maxi n. sp. but the ranges of measurements for M. maxi n. sp. are greater in all instances.
Myxidium maxi n. sp. is also morphologically similar in shape to species belonging to the genus Sigmomyxa, S. sphaerica (Thélohan, 1895) and S. elmatboulii (Ali, Abdel-Baki & Sakran, 2006). Furthermore, S. sphaerica has been recorded previously (as M. sphaericum) in Istiblennius meleagris from off Heron Island (Moser et al., 1989). The isolate of S. sphaerica reported by Moser et al., 1989 is distinguished by its longer spores (15–20 vs 11.6–15.5 μm) and wider polar capsules (3–5 vs 2.5–3.5 μm) than M. maxi n. sp. Both Sigmomyxa spp. can be distinguished from M. maxi n. sp. by their larger spores (10.2–12.8 × 16.7–19.4 μm and 9–12 × 19–23 μm for S. sphaerica and S. elmatboulii, respectively) and polar capsules (3.5–4.9 × 6.3–9.3 μm and 3.5–4 × 8–10 μm, respectively). Myxidium maxi n. sp. is further distinguished by fewer polar filament coils (5–6) than S. sphaerica (9–12) and S. elmatboulii (9–10). Furthermore, M. maxi n. sp differs by 108 nucleotides from S. sphaerica (GenBank accession number JN033225).
Two SSU rDNA sequences of 1,431 and 1,420 bases were generated from two individual hosts for M. maxi n. sp. (GenBank accession numbers KF179054 and KF179055). There are 2 nucleotides difference (99.9% sequence identity) between the SSU rDNA sequences. The sequence differs from the aligned sequences of other bivalvulidan species by 86–108 nucleotides (Table 3). Myxidium maxi n. sp. is genetically most similar to Myxidium gadi (DQ377711) and Myxidium bergense (JN033231) with 92.6% maximum sequence identity.
Myxidium milleri n. sp.
Type-host: Corythoichthys schultzi Herald (Syngnathidae).
Type-locality: Lagoon, off Heron Island (23°26′S, 151°54′E), Great Barrier Reef, Queensland, Australia.
Site of infection: Gall bladder.
Prevalence: 1 of 1 fish (100%).
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465671).
Representative sequence: GenBank accession number KF179053.
Etymology: The species epithet milleri is in honour of Dr Terry Miller, in recognition of his contribution to the research of Australian parasitology.
Description (Figs. 1O, 2)
[Mature spore measurements (n = 30) are shown in Table 1.] Main body of mature spores ellipsoidal, but valvular protrusions associated with the tips of the polar capsules give spores a sigmoid appearance in sutural view. Longitudinal suture line faint. Polar capsules equal, pyriform, opening terminally in opposite directions and on opposite sides of the suture line. Polar capsules lie along longest axis of the spore in valvular view and diagonally opposed in sutural view. Sporoplasm fills cavity between polar capsules.
Remarks
Myxidium milleri n. sp. is morphologically similar to M. cochleatum Yurakhno, 1991, M. glossogobi Chakravarty, 1939, M. glutinosum Davis, 1917, M. incurvatum, M. maxi n. sp. and M. queenslandicus. Myxidium milleri n. sp. has smaller spores (4.7–6 × 8.1–10.1 μm) than M. cochleatum (6.3–6.9 × 11.3–13.1 μm), M. glossogobi (8.5–10 × 12–15 μm), M. queenslandicus (6.1–9.7 × 13.4–21.6 μm) and M. maxi n. sp. Myxidium queenslandicus, M. cochleatum and M. maxi n. sp. are further distinguished by their larger polar capsules. Myxidium glutinosum has longer spores (10–11 vs 8.1–10.1 μm) than M. milleri n. sp. and is further distinguished by the presence of a gelatinous envelope surrounding the spore. Myxidium incurvatum has spores of similar size (4–5 × 8–9 vs 4.7–6 × 8.1–10.1 μm) to M. milleri n. sp. but the ranges are greater for M. milleri n. sp.
Myxidium milleri n. sp. is genetically distinct from M. incurvatum, M. queenslandicus and M. maxi n. sp. with 57, 109 and 99 nucleotides difference, respectively (Table 3). Interestingly, M. milleri n. sp. shows a close phylogenetic relationship with Sinuolinea phyllopteryxa Garner, Atkinson, Hallett, Bartholomew, Nordhausen, Reed, Adams & Whitaker, 2008. Sinuolinea phyllopteryxa is morphologically distinguished from M. milleri n. sp. by its spherical spores and polar capsules. Furthermore, S. phyllopteryxa infects the kidney and renal tubules of its host whereas M. milleri n. sp. infects the gall bladder.
A single SSU rDNA sequence of 1,343 bases was generated for M. milleri n. sp. (GenBank accession number KF179053). The sequence differs from the aligned sequences of other bivalvulidan species by 1–109 nucleotides (Table 3). Myxidium milleri n. sp. is genetically most similar to Sinuolinea phyllopteryxa (DQ645952) with 99.9% sequence identity (4 nucleotides, 99.7% similarity over 100% of sequence according to BLAST search).
-
Genus Zschokkella Auerbach, 1910
Diagnosis
Spores ellipsoidal in sutural view and slightly bent or semicircular in valvular view, with rounded or bluntly pointed ends. Shell valves smooth or with ridges. Suture straight, curved or sinuous. Polar capsules almost spherical, open slightly subterminally and both to one side; sporoplasm binucleate (Lom & Dyková, 2006).
Zschokkella cardinalis n. sp.
Type-host: Cheilodipterus quinquelineatus Cuvier (Apogonidae).
Type-locality: Lagoon, off Heron Island (23°26′S, 151°54′E), Great Barrier Reef, Queensland, Australia.
Other locality: Off Lizard Island.
Site of infection: Gall bladder.
Prevalence: 15 of 28 fishes (54%) off Heron Island; 2 of 34 fishes (6%) off Lizard Island.
Other hosts and localities: Apogonidae - Archamia fucata (Cantor), 7 of 20 fishes (35%) off Heron Island, 0 of 26 fishes off Lizard Island; Nectamia fusca (Quoy & Gaimard), 3 of 32 fishes (9%) off Heron Island, 0 of 21 fishes off Lizard Island.
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465672–G465675); voucher - Giemsa-stained, air-dried spores (QM G465676–G465679).
Representative sequence: GenBank accession number KF179056.
Etymology: The species epithet cardinalis is in reference to the common name of the type-host family, Cardinalfishes (Apogonidae).
Description (Figs. 1P–S, 2)
[Mature spore measurements (n = 163) are shown in Table 1.] Mature spores ellipsoidal in sutural view, ovoid in valvular view. Suture line distinct, S-shaped, bisecting the spore in sutural view. Small valvular protrusions associated with the tips of the polar capsules present in sutural view. Spore valves with 6–11 longitudinal striations. Polar capsules equal, pyriform, opening terminally in opposite directions on opposite sides of suture. Polar capsules lie along longest axis of the spore in valvular view and diagonally opposed in sutural view. Polar filament with 2–4 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Zschokkella cardinalis n. sp. is morphologically similar to Z. ohlalae n. sp., Z. heronensis Moser, Kent & Dennis, 1989, Z. soleae Yemmen, Marton, Bahri & Eszterbauer, 2013, Z. nilei Abdel-Ghaffar, El-Toukhy, Al-Quraishy, Al-Rasheid, Abdel-Baki, Hegazy & Bashtar, 2008 and Z. helmii Abdel-Ghaffar, Ali, Al-Quraishy, Entzeroth, Abdel-Baki, Al Farraj & Bashtar, 2008. Zschokkella soleae, Z. helmii and Z. nilei all differ from Z. cardinalis n. sp. in having spherical polar capsules. Furthermore, Z. helmii and Z. nilei differ in their site of infection, Z. helmii is histozoic in the gall bladder wall and Z. nilei infects the kidney of its freshwater hosts. The spore measurements of Z. ohlalae n. sp. overlap with those of Z. cardinalis n. sp. but are smaller for spore length (8.9–10.7 vs 9–11.6 μm) and polar capsule size (1.9–3 × 2.6–4 vs 2.1–3.4 × 3–4.7 μm). Zschokkella heronensis has smaller polar capsules (2 × 2.5–3 vs 2.1–3.4 × 3–4.7 μm) with a greater number of polar filament coils (4–6 vs 3) than Z. cardinalis n. sp. Zschokkella soleae has larger spores (10.4–11.2 × 13.6–14.4 vs 5–8.5 × 9–11.6 μm) than Z. cardinalis n. sp. and is genetically distinct with 70 nucleotides difference.
Identical SSU rDNA sequences of 829 bases were generated from Cheilodipterus quinquelineatus, Archamia fucata and Nectamia fusca (all from off Heron Island) for Z. cardinalis n. sp. (five isolates from individual hosts with 100% sequence identity) (GenBank accession number KF179056). Due to mixed infections with Ceratomyxa spp., only the primer combination of MyxospecF/Myxid1R could successfully amplify clean, uncontaminated SSU rDNA sequences. The sequence differs from the aligned sequences of other bivalvulidan species by 11–72 nucleotides (Table 4). Zschokkella cardinalis n. sp. is genetically most similar to Z. jaimeae n. sp. with 98.5% maximum sequence identity. BLAST analysis revealed that Z. cardinalis n. sp. is genetically closest to a sequence of an undescribed species of Zschokkella taken from the gall bladder of Pseudanthias squamipinnis from the Red Sea (GenBank accession DQ333435) with maximum sequence identity of 99% over 100% coverage of the sequence.
Zschokkella ohlalae n. sp.
Type-host: Ostorhinchus cyanosoma (Bleeker) (Apogonidae).
Type-locality: Off Point Cloates (22°40′S, 113°41′E), Ningaloo Reef, Western Australia, Australia.
Site of infection: Gall bladder.
Prevalence: 4 of 23 fishes (17%) at Ningaloo Reef, 0 of 4 fishes off Lizard Island.
Material deposited: Syntype - Giemsa-stained, air-dried spores (WAM Z27998).
Representative sequence: GenBank accession number KF179057.
Etymology: The species epithet ohlalae is for Sailing Vessel (SV) Oh Là Là on which HH further developed her passion for coral reef fauna.
Description (Figs. 1T, 2)
[Mature spore measurements (n = 27) are shown in Table 1.] Mature spores ellipsoidal in sutural view, ovoid in valvular view. Small valvular protrusions associated with the tips of the polar capsules present in sutural view. Spore valves with 9–10 longitudinal striations. Suture line distinct, straight, bisecting the spore in sutural view. Polar capsules equal, pyriform, opening in opposite directions on opposite sides of the suture. Polar capsules lie along longest axis of the spore in valvular view and diagonally opposed in sutural view. Polar filament with 2 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Zschokkella ohlalae n. sp. is morphologically similar to Z. heronensis, Z. soleae, Z. cardinalis n. sp., Z. jaimeae n. sp. and Z. balistoidi n. sp. Zschokkella heronensis has fewer spore striations (6–8 vs 9–10) and a greater number of polar filament coils (4–6 vs 2) than Z. ohlalae n. sp. Zschokkella soleae differs from the present species by its larger spores (10.4–11.2 × 13.6–14.4 vs 6.4–7.9 × 8.9–10.7 μm) and spherical polar capsules. Differences between Z. ohlalae n. sp. and Z. cardinalis n. sp. are discussed above. Zschokkella jaimeae n. sp. and Z. balistoidi n. sp. differ in their polar capsule orientation, both to one side, rather than in opposite directions as observed for Z. ohlalae n. sp. Furthermore, Z. soleae, Z. jaimeae n. sp., Z. cardinalis n. sp. and Z. balistoidi n. sp. are all genetically distinct from Z. ohlalae n. sp. (see Table 4).
A single SSU rDNA sequence of 811 bases was generated for Z. ohlalae n. sp. (GenBank accession number KF179057). Due to mixed infections with Ceratomyxa species, only the primer combination of MyxospecF/Myxid1R could successfully amplify clean, uncontaminated SSU rDNA sequences. The sequence differs from the aligned sequences of other bivalvulidan species by 4–74 nucleotides (Table 4). Zschokkella ohlalae n. sp. is genetically most similar to Z. jaimeae n. sp. with 99.5% sequence identity. BLAST analysis revealed that Z. ohlalae n. sp. is genetically closest to a sequence of an undescribed species of Zschokkella taken from the gall bladder of Pseudanthias squamipinnis from the Red Sea (GenBank accession DQ333435) with maximum sequence identity of 99% over 100% coverage of the sequence.
Zschokkella compressis n. sp.
Type-host: Ostorhinchus compressus (Smith & Radcliffe) (Apogonidae).
Type-locality: Lagoon, off Lizard Island (14°40′S, 145°27′E), Great Barrier Reef, Queensland, Australia.
Site of infection: Gall bladder.
Prevalence: 2 of 5 fishes (40%).
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465680). DNA voucher (QM G465681).
Representative sequence: GenBank accession number KF179058.
Etymology: The species epithet compressis is in reference to the type-host species, O. compressus.
Description (Figs. 1U–V, 2)
[Mature spore measurements (n = 30) are shown in Table 1.] Mature spores ellipsoidal in sutural view, ovoid in valvular view. Small valvular protrusions associated with the tips of the polar capsules present in sutural view. Spore valves smooth. Suture line distinct, straight, bisecting the spore in sutural view. Polar capsules equal, pyriform, opening terminally in opposite directions on opposite sides of suture. Polar capsules lie along longest axis of the spore in valvular view and diagonally opposed in sutural view. Polar filament with 3 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Zschokkella compressis n. sp. is morphologically similar to Z. parasiluri Fujita, 1927, Z. mugilidae Kpatcha, Diebakate & Toguebaye, 1996, Z. psuedosciaena Sarkar, 1996, Z. embiotocidis Moser & Haldorson, 1976 and Z. bicarinatis n. sp. Zschokkella parasiluri, Z. mugilidae and Z. pseudosciaena are all distinguished from Z. compressis n. sp. by their spherical polar capsules. Zschokkella pseudosciaena is further distinguished by its site of infection (kidney tubules). Zschokkella parasiluri is recorded from a freshwater environment whereas the present species is from a marine environment. Furthermore, Z. compressis n. sp. is genetically distinct from Z. parasiluri with 88% sequence identity (BLAST results over 100% sequence coverage). Zschokkella compressis n. sp. has shorter spores than Z. mugilidae (8.5–11.3 vs 13.5–18 μm), Z. embiotocidis (8.5–11.3 vs 13–17 μm) and Z. bicarinatis n. sp. (8.5–11.3 vs 10.3–12.1 μm).
A single SSU rDNA sequence of 790 bases was generated for Z. compressis n. sp. (GenBank accession number KF179058). Due to mixed infections with Ceratomyxa spp., only the primer combination of MyxospecF/Myxid1R could successfully amplify clean, uncontaminated SSU rDNA sequences. The sequence differs from the aligned sequences of other bivalvulidan species by 7–71 nucleotides (Table 4). Zschokkella compressis n. sp. is genetically most similar to Z. jaimeae n. sp. and Z. bicarinatis n. sp. with 99% sequence identity. BLAST analysis revealed that Z. compressis n. sp. is genetically closest to a sequence of an undescribed species of Zschokkella taken from the gall bladder of Pseudanthias squamipinnis from the Red Sea (GenBank accession DQ333435) with maximum sequence identity of 99% over 100% coverage of the sequence.
Zschokkella jaimeae n. sp.
Type-host: Tylosurus gavialoides (Castelnau) (Belonidae).
Type-locality: Off Stradbroke Island (27°11′S, 153°15′E), Queensland, Australia.
Site of infection: Gall bladder.
Prevalence: 4 of 8 fishes (50%).
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465682).
Representative sequence: GenBank accession number KF179059.
Etymology: The species epithet jaimeae is in honour of Miss Jaime Heiniger in recognition of her support and encouragement of this work.
Description (Figs. 1W–X, 2)
[Mature spore measurements (n = 30) are shown in Table 1.] Mature spores ellipsoidal or ovoid. Longitudinal suture line slightly curved. Spore valves with 9–11 striations parallel to the suture line. Polar capsules equal, subspherical to pyriform, opening terminally on opposite sides of suture. Usually both polar capsules open to the anterior (or posterior) side of the spore. Polar filament with 4 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Zschokkella jaimeae n. sp. is morphologically similar to Z. balistoidi n. sp., Z. icterica Diamant & Paperna, 1992, Z. nova Klokacewa, 1914, Z. acheilognathi Kudo, 1916, Z. russelli Tripathi, 1948, Z. egyptica Ali, Abdel-Baki & Abdel-Ghaffar, 2007 and Z. australis Kovaleva, Rodjuk & Grudnev, 2002. Zschokkella jaimeae n. sp. has smaller polar capsules (1.6–2.8 × 1.7–3.6 μm) than Z. icterica (2.7–3.2 × 3.4–4.0 μm) and Z. australis (5.3–6.5 μm). Zschokkella australis is further distinguished by its smooth spore valves and greater number of polar filament coils (8–9 vs 4). Zschokkella icterica has a greater number of spore valve striations (12–13 vs 9–11) than Z. jaimeae n. sp. Zschokkella egyptica has larger spores (9.5–11 × 12.2–15.4 vs 6.4–7.8 × 9.7–13.8 μm) and larger, spherical polar capsules (4.2–5.2 vs 1.7–3.6 μm) than the present species. Zschokkella nova and Z. acheilognathi have spherical polar capsules while Z. jaimeae n. sp. has subspherical to pyriform polar capsules. Zschokkella russelli has larger spores (8.6–9.9 × 13.2–16.6 vs 6.4–7.8 × 9.7–13.8 μm) than Z. jaimeae n. sp. The spore measurements of Z. balistoidi n. sp. overlap with those of Z. jaimeae n. sp. but are slightly smaller for Z. balistoidi n. sp. Zschokkella balistoidi n. sp., Z. nova and Z. icterica are genetically distinct from Z. jaimeae n. sp. with 16, 48 and 141 nucleotides difference, respectively.
A single SSU rDNA sequence of 1,572 bases was generated for Z. jaimeae n. sp. (GenBank accession number KF179059). The sequence differs from the aligned sequences of other bivalvulidan species by 4–141 nucleotides (Table 4). Zschokkella jaimeae n. sp. is genetically most similar to Z. ohlalae n. sp. with 99.5% sequence identity.
Zschokkella bicarinatis n. sp.
Type-host: Grammatorcynus bicarinatus (Quoy & Gaimard) (Scombridae).
Type-locality: Lagoon, off Lizard Island (14°40′S, 145°27′E), Great Barrier Reef, Queensland, Australia.
Site of infection: Gall bladder.
Prevalence: 2 of 11 fishes (18%) off Lizard Island, 0 of 5 fishes off Heron Island.
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465683). DNA voucher (QM G465684).
Representative sequence: GenBank accession number KF179060.
Etymology: The species epithet bicarinatis is in reference to the type-host species, G. bicarinatus.
Description (Figs. 1Y–Z, 2)
[Mature spore measurements (n = 30) are shown in Table 1.] Mature spores ellipsoidal or ovoid. Small valvular protrusions associated with the tips of the polar capsules present. Spore valves smooth. Longitudinal suture line sometimes slightly curved. Polar capsules equal, pyriform, opening terminally in opposite directions. Polar filament with 3 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Zschokkella bicarinatis n. sp. is morphologically similar to Z. parasiluri, Z. mugilidae, Z. psuedosciaena, Z. embiotocidis and Z. compressis n. sp. Zschokkella bicarinatis n. sp. has smaller spores than Z. mugilidae (6.8–8.7 × 10.3–12.1 vs 9–13.5 × 13.5–18 μm) and Z. embiotocidis (6.8–8.7 × 10.3–12.1 vs 9.5–13 × 13–17 μm). Zschokkella embiotocidis is further distinguished by its wider polar capsules (3–4 vs 1.7–2.7 μm) and greater number of polar filament coils (6–8 vs 3). Zschokkella parasiluri has longer spores (15 vs 10.3–12.1 μm), greater number of polar filament coils (6–7 vs 3) and is genetically distinct from Z. bicarinatis n. sp. (96% sequence identity over 95% sequence coverage). Zschokkella pseudosciaena has narrower spores (5–7 vs 6.8–8.7 μm) and a different site of infection (kidney tubules) to Z. bicarinatis n. sp. Differences between Z. bicarinatis n. sp. and Z. compressis n. sp. are discussed above.
A single SSU rDNA sequence of 1,549 bases was generated for Z. bicarinatis n. sp. (GenBank accession number KF179060). The sequence differs from the aligned sequences of other bivalvulidan species by 7–141 nucleotides (Table 4). Zschokkella bicarinatis n. sp. is genetically most similar to Z. compressis n. sp. with 99% sequence identity.
Zschokkella balistoidi n. sp.
Type-host: Balistoides viridescens (Bloch & Schneider) (Balistidae).
Type-locality: Lagoon, off Lizard Island (14°40′S, 145°27′E), Great Barrier Reef, Queensland, Australia.
Site of infection: Gall bladder.
Prevalence: 1 of 1 fish (100%).
Material deposited: Syntype - Giemsa-stained, air-dried spores (QM G465685). DNA voucher (QM G465686).
Representative sequence: GenBank accession number KF179061.
Etymology: The species epithet balistoidi is in reference to the type-host genus, Balistoides.
Description (Figs. 1.1–2, 2)
[Mature spore measurements (n = 30) are shown in Table 1.] Mature spores ellipsoidal or elongate ovoid. Small valvular protrusions associated with the tips of the polar capsules. One spore valve often slightly flattened or concave. Longitudinal suture line slightly curved. Spore valves with 10–11 striations parallel to the suture line. Polar capsules equal, subspherical, opening terminally on opposite sides of suture. Polar capsules open towards the flattened or concave spore valve. Polar filament with 3 coils perpendicular to the long axis of the polar capsule. Sporoplasm fills cavity between polar capsules.
Remarks
Zschokkella balistoidi n. sp. is morphologically similar to Z. australis, Z. icterica, Z. pleomorpha Lom & Dyková, 1995, Z. russelli, Z. leptatherinae Su & White, 1995 and Z. jaimeae n. sp. Zschokkella leptatherinae is reported in fishes from off Tasmania, Australia, but differs from the present species in spore size (larger in all dimensions), spore shape (pointed ends) and also site of infection (hepatic ducts). Zschokkella pleomorpha has longer spores (14.4–17.6 vs 9.8–12.3 μm) and larger spherical polar capsules (4.2–5.2 vs 1.8–3.8 μm) than Z. balistoidi n. sp. Zschokkella australis has larger spores (10.6–12 × 16–20 vs 5.4–7.8 × 9.8–12.3 μm) and larger spherical polar capsules (5.3–6.7 vs 1.8–3.8 μm) than Z. balistoidi n. sp. Furthermore, Z. pleomorpha and Z. australis both have smooth spore valves. The spores of Z. russelli are larger (8.6–9.9 × 13.2–16.6 vs 5.4–7.8 × 9.8–12.3 μm) than Z. balistoidi n. sp. Zschokkella icterica is distinguished from Z. balistoidi n. sp. by a greater number of valve striations (12–13 vs 10–11) and pyriform polar capsules. Zschokkella icterica is also genetically distinct with 48 nucleotides difference with Z. balistoidi n. sp. Differences between Z. balistoidi n. sp. and Z. jaimeae n. sp. are discussed above.
A single SSU rDNA sequence of 1,530 bases was generated for Z. balistoidi n. sp. (GenBank accession number KF179061). The sequence differs from the aligned sequences of other bivalvulidan species by 12–139 nucleotides (Table 4). Zschokkella balistoidi n. sp. is genetically most similar to Z. jaimeae n. sp. with 98.9% sequence identity.
Phylogenetic analyses
Bayesian inference and maximum likelihood analyses resulted in phylograms of similar topology but with varying levels of clade support (see Fig. 3). The novel species of Ellipsomyxa formed a well-supported monophyletic clade with all other species of Ellipsomyxa. This relationship was also reflected in the neighbour-joining and parsimony analyses with moderate support (Fig. 4A). In all analyses the novel species of Zschokkella showed a close relationship with each other and formed a well-supported clade with all other species of Zschokkella reported from marine environments as well as another marine, gall bladder infecting species, Myxobolus spirosulcatus Maeno, Sorimachi, Ogawa & Kearn, 1995 (Figs. 3 and 4B). Myxidium milleri n. sp. showed a close relationship to M. incurvatum and Sinuolinea phyllopteryxa with high nodal support in all analyses (Figs. 3 , 4A). Isolates of M. maxi n. sp. clustered together and are sister to M. milleri n. sp., M. incurvatum and S. phyllopteryxa but this position is only comprehensively resolved in the Bayesian analyses (Figs. 3, 4A). Similarly, the position of M. scomberomori n. sp. is unresolved in all analyses (Figs. 3, 4A).
Phylogenetic tree resulting from Bayesian analysis inferred from the SSU rDNA dataset. Support values at branching points are listed as posterior probabilities from Bayesian analysis/Bootstrap values from Maximum likelihood analysis. Values above 95% for all analyses are indicated by a star. Values below 50% are indicated by a dash. Species from this study are shown in bold. Type-species of the genera Ellipsomyxa, Myxidium and Zschokkella are indicated by (Type) shown in bold
Phylogenetic trees resulting from Bayesian analysis inferred from the SSU rDNA datasets for novel species of Myxidium and Ellipsomyxa (A) and Zschokkella (B). Support values at branching points are listed as posterior probabilities from Bayesian analysis/Bootstrap values from Neighbour-joining analysis/Bootstrap values from Maximum Parsimony analysis. Values above 95% for all analyses are indicated by a star. Values below 50% are indicated by a dash. Species from this study are shown in bold
Discussion
The 13 novel species described here are distinct from all previously described species of Ellipsomyxa, Myxidium and Zschokkella. Furthermore, these descriptions provide the first record of Ellipsomyxa from Australian waters. Only two species of Myxidium and three species of Zschokkella have been previously described from fishes from Australian waters (O’Donoghue & Adlard, 2000; Gunter & Adlard, 2008). Myxidium queenslandicus was described from two species of Abudefduf Forsskål and M. therapon Johnston & Bancroft, 1918 from two species of Terapontidae (see O’Donoghue & Adlard, 2000; Gunter & Adlard, 2008). Zschokkella heronensis was described from species of Labridae and Chaetodontidae, Z. macrocapsula and Z. leptatherinae were both described from species of Atherinidae (see Moser et al., 1989; Su & White, 1994, 1995). New host records for a further three species and six unidentified species of Myxidium and a single unidentified species of Zschokkella have been recorded from Australian waters (see O’Donoghue & Adlard, 2000). The results from the current study indicate that species of Myxidium, Zschokkella and Ellipsomyxa are present in a diverse range of hosts but the overall incidence in Australian waters appears to be relatively low compared with that seen in other genera of myxosporeans (e.g. species of Ceratomyxa and Kudoa Meglitsch, 1947).
A combination of morphological and molecular data were used when assessing the generic placement of the novel species described here. The lack of clear taxonomic boundaries between Myxidium and Zschokkella is well documented (Diamant et al., 1994; Lom & Dyková, 2006). However, the three novel species of Myxidium described here are united by spores with a combination of morphological characters unique to the genus, i.e. sigmoid spores with pyriform polar capsules that open in opposite directions. The generic assignment of these species to Myxidium is further supported by a close phylogenetic relationship to other members of the genus. The six novel species of Zschokkella are all described as having ellipsoidal spores, a diagnostic character of the genus. Furthermore, the polar capsule orientation (both to one side) of two of the novel species, Z. balistoidi n. sp. and Z. jaimeae n. sp., is unique to Zschokkella (but a character that we consider polymorphic within the genus having some shared states with other genera). The remaining novel species assigned to Zschokkella have polar capsules oriented in opposite directions, a character shared by other species assigned to the genus and also a diagnostic character of other bivalvulidan genera (i.e. Myxidium, Sigmomyxa, Enteromyxum and Sphaeromyxa). The placement of these species into Zschokkella was on the basis of these morphological characters combined with the phylogenetic relatedness to already described species from the gall bladders of marine fishes.
The generic diagnosis of Ellipsomyxa has undergone several revisions since its proposal by Køie (2003). Unfortunately, the most recent revisions made by Gunter & Adlard (2010) and Køie & Karlsbakk (2009) were made independently of each other (the papers were accepted by their respective journals within four days of each other) resulting in neither revision comprehensively encompassing all species assigned to the genus. If both revisions are considered, species of Ellipsomyxa are defined by myxospores that are ellipsoidal or elongate in the direction perpendicular to the suture and thin-walled. The suture line may be straight, central, perpendicular, sinuous, or forming an acute angle to the thickness (longitudinal) axis. Mature spores contain two equal-sized spherical to pyriform polar capsules located on opposite sides of the spore and discharging in opposite directions. In lateral view, polar capsules are positioned either close to or at some distance from the sutural plane. In apical view, polar capsules are positioned at some distance from each other and either close to or at some distance from the spore surface. Plasmodia are monosporic or disporic in the gall bladders of marine fishes including elasmobranchs (Gunter & Adlard, 2010; Køie & Karlsbakk, 2009).
The three novel species of Ellipsomyxa described here are consistent with the diagnosis of the genus. However, most of the diagnostic characters of the genus now overlap with those of Zschokkella. Furthermore, in a recent study Whipps & Font (2013) questioned the nature of the suture line (a distinguishing character between species of Zschokkella and Ellipsomyxa) in previously described species and after close examination found that descriptions of the suture line were not consistent with the supplied photomicrographs. The close phylogenetic relationship of the novel species with all other species of Ellipsomyxa recorded from the gall bladder of marine fish supports the taxonomic placement of these species within the monophyletic Ellipsomyxa.
In phylogenetic analyses, species of Ellipsomyxa are sister to Myxidium queenslandicus and Sigmomyxa sphaerica. Whipps & Font (2013) suggested the possibility of future synonymy of Sigmomyxa and transfer of M. queenslandicus to Ellipsomyxa. Indeed, Sigmomyxa was originally proposed based on morphological and molecular distinction from Myxidium sensu stricto and no explanation of the distinction to Ellipsomyxa was given at the time of proposal, despite a close phylogenetic relationship between the genera (Karlsbakk & Køie, 2012). Furthermore, both species of Ellipsomyxa and Sigmomyxa use Nereis spp. as invertebrate hosts to complete their life-cycles (Køie et al., 2004; Rangel et al., 2009; Karlsbakk & Køie, 2012). Our results further question the validity of Sigmomyxa. The isolates identified here as M. maxi n. sp. show a close morphological affinity to the spore shape and polar capsule arrangement of spores of S. sphaerica, and, with the exception of the number of polar filament coils, fit the generic diagnosis of Sigmomyxa. However, these species are phylogenetically distinct (90.5% sequence identity) from sequences of M. maxi n. sp., which shows a close phylogenetic relationship with M. incurvatum and M. milleri n. sp., two species also with similar spore morphology. Clearly, the morphological characters used to define species of Sigmomyxa are not unique to the genus. As additional species are described and more sequences become available, including S. elmatboulii (Ali, Abdel-Baki & Sakran, 2006), the validity of genus Sigmomyxa should be re-evaluated. At such a time the generic status of Myxidium queenslandicus should also be reconsidered.
Molecular data are now an essential tool for taxonomic identification of closely related species. Bivalvulidan classification based on spore morphology alone to distinguish between closely related species is difficult due to the paucity of informative characters at the light microscopy level and the morphological plasticity of spores (Meglitsch, 1960; Sitja-Bobadilla & Alvarez-Pellitero, 1993; Heiniger et al., 2008; Gunter et al., 2009). Molecular data are usually combined with morphological and biological characters to provide evidence for the recognition of novelty of a species. However, several of the species presented here are truly cryptic (in that they are biologically and/or morphologically indistinguishable) and molecular data were critical for the taxonomic identification of these isolates.
Ellipsomyxa apogoni n. sp. and Zschokkella ohlalae n. sp. are biologically (both recorded from O. cyanosoma) and morphologically similar but are geographically (off Lizard Island and Ningaloo Reef, respectively) and genetically (see Fig. 3) distinct. Similarly, E. nigropunctatis n. sp. and E. arothroni n. sp. are biologically (both recorded from species of Arothron spp. off Lizard Island) and morphologically similar but genetically (three nucleotides difference, 99.8% identity) distinct. There is no set percentage of nucleotide difference which equates to the proposal of a species. Such a proposal clearly needs to be assessed for each individual isolate from a range of characters including morphological, biological and genetic data. As such, the genetic variation observed between these two Ellipsomyxa species, combined with the 0% intra-specific genetic variation observed for isolates of E. arothroni n. sp. (five isolates from two host species from off Lizard Island and Ningaloo Reef) and subtle morphological differences, provide compelling evidence for the discrimination of these two species. Similar levels of inter-specific genetic variation were observed between E. nigropunctatis n. sp. and E. apogoni n. sp. (two nucleotides difference, 99.7% identity), and Z. ohlalae n. sp. and Z. jaimeae n. sp. (four nucleotides difference, 99.5% identity) but these species are biologically (host) and morphologically distinct. Similar levels of inter-specific genetic variation have been previously reported for bivalvulidan species (0.5% in Ellipsomyxa and 1% in Ceratomyxa) (Gunter & Adlard, 2009; Køie & Karlsbakk, 2009). Interestingly, two morphologically and biologically (host and site of infection) distinct species assigned to different genera (and families), M. milleri n. sp. and Sinuolinea phyllopteryxa, are genetically the most similar with 99.9% sequence identity (one nucleotide difference). The genetic relatedness of these two otherwise distinct species suggests that SSU rDNA is not a suitable genetic marker for the distinction of these species and other genetic regions should be explored to provide clearer taxonomic resolution (e.g. LSU rDNA, elongation factor 2).
Phylogenetic analyses included 109 myxosporean sequences representing species from the majority of the clades identified by Fiala (2006), with the exclusion of Kudoa, Henneguya Thélohan, 1892 and Ceratomyxa. Previous myxosporean phylogenies have identified a major division between marine and freshwater lineages (Kent et al., 2001; Fiala, 2006; Bartošová et al., 2009). The novel species of Ellipsomyxa and Myxidium fell within the ‘marine’ clade of Fiala (2006). However, the novel species of Zschokkella fell within the ‘freshwater’ clade of Fiala (2006) and formed a well-supported clade with all other species of Zschokkella from the gall bladders of marine fish plus Myxobolus spirosulcatus, a species also found in the gall bladders (and central nervous system) of marine fishes. Fiala & Bartošová (2010) hypothesise that the ancestor of all myxosporeans was a marine species, with the entry into the freshwater environment occuring three times independently. They further hypothesise that some freshwater species then re-entered the marine environment (e.g. species of Sphaeromyxa) (Fiala & Bartošová, 2010). The phylogenetic placement of the novel species of Zschokkella described here within the ‘freshwater’ clade provide evidence consistent with a second group of species re-entering the marine environment.
Phylogenetic relationships among myxosporeans based on genetic data largely disagree with the current taxonomic classification based on spore morphology. The incongruence of morphological and biological characters, such as host, geographical location and tissue tropism, with phylogenies derived from genetic data is well documented (Kent et al., 2001; Holzer et al., 2004; Burger et al., 2007; Fiala & Bartošová, 2010). This incongruence is particularly evident for species of Myxidium and Zschokkella. Based on the present phylogenetic analyses no morphological or biological character was consistently correlated with genetic data. Previous studies have suggested the future synonymy of Zschokkella with Myxidium (see Diamant et al., 1994). However, based on the current phylogenies this would not resolve the paraphyly observed for both genera. Furthermore, current phylogenetic relationships reveal that the majority of the assigned species of Myxidium and Zschokkella are clearly distant from the type-species of their respective genera. Both type-species are described from the urinary bladders of their hosts, M. lieberkuehni from a freshwater fish and Z. hildae from a marine fish. To date, only one other sequence is available for a species of Myxidium or Zschokkella (Z. lophii) described from the urinary bladder of its host. Zschokkella lophii shows a close phylogenetic relationship with Z. hildae. As additional species are described and sequences made available (especially for species from the urinary bladder) the nomenclature of currently assigned species of these genera should be re-evaluated.
References
Apte, A., & Singh, S. (2007). AlleleID: A pathogen detection and identification system. Methods in Molecular Biology, 402, 329–345.
Auerbach, M. (1910). Die Cnidosporidien (Myxosporidien, Actinomyxidien, Microsporidien). Leipzig: Verlag von Dr. Werner Klinkhardt.
Bartošová, P., Fiala, I., & Václav, H. (2009). Concatenated SSU and LSU rDNA data confirm the main evolutionary trends within myxosporeans (Myxozoa: Myxosporea) and provide an effective tool for their molecular phylogenetics. Molecular Phylogenetics and Evolution, 53, 81–93.
Burger, M. A. A., Cribb, T. H., & Adlard, R. D. (2007). Patterns of relatedness in the Kudoidae with descriptions of Kudoa chaetodoni n. sp. and K. lethrini n. sp. (Myxosporea: Multivalvulida). Parasitology, 134, 669–681.
Bütschli, O. (1882). Myxosporidia. In: Bronn’s Klassen und Ordnungen des Tierreichs. 1. Protozoa. Leipzig: C. F. Winter, pp. 590–603.
Canning, E. U., Curry, A., Anderson, C. L., & Okamura, B. (1999). Ultrastructure of Myxidium trachinorum sp. nov. from the gallbladder of the lesser weever fish Echiichthys vipera. Parasitology Research, 85, 910–919.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17, 540–552.
Chen, C. L., & Ma, C. L. (1998). Fauna Sinica. Myxozoa. Myxosporea. Beijing: Science Press.
Diamant, A., Lom, J., & Dyková, I. (1994). Myxidium leei n. sp., a pathogenic myxosporean of cultured sea bream Sparus aurata. Diseases of Aquatic Organisms, 20, 137–141.
Drummond, A. J., & Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214.
Drummond, A. J., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Duran, C., et al. (2010). Geneious v5.6. Available from http://www.geneious.com.
Edgar, R. C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5, 113.
Eiras, J. C., Saraiva, A., Cruz, C. F., Santos, M. J., & Fiala, I. (2011). Synopsis of the species of Myxidium Bütschli, 1882 (Myxozoa: Myxosporea: Bivalvulida). Systematic Parasitology, 80, 81–116.
Fiala, I. (2006). The phylogeny of the Myxosporea (Myxozoa) based on small subunit ribosomal RNA gene analysis. International Journal for Parasitology, 36, 1,521–1,534.
Fiala, I., & Bartošová, P. (2010). History of myxozoan character evolution on the basis of rDNA and EF-2 data. BMC Evolutionary Biology, 10, 228.
Freeman, M. A., & Shinn, A. P. (2011). Myxosporean hyperparasites of gill monogeneans are basal to the Multivalvulida. Parasites & Vectors, 4, 220–231.
Gong, X., Lu, Y., & Wang, J. (2003). Description of two new Myxosporean species parasitic in freshwater fishes from the Yangtze River in China. Acta Protozoologica, 42, 239–243.
Gunter, N. L., & Adlard, R. D. (2008). Bivalvulidan (Myxozoa: Myxosporea) parasites of damselfishes with description of twelve novel species from Australia’s Great Barrier Reef. Parasitology, 135, 1,165–1,178.
Gunter, N. L., & Adlard, R. D. (2009). Seven new species of Ceratomyxa Thelohan, 1892 (Myxozoa) from the gall-bladders of serranid fishes from the Great Barrier Reef, Australia. Systematic Parasitology, 73, 1–11.
Gunter, N. L., & Adlard, R. D. (2010). The demise of Leptotheca Thélohan, 1895 (Myxozoa: Myxosporea: Ceratomyxidae) and assignment of its species to Ceratomyxa Thélohan, 1892 (Myxosporea: Ceratomyxidae), Ellipsomyxa Køie, 2003 (Myxosporea: Ceratomyxidae), Myxobolus Bütschli, 1882 and Sphaerospora Thélohan, 1892 (Myxosporea: Sphaerosporidae). Systematic Parasitology, 75, 81–104.
Gunter, N. L., Whipps, C. M., & Adlard, R. D. (2009). Ceratomyxa (Myxozoa: Bivalvulida): Robust taxon or genus of convenience? International Journal for Parasitology, 39, 1,395–1,405.
Hartigan, A., Fiala, I., Dyková, I., Rose, K., Phalen, D. N., & Šlapeta, J. (2012). New species of Myxosporea from frogs and resurrection of the genus Cystodiscus Lutz, 1889 for species with myxospores in gallbladders of amphibians. Parasitology, 139, 478–496.
Heiniger, H., & Adlard, R. D. (2012). Host specificity and local infection dynamics of Kudoa leptacanthae n. sp. (Multivalvulida: Kudoidae) from the pericardial cavity of two Zoramia spp. (Perciformes: Apogonidae) at Lizard Island lagoon, Queensland, Australia. Parasitology International, 61, 697–706.
Heiniger, H., Cribb, T. H., & Adlard, R. D. (2013). Intra-specific variation of Kudoa spp. (Myxosporea: Multivalvulida) from apogonid fishes (Perciformes), including the description of two new species, K. cheilodipteri n. sp. and K. cookii n. sp., from Australian waters. Systematic Parasitology, 84, 193–215.
Heiniger, H., Gunter, N. L., & Adlard, R. D. (2008). Relationships between four novel ceratomyxid parasites from the gall bladders of labrid fishes from Heron Island, Australia. Parasitology International, 57, 158–165.
Heiniger, H., Gunter, N. L., & Adlard, R. D. (2011). Re-establishment of the family Coccomyxidae and description of five novel species of Auerbachia and Coccomyxa (Myxosporea: Bivalvulida) parasites from Australian fishes. Parasitology, 138, 501–515.
Hine, P. M. (1975). Three new species of Myxidium (Protozoa: Myxosporidia) parasitic in Anguilla australis Richardson, 1848 and A. dieffenbachii Gray, 1842 in New Zealand. Journal of the Royal Society of New Zealand, 5, 153–161.
Holzer, A. S., Sommerville, C., & Wootten, R. (2004). Molecular relationships and phylogeny in a community of myxosporeans and actinosporeans based on their 18S rDNA sequences. International Journal for Parasitology, 34, 1,099–1,111.
Karlsbakk, E. (2001). Myxozoa. In: Costello, M. J., Emblow, C., & White, R. (Eds.), European register of marine species: A checklist of the marine species in Europe and a bibliography of guides to their identification, Vol. 50. Paris: Patrimoines Naturels, pp. 80–84.
Karlsbakk, E., & Køie, M. (2012). The marine myxosporean Sigmomyxa sphaerica (Thélohan, 1895) gen. n., comb. n. (syn. Myxidium sphaericum) from garfish (Belone belone (L)) uses the polychaete Neris pelagica L. as invertebrate host. Parasitology Research, 110, 211–218.
Kent, M. L., Andree, K. B., Bartholomew, J. L., El-Matbouli, M., Desser, S. S., Devlin, R. H., et al. (2001). Recent advances in our knowledge of the Myxozoa. Journal of Eukaryotic Microbiology, 48, 395–413.
Køie, M. (2003). Ellipsomyxa gobii gen. et sp. n. (Myxozoa: Ceratomyxidae) in the common goby Pomatoschistus microps (Teleostei: Gobiidae) from Denmark. Folia Parasitologica, 50, 269–271.
Køie, M., & Karlsbakk, E. (2009). Ellipsomyxa syngnathi sp. n. (Myxozoa, Myxosporea) in the pipefish Syngnathus typhle and S. rostellatus (Teleostei, Syngnathidae) from Denmark. Parasitology Research, 105, 1,611–1,616.
Køie, M., Whipps, C. M., & Kent, M. L. (2004). Ellipsomyxa gobii (Myxozoa: Ceratomyxidae) in the common goby Pomatoschistus microps (Teleostei: Gobiidae) uses Nereis spp. (Annelida: Polychaeta) as invertebrate hosts. Folia Parasitologica, 51, 14–18.
Kristmundsson, Á., & Freeman, M. A. (2013). Sphaeromyxids form part of a diverse group of myxosporeans infecting the hepatic biliary systems of a wide range of host organisms. Parasites & Vectors, 6, 51.
Lom, J., & Arthur, J. R. (1989). A guidline for the preparation of species descriptions in Myxosporea. Journal of Fish Diseases, 12, 151–156.
Lom, J., & Dyková, I. (2006). Myxozoan genera: Definition and notes on taxonomy, life cycle terminology and pathogenic species. Folia Parasitologica, 53, 1–36.
Maddison, D. R., & Maddison, W. P. (2005). MacClade 4: Analysis of phylogeny and character evolution. Version 4.08. Sunderland, MA: Sinauer Associates.
Meglitsch, P. A. (1960). Some coelozoic Myxosporidia from New Zealand fishes I. General, and Family Ceratomyxidae. Transactions of the Royal Society of New Zealand, 88, 265–356.
Miller, M. A., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (CGE). San Diego Supercomputer Centre, La Jolla, CA, USA, pp. 1–8.
Moser, M., Kent, M. L., & Dennis, D. (1989). Gall-bladder Myxosporea in coral-reef fishes from Heron Island, Australia. Australian Journal of Zoology, 37, 1–13.
O’Donoghue, P. J., & Adlard, R. D. (2000). Catalogue of protozoan parasites recorded in Australia. Memoirs of the Queensland Museum, 45, 1–163.
Palenzuela, O., Redondo, M. J., & Álvarez-Pellitero, P. (2002). Description of Enteromyxum scophthalmi gen. nov., sp. nov. (Myxozoa), an intestinal parasite of turbot (Scophthalmus maximus L.) using morphological and ribosomal RNA sequence data. Parasitology, 124, 369–379.
Posada, D. (2008). jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution, 25, 1,253–1,256.
Prunescu, C. C., Prunescu, P., Pucek, Z., & Lom, J. (2007). The first finding of myxosporean development from plasmodia to spores in terrestrial mammals: Soricimyxum fegati gen. et sp. n. (Myxozoa) from Sorex araneus (Soricomorpha). Folia Parasitologica, 54, 159–164.
Rangel, L. F., Santos, M. J., Cech, G., & Székely, C. (2009). Morphology, molecular data, and development of Zschokkella mugilis (Myxosporea, Bivalvulida) in a polychaete alternate host, Nereis diversicolor. Journal of Parasitology, 95, 561–569.
Ronquist, F., & Huelsenbeck, J. P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1,572–1,574.
Schulman, S. S. (1966). Miksosporidii fauny USSR. Moscow-Leningrad: Nauka (In Russian).
Sitja-Bobadilla, A., & Alvarez-Pellitero, P. (1993). Light and electron-microscopic description of Ceratomyxa labracis n. sp. and a redescription of C. diplodae (Myxosporea, Bivalvulida) from wild and cultered Mediterranean sea bass Dicentrarchus labrax (L) (Teleostei, Serranidae). Systematic Parasitology, 26, 215–223.
Stamatakis, A., Hoover, P., & Rougemont, J. (2008). A rapid bootstrapping algorithm for the RAxML web-servers. Systematic Biology, 57, 758–771.
Su, X. Q., & White, R. W. G. (1994). New myxosporeans (Myxozoa, Myxosporea) from marine fishes of Tasmania, Australia. Acta Protozoologica, 33, 251–259.
Su, X. Q., & White, R. W. G. (1995). A new myxosporean, Zschokkella leptatherinae n. sp. (Myxozoa: Myxiidae), from the hepatic ducts and gall bladder of Australian marine fishes. Systematic Parasitology, 32, 125–129.
Swofford, D. L. (2002). PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4,673–4,680.
Whipps, C. M., Adlard, R. D., Bryant, M. S., Lester, R. J. G., Findlay, V., & Kent, M. L. (2003). The first report of three Kudoa species from Eastern Australia: Kudoa thyrsites from mahi mahi (Coryphaena hippurus), Kudoa amamiensis and Kudoa minithyrsites n. sp. from sweeper (Pempheris ypsilychnus). Journal of Eukaryotic Microbiology, 50, 215–219.
Whipps, C. M., & Font, W. F. (2013). Interaction of two myxozoan parasites from naked goby Gobiosoma bosc, in Lake Pontchartrain, Louisiana. Journal of Parasitology, 99, 441–447.
Yemmen, C., Marton, S., Bahri, S., & Eszterbauer, E. (2013). Morphology, seasonality and phylogeny of Zschokkella solea sp. n. (Myxozoa, Myxosporea) parasite of Solea solea (L.) (Pleuronectiformes, Solidae) from Ghar El Melh Lagoon. Tunisia. Journal of Fish Diseases, 36, 871–879.
Acknowledgements
This study was funded by, and is a contribution from, the Australian node of the CReefs global research initiative (grant number 209/29), a project generously sponsored by BHP Billiton in partnership with The Great Barrier Reef Foundation, the Australian Institute of Marine Science, the Australian Biological Resources Study and the Alfred P. Sloan Foundation. CReefs is a field program of the Census of Marine Life. We thank Dr Terrence Miller for technical advice, phylogenetic discussions and assistance in field collections, Jeff Johnson for assistance with fish identification, Dr Tom Cribb for discussion on species boundaries and phylogenetics and assistance in field collections and Ricky Gleeson who assisted in field collection, and the staff at Heron Island and Lizard Island Research Stations for logistical support in the field.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Heiniger, H., Adlard, R.D. Relatedness of novel species of Myxidium Bütschli, 1882, Zschokkella Auerbach, 1910 and Ellipsomyxa Køie, 2003 (Myxosporea: Bivalvulida) from the gall bladders of marine fishes (Teleostei) from Australian waters. Syst Parasitol 87, 47–72 (2014). https://doi.org/10.1007/s11230-013-9454-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-013-9454-3