Abstract
Kudoa spp. from the musculature and intestinal mucosa of species of the teleost family Apogonidae were examined for their taxonomic identity. Two novel species are characterised: Kudoa cheilodipteri n. sp. from the musculature of Cheilodipterus quinquelineatus Cuvier, Ostorhinchus cyanosoma (Bleeker) and O. aureus (Lacépède); and Kudoa cookii n. sp. from the submucosa of the intestines of O. cookii (Macleay) only. Both species are characterised using morphology, small subunit ribosomal DNA (SSU rDNA), large subunit ribosomal DNA (LSU rDNA), and biological characters. Three new host records, O. cyanosoma, O. aureus and Apogon doederleini, and associated geographical, morphological and genetic data are also provided for Kudoa whippsi Burger & Adlard, 2010. Morphological and molecular intra-specific variation of all isolates assigned to K. whippsi is also examined. Phylogenetic analyses further support the idea that tissue tropism is a distinguishing character between morphologically similar species; species reported here display close relatedness to morphologically similar species infecting the same tissue within their hosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The teleost family Apogonidae (Cardinalfishes) forms a major component of coral reef fish assemblages both in their species richness and in their numerical abundance (Marnane, 2000). Apogonids are mainly marine, inshore reef inhabitants, feeding nocturnally on benthic and pelagic plankton, invertebrates and small fish (Kuiter & Kozawa, 1999). Day resting sites for apogonids include either caves and crevices (mostly solitary or small groups of a single species) or around or above branching corals (often large multi-species aggregations) (Greenfield & Johnson, 1990). Apogonids are generally small (30–250 mm standard length), laterally compressed fish with relatively large mouths. The family is one of few to provide extended parental care through mouth brooding (Thacker & Roje, 2009).
A diverse fauna of parasitic infections has been reported in apogonids from Australian waters, including microsporidians (Vagelli et al., 2005), monogeneans (Ernst et al., 2001), digeneans (Bott & Cribb, 2005; Aken’Ova et al., 2006; Bray & Cribb, 1989) and isopods (Fogelman & Grutter, 2008; Fogelman et al., 2009; Jones et al., 2007; Nagel & Grutter, 2007; Bruce, 1987). A cymothoid isopod, Anilocra apogonae Bruce, 1986, has been reported to cause castration of its host, Cheilodipterus quinquelineatus (see Fogelman et al., 2009). Within the freshwater genus, Glossamia, a gorgoderid digenean has also been reported (Cribb, 1987).
Only five myxosporean infections have been reported in apogonids, four belonging to the Kudoidae Meglitsch, 1960 and one to the Ceratomyxidae Doflein, 1899. Kudoa iwatai Egusa & Shiomitsu, 1983 has been recorded from Apogon fleurieu in the Red Sea off Israel (Diamant et al., 2005), K. gunterae Burger & Adlard, 2010 and K. whippsi Burger & Adlard, 2010 were both recorded from Ostorhinchus properuptus (as Apogon properuptus) on the Great Barrier Reef, Australia (Burger & Adlard, 2010a) and K. thalassomi Adlard, Bryant, Whipps & Kent, 2005 was recorded from Cheilodipterus macrodon also on the Great Barrier Reef, Australia (Burger & Adlard, 2011). Ceratomyxa apogoni (Narasimhamurti, Kalavati, Anuradha & Padma Dorothy, 1990) [as Leptotheca apogoni] has been recorded from Ostorhinchus aureus (as Apogon aureus) from the Bay of Bengal, India (Narasimhamurti et al., 1990).
The family Kudoidae consists of a single genus, Kudoa Meglitsch, 1947, characterised by four or more spore valves and polar capsules (Whipps et al., 2004). Currently there are 86 species of Kudoa described from diverse marine and estuarine fishes worldwide (e.g. Moran et al., 1999; Lom & Dykova, 2006; Kent et al., 2001). Species are mainly histozoic in the somatic musculature of their hosts; however, they can also be found in the heart, intestines, gills, brain, kidney and gall-bladder. Although most species appear to have relatively little effect on their hosts, others are significant pathogens, causing encephalomyelitis (Grossel et al., 2003), or have significant commercial impact through post-mortem myoliquefaction and the production of macroscopic cysts (Langdon, 1991; Yokoyama et al., 2004; Egusa & Nakajima, 1980; Matsumoto, 1954; Iversen & Van-Meter, 1967). This paper describes two new species of Kudoa infecting four species of apogonids from Australian waters. It also reports new host records for K. whippsi from the muscle of apogonids.
Materials and methods
Sample collection
Apogonids were collected using localised sprays of clove oil anaesthetic and microspear off Heron Island (23°26′S, 151°54′E) and Lizard Island (14°40′S, 145°27′E), Great Barrier Reef, Queensland, and off Point Cloates (22°40′S, 113°41′E), Ningaloo Reef, Western Australia. Fish were killed by neural pithing. Apogonid identification was performed with reference to Kuiter & Kozawa (1999) and Fishbase (www.fishbase.org). Currently valid host names were taken from the California Academy of Sciences Catalogue of Fishes (http://research.calacademy.org/ichthyology/catalog/fishcatmain.asp). The somatic muscle of each fish was examined for the presence of kudoid infections using the preparation methods described by Burger & Adlard (2010a). Intestinal walls were examined by first excising the intestine, which was then sliced open and examined for cysts or abnormalities under a dissecting microscope. Any cysts or abnormalities were removed and placed on a glass microscope slide and covered with a glass coverslip. Intestinal and muscle preparations were examined under an Olympus BH2 microscope at ×400 magnification for the presence of kudoid spores. Infected tissue samples were subdivided and preserved in 95% ethanol for molecular analysis, 10% buffered neutral formalin for histological sectioning (samples were transferred to 70% ethanol before sectioning) and frozen for morphological analysis. Where material was limited, imaging for morphological analysis (details below) was performed in the field and priority went to ethanol preserved material for molecular analysis.
Morphological analysis
Preparation of slides was performed as described above. Measurements of spores followed the guidelines of Lom & Arthur (1989) and also followed further recommendations by Burger & Adlard (2010a). Images of 30 spores (unless otherwise stated) were taken with an Olympus BH2 microscope at ×400 or ×1,000 magnification using a Nikon Digital Sight DS-LI digital camera (Nikon Corporation, Japan). Measurements were taken from microphotographs using the measuring tool in the Nikon NIS Elements software (Nikon Corporation, Japan) calibrated against a stage micrometer. Mean measurements and their standard deviation were calculated for each spore dimension, allowing characterisation of each isolate. All measurements are given in micrometres.
Principal component analyses (PCA) were performed on all morphometric data using the software PAST version 2.10 (Hammer et al., 2001). Scatterplots with 95% confidence ellipses were generated using variant-covariant matrices.
DNA analysis
Spore suspensions from ethanol preserved material were performed as for the frozen material described above. Spore suspensions were first examined under a microscope at ×400 magnification for the presence of spores. Suspensions were then concentrated by centrifugation at 15,700 g for 10 min and the ethanol supernatant removed. DNA was extracted from the resulting pellet as per the recommended protocol accompanying the QIAgen DNeasy Kit (QIAGEN Inc., Valencia, CA, USA). Small subunit ribosomal DNA (SSU rDNA) was amplified by PCR using the primers 18e 5′ CTG GTT GAT CCT GCC AGT (Hillis and Dixon 1991), MbSeq1r 5′ CAA TCC TAT CAA TGT CTG GAC CTG (Burger et al. 2007), Kud2f 5′ TGA ATG TTA TAG CAT GGA A (Whipps et al., 2003b) and 18R 5′ CTA CGG AAA CCT TGT TACG (Whipps et al., 2003a). Large subunit rDNA (LSU rDNA) was amplified by PCR using the primers Kt28S1F5′ CAA GAC TAC CTG CTG ACC (Whipps et al., 2004) and 28S1R 5′ GTG TTT CAA GAC GGG TCG (Whipps et al., 2004). Reaction mixtures (25 μl) were prepared as described by Heiniger et al. (2008). PCR amplification was performed in a cp2-01 thermocycler (Corbett Research, Australia) using the cycling parameters of an initial denaturation at 94°C for 2 min 30 s, followed by 30 cycles of denaturation at 94°C for 20 s, annealing at 50°C for 30 s and extension at 72°C for 60 s with a final extension at 72°C for 10 min for SSU rDNA reactions. Cycling parameters for LSU rDNA reactions were as described above, with the following exceptions: performed using 25 cycles and an annealing temperature of 56°C. Amplified PCR products were purified using a QIAgen PCR purification kit as per the recommended protocol (QIAgen Inc., Valencia, CA, USA). Purified DNA was sent to the Australian Genome Research Facility, The University of Queensland, Australia, for sequence determination.
Phylogenetic analysis
The rDNA regions from the taxa sequenced in this study were edited using Geneious Pro version 5.4.6 (Drummond et al., 2010). Selected SSU and LSU rDNA sequences belonging to species of Kudoa were downloaded from GenBank for use in phylogenetic analyses. Unicapsula spp. were used as outgroup taxa in all analyses. All sequences generated in this study were lodged in GenBank. An alignment of all taxa included here was produced using Muscle version 3.7 (Edgar, 2004) using the Clustal W algorithm (Thompson et al., 1994) with UPGMB parameters for all iterations on the CIPRES portal (Miller et al., 2010). The resulting alignment was exported as fasta and nexus files, edited by eye and trimmed using MacClade version 4.08 (Maddison & Maddison, 2005). This produced alignments of 1,542 and 903 bases for SSU and LSU rDNA, respectively. These alignments were used to conduct all phylogenetic analyses.
Neighbour-joining and parsimony analyses were conducted using the default parameters to construct trees using PAUP* 4.0b10 (Swofford, 2002). The strength of resultant relationships was tested by bootstrap analyses with 10,000 replicates. Parsimony analyses employed a heuristic search with 50 repetitions of random sequence addition and tree bisection and reconnection branch swapping. Maximum likelihood analyses were conducted using the RAxML algorithm (Stamatakis et al., 2008) on the CIPRES portal with the gamma rate model of heterogeneity and maximum likelihood search estimating the proportion of invariable sites parameters. Nodal support was inferred based on 100 bootstrap replicates. Bayesian analyses were conducted using MrBayes version 3.1.2 (Ronquist & Huelsenbeck, 2003). The software jModelTest version 0.1.1 (Posada, 2008) was used to estimate the best substitution model for the SSU and LSU rDNA datasets. Bayesian inference analysis was conducted on the SSU rDNA dataset using the TVMef+I+G model and on the LSU rDNA dataset using the TVM+G model predicted as the best estimator by the Akaike Information Criterion (AIC) in jModelTest. Bayesian inference analyses were run over 10,000,000 generations (ngen = 10,000,000) with 2 runs each containing 4 simultaneous Markov Chain Monte Carlo (MCMC) chains (nchains = 4) and every 1,000th tree saved (samplefreq = 1,000). Bayesian analyses used the following parameters: nst = 6; rates = invgamma and gamma for SSU and LSU rDNA datasets respectively; ngammacat = 4; the MCMC parameters were left at the default settings; and the priors parameters of the combined dataset were set to ratepr = fixed. Samples of substitution model parameters, and tree and branch lengths were summarised using the parameters ‘sump burnin = 3000’ and ‘sumt burnin = 3000’. These ‘burnin’ parameters were chosen because the log likelihood scores ‘plateaued’ well before 3,000,000 replicates in the Bayesian inference analyses.
Neighbour-joining analysis was also performed as described above on a separate dataset containing all LSU rDNA isolates of Kudoa whippsi. This analysis was to examine the relatedness of isolates and also to explore possible biological correlates within this clade. This dataset was aligned, edited and trimmed as described above, resulting in an alignment of 705 bases.
Pairwise differences were also analysed using PAUP* 4.0b10 to determine total nucleotide distance and percentage differences. Pairwise difference analyses were performed using datasets containing the new species from this study and the most closely related sequences as determined by BLAST analyses. These datasets were aligned, edited and trimmed as described above. The resulting alignments were 1,468 and 756 bases for the SSU and LSU rDNA datasets for comparison of muscle infecting species, and 1,541 and 683 bases for the SSU and LSU rDNA datasets for comparison of Kudoa cookii n. sp.
Histology
Sections (5 μm thick) were cut from 10% formalin-fixed samples using standard histological techniques. Slides were stained with either haematoxylin and eosin (H & E), to assist in identifying the presence of any immunological response, or Giemsa and eosin, to differentially stain the polar capsules. Coverslips were applied using DePeX (BDH, England). Digital, light microphotographs of the sections were taken at ×100, ×200 and ×400 magnification, as described above.
Results
Since 2005, 725 individual apogonids were examined for the presence of kudoid infections at four localities, Heron and Lizard Islands on the Great Barrier Reef, Ningaloo Reef off Western Australia and Moreton Bay off Southern Queensland (Table 1). A total of 24 apogonid species belonging to nine genera were sampled. This survey identified 29 isolates (eight host/parasite/location combinations) of Kudoa infecting either the skeletal muscle or intestinal wall from three locations, off Heron and Lizard Islands and Ningaloo Reef. Due to the low intensity of infection and limited material, the two infections of Kudoa in Cheilodipterus intermedius are not described in this publication. The survey of apogonid fish from Australian waters revealed that only 4% were infected with muscle or intestinal dwelling kudoids (Table 1).
Phylum Myxozoa
Class Myxosporea
Order Multivalvulida
Family Kudoidae Meglitsch, 1960
Genus Kudoa Meglitsch, 1947
Kudoa whippsi Burger & Adlard, 2010
Type-host: Pomacentridae: Abudefduf bengalensis (Bloch) (Bengal sergeant).
Type-locality: Off Heron Island, the Great Barrier Reef, Queensland, Australia.
Other hosts: Pomacentridae: Abudefduf whitleyi Allen & Robertson (Whitley’s sergeant); Acanthochromis polyacanthus (Bleeker) (Spiny puller); Amphiprion akindynos Allen (Barrier reef anemonefish); A. melanopus Bleeker (Fire clownfish); Chromis viridis (Cuvier) (Blue green damselfish); Neoglyphidodon melas (Cuvier) (Bowtie damselfish); Pomacentrus chrysurus Cuvier (Whitetail damsel). Apogonidae: Ostorhinchus properuptus (Whitley) [as Apogon properuptus] (Southern orange-lined cardinalfish).
Other localities: Off Lizard Island, the Great Barrier Reef, Queensland, Australia.
Site of infection: Histozoic, intracellular within myofibrils of skeletal muscle cells.
New hosts and localities: Apogonidae: Ostorhinchus cyanosoma (Bleeker) (Yellowstriped cardinalfish), 2 of 4 (50%) off Lizard Island, 1 of 23 (4.35%) off Ningaloo Reef; O. aureus (Lacépède) (Ring-tailed cardinalfish), 1 of 14 (7.14%) off Ningaloo Reef; and Apogon doederleini Jordan & Snyder (Doederlein’s cardinalfish), 1 from 2 (50%) off Lizard Island, none from 14 off Heron Island.
Site: Histozoic, intracellular within myofibrils of skeletal muscle cells. Not found in other tissues examined (brain, gall-bladder, heart, intestinal wall). No inflammatory response evident.
New material: Giemsa and eosin stained histological sections (ex Ostorhinchus cyanosoma, G465514), haematoxylin and eosin stained histological sections (ex Apogon doederleini, G465515; ex O. cyanosoma, G465516), Giemsa-stained, air-dried spores (ex A. doederleini, G465517–G465518; ex O. cyanosoma, G465519; ex O. aureus, G465520) and DNA vouchers (ex A. doederleini, G465521–G465522; ex O. cyanosoma, G465523) deposited in the collections of the Queensland Museum, Brisbane, Australia. GenBank Accession numbers: ex O. cyanosoma and O. aureus from Ningaloo Reef, JX090292 and ex O. cyanosoma and A. doederleini from Lizard Island, JX090293 for SSU; and ex O. cyanosoma and O. aureus from Ningaloo Reef, JX090296, ex A. doederleini from Lizard Island, JX090297 and ex O. cyanosoma from Lizard Island, JX090298 for LSU (Figs. 5–7).
Remarks (Figs. 1C–F,H, 2, 3–7; Tables 2, 3)
Morphological affinities
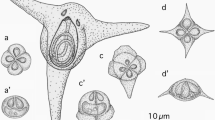
A–D. Photomicrographs of spores: A,B. Kudoa cheilodipteri n. sp. ex Cheilodipterus quinquelineatus in apical view (A) and side view (B); C,D. K. whippsi ex Apogon doederleini in apical view (C) and side view (D). E,F. Photomicrographs of histological sections stained with Giemsa and eosin: E. Kudoa cheilodipteri n. sp. ex Cheilodipterus quinquelineatus; F. K. whippsi ex Ostorhinchus cyanosoma. Scale-bars: A–D, 5 μm; E–F, 50 μm
Principal component analysis comparison of morphometric data in apical view with 95% confidence ellipses from: Kudoa cheilodipteri n. sp. ex Cheilodipterus quinquelineatus and Ostorhinchus cyanosoma (data points = filled triangles); K. whippsi ex Apogon doederleini (data points = crosses); and K. whippsi ex O. cyanosoma from off Lizard Island (data points = filled squares)
Principal component analysis comparison of morphometric data in apical view with 95% confidence ellipses from: Kudoa whippsi isolates from this study ex Apogon doederleini (data points = crosses) and ex Ostorhinchus cyanosoma from off Lizard Island (data points = filled squares); K. whippsi isolates from the original description by Burger & Adlard (2010a) ex Acanthochromis polyacanthus, Amphiprion akindynos and O. properuptus (data points = hollow ovals)
Phylogenetic tree resulting from Neighbour-joining analysis inferred from the LSU rDNA dataset of all available Kudoa whippsi sequences. Bootstrap support values are shown at branching points. Species from this study are shown in bold. GenBank accession number follows each taxon. Locality of each K. whippsi isolate are listed to the right with the following abbreviations: LI = Lizard Island; HI = Heron Island; NR = Ningaloo Reef
Phylogenetic tree resulting from Bayesian analysis inferred from the SSU rDNA dataset. Support values at branching points are listed as: Posterior probabilities (PP) from Bayesian analysis/Bootstrap values from Maximum likelihood analysis/Bootstrap values from parsimony analysis/Bootstrap values from Neighbour-joining analysis. Values below 50% are indicated by dashes. Values of 100% for all analyses are indicated by a star. Species from this study are shown in bold. GenBank accession number follows each taxon
Phylogenetic tree resulting from Bayesian analysis inferred from the LSU rDNA dataset. Support values at branching points are listed as: Posterior probabilities (PP) from Bayesian analysis/Bootstrap values from Maximum likelihood analysis/Bootstrap values from parsimony analysis/Bootstrap values from Neighbour-joining analysis. Values below 50% are indicated by dashes. Values of 100% for all analyses are indicated by a star. Species from this study are shown in bold. GenBank accession number follows each taxon
Plasmodia and spore shape fit the original description of the species Kudoa whippsi Burger & Adlard, 2009. Mature spore measurements taken from infections in Apogon doederleini (n = 30), and Ostorhinchus cyanosoma from Lizard Island (n = 30) and Ningaloo Reef (n = 2 and 25 in apical and side view respectively) are given in Table 2. There is broad morphological overlap between isolates of this study and those from the original description (isolates from three hosts are pooled as there was negligible differences found in analyses not shown here) (Table 2; Fig. 4). Small morphological differences between the isolates reported here and those from the original description include a longer large polar capsule and extended ranges for spore length for the new material. The isolates from A. doederleini and O. cyanosoma from Lizard Island also have a slightly smaller spore width and the isolate from O. cyanosoma from Ningaloo Reef has a thicker spore.
Molecular affinities
SSU rDNA sequences were generated for four host/parasite/locality combinations, O. cyanosoma from both Lizard Island and Ningaloo Reef, A. doederleini from Lizard Island and O. aureus from Ningaloo Reef. The two Lizard Island sequences were identical as were the two Ningaloo Reef sequences, and these differed from each other at five nucleotides (99.7% sequence identity) (Table 3A). LSU rDNA sequences were generated for the same four host/parasite/combinations as for SSU rDNA. Again the two Ningaloo Reef isolates were identical. There are 3–11 nucleotide differences (98.3–99.6% sequence identity) between the three LSU rDNA sequences (Table 3B). In SSU rDNA there is 99.2–100% genetic similarity (0–12 nucleotide differences) between the sequences generated here and previously reported sequences of K. whippsi (see Burger & Adlard, 2010a). Maximum genetic sequence identity (100%) was found between the SSU rDNA sequence already available on GenBank from Ostorhinchus properuptus and the consensus sequence generated in this study from O. cyanosoma and A. doederleini from Lizard Island. In LSU rDNA there is 98.2–99.9% genetic similarity (1–12 nucleotide differences) between the sequences generated here and sequences of K. whippsi already available on GenBank. These levels of variation double those originally reported for intra-specific variation of K. whippsi isolates in SSU rDNA (0–6 nucleotides), but are within the level reported for LSU rDNA (0–13 nucleotides) (Burger & Adlard, 2010a). Neighbour-joining analysis of all LSU rDNA sequences of K. whippsi show no host (species or family) or geographical patterns within the ‘whippsi’ clade (Fig. 5). In all phylogenetic analyses the sequences generated in this study formed a well-supported clade with all previously reported sequences of K. whippsi (Figs. 6, 7).
Kudoa cheilodipteri n. sp.
Type-host: Cheilodipterus quinquelineatus Cuvier (Five-lined cardinalfish), Apogonidae.
Type-locality: Off Lizard Island, Great Barrier Reef, Queensland (14°40′S, 145°27′E).
Prevalence: 7 of 34 (20.59%), off Lizard Island; none of 28, off Heron Island.
Other hosts: Apogonidae: Ostorhinchus cyanosoma (Bleeker) (Yellowstriped cardinalfish), 2 of 23 (8.7%) off Ningaloo Reef, none of 4 off Lizard Island; O. aureus (Lacépède) (Ring-tailed cardinalfish), 2 of 14 (14.29%), off Ningaloo Reef.
Site: Histozoic, intracellular within myofibrils of skeletal muscle cells. Not found in other organs examined (brain, gall-bladder, heart, intestinal wall). No inflammatory response evident.
Type-material: Syntypes - Giemsa and eosin stained histological sections (ex C. quinquelineatus, G465524), haematoxylin and eosin stained histological sections (ex C. quinquelineatus, G465525). Vouchers - Giemsa-stained, air-dried spores (ex C. quinquelineatus, G465526; ex O. aureus, G465527–G465528; ex O. cyanosoma, G465529–G465530), and DNA vouchers (ex C. quinquelineatus, G465531; ex O. cyanosoma, G465532) deposited in the collections of the Queensland Museum, Brisbane, Australia. GenBank Accession numbers: ex C. quinquelineatus from Lizard Island, O. cyanosoma and O. aureus from Ningaloo Reef, JX090295 for SSU; and ex C. quinquelineatus from Lizard Island, JX090299 and ex O. cyanosoma and O. aureus from Ningaloo Reef, JX090300 for LSU (see Figs. 6, 7).
Etymology: Named after the genus of the type-host, Cheilodipterus; used as a substantive in the genitive case.
Description (Figs. 1A–B, 2, 3, 6, 7; Tables 2, 3)
Plasmodia. Microscopic, polysporic. Intracellular within muscle fibres. Spores of uniform development. Spores. Spores typical of Kudoa. Mature spores stellate in apical view, with bilateral symmetry in sagittal plane only, pyramidal in side view. Four polar capsules convergent, pyriform, with 1 much larger than other 3. No other morphotypes present. Polar filament indistinct. Spore measurements taken from infections in C. quinquelineatus (n = 73 and 55 in apical and side view, respectively) and O. cyanosoma (n = 63 and 65 in apical and side view, respectively) given in Table 2.
Remarks
Morphological affinities
Kudoa cheilodipteri n. sp. resembles K. whippsi, and both species have been found in overlapping host (Ostorhinchus cyanosoma and O. aureus) and geographical ranges (off Ningaloo Reef and Lizard Island) (Table 2; Fig. 3). The new species can be differentiated from K. whippsi by having thicker and longer spores and wider and larger polar capsules. It is also superficially similar in size to K. gunterae Burger & Adlard, 2010, K. lateolabracis Yokoyama, Whipps, Kent, Mizuno, & Kawakami, 2004, K. cruciformum (Matsumoto, 1954), K. thyrsites (Gilchrist, 1923) and K. minithyrsites Whipps, Adlard, Bryant, Lester, Findlay & Kent 2003 (Table 2). Spores of K. cheilodipteri are thicker and shorter and with wider polar capsules than those of K. gunterae, and are narrower in width than those of K. lateolabracis, K. thyrsites and K. cruciformum. Both K. lateolabracis and K. thyrsites can further be distinguished by having thicker spores. The longer spores and longer large polar capsule are additional diagnostic features of K. cruciformum. The new species is differentiated from K. minithyrsites by its longer spore and larger polar capsules.
Molecular affinities
Identical SSU rDNA sequences of 1,636 bases were generated from three fish species (9 isolates from individual fish with 100% identity) for Kudoa cheilodipteri n. sp. Two LSU rDNA sequences (773 and 774 bases) from three fish species (7 isolates from individual fish) were also generated. There are nine nucleotide differences (98.7% sequence identity) between both LSU rDNA sequences. The most genetically similar species, K. thalassomi Adlard, Bryant, Whipps & Kent, 2005, differs from K. cheilodipteri by 46 nucleotides (96.8% sequence identity) in SSU rDNA and by 59–61 nucleotides (89.7–90.1% sequence identity) in LSU rDNA (Tables 3). The phylogenetic position of K. cheilodipteri is unresolved in all SSU rDNA analyses. In LSU rDNA analyses both K. cheilodipteri sequences cluster together with high posterior probability (1.00) and bootstrap support (100%) in all analyses, but once again the position of this species is unresolved in all analyses, with the exception of the Bayesian analysis, where it showed a close relationship with a clade made up of K. thalassomi and brain infecting kudoid species (Fig. 7).
Kudoa cookii n. sp.
Type-host: Ostorhinchus cookii (Macleay) (Cook’s cardinalfish), Apogonidae.
Type-locality: Off Heron Island, Great Barrier Reef, Queensland (23°26′S, 151°54′E).
Prevalence: 11 from 35 (31.42%) Heron Island; none from 8 Lizard Island; none from 2 Ningaloo Reef.
Site: Submucosa of the intestines. Not found in other organs examined (muscle, brain, gall-bladder, heart). No inflammatory response evident.
Type-material: Syntypes - Giemsa and eosin stained histological sections (G465538), haematoxylin and eosin stained histological sections (G465539); Vouchers - Giemsa-stained, air-dried spores (G465533–G465535), and DNA vouchers (G465536–G465537) deposited in the collections of the Queensland Museum, Brisbane, Australia. GenBank Accession numbers: JX090294 for SSU; and JX090301, JX090302 and JX090303 for LSU (see Figs. 6, 7).
Etymology: Named after the type-host species, cookii, used as a substantive in the genitive case.
Description (Figs. 2, 6, 7, 8A–D; Tables 4, 5)
Plasmodia. Microscopic, polysporic. Attached to submucosa of intestines. Cysts white and ovoid, 1.3–1.5 × 1 mm in size. Spores. Spores typical of Kudoa. Mature spores quadrate with 4 equal rounded spore valves in apical view and umbonate ellipsoidal in side view. Spores with tetramerous radial symmetry. Four polar capsules equal, convergent and pyriform. No other morphotypes present. Polar filament indistinct. Spore measurements (n = 67 and 90 in apical and side views, respectively) given in Table 4.
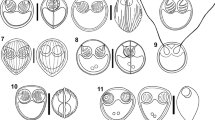
Light photomicrographs of Kudoa cookii n. sp. from the submucosa of the intestines of Ostorhinchus cookii: A. Microscopic cysts (indicated by arrows) visible on the submucosa of the intestines; B,C. Histological sections stained with Giemsa and eosin at ×40 (B) and ×10 (C); D,E. Fresh spores in apical view (D) and side view (E). Scale-bars: A, 5 mm; B, 10 μm; C, 100 μm; D,E, 5 μm
Remarks
Morphological affinities
Kudoa cookii n. sp. closely resembles K. ciliatae Lom, Rohde & Dyková, 1992, K. kenti Burger & Adlard, 2010, K. sphyraeni Narasimhamurti & Kalavati, 1979, K. cascasia Sarkar & Chaudhury, 1996, K. iwatai Egusa & Shiomitsu, 1983 and K. crumena Iversen & Van Meter, 1967 (Table 4). The new species can be distinguished from K. ciliatae by differences in the spore width and thickness. It can be distinguished from K. kenti, K. sphyraeni and K. iwatai by its longer spore, and differentiated further by its longer polar capsules from K. sphyraeni and K. iwatai. The latter species also has narrower polar capsules. Both K. kenti and K. crumena can also be distinguished from K. cookii by the site of infection (skeletal muscle). As compared with K. cookii, K. crumena has shorter and narrower polar capsules.
Molecular affinities
Identical SSU rDNA sequences of 1,640 bases were generated from four individual fish for K. cookii n. sp. Three LSU rDNA sequences (747, 751 and 754 bases) were also generated from four individual fish. There are 1–3 nucleotide differences (99.6–99.9% sequence identity) between the LSU rDNA sequences. The new species is genetically most similar to K. ciliatae with 39 nucleotide differences (97.4% sequence identity) in SSU and 35–36 nucleotide differences (94.2–94.4% sequence identity) in LSU rDNA (Tables 5A,B). In all phylogenetic analyses K. cookii forms a well-supported monophyletic clade with K. ciliatae, the only other species for which rDNA sequences are available and found forming cysts solely in the intestinal musculature with quadrate spores and equal polar capsules (Figs. 6, 7).
Principal component analyses
Principal component analysis (PCA) was used to explore the discrimination of the morphologically similar species found in the musculature of apogonids, Kudoa cheilodipteri n. sp. and K. whippsi (Fig. 3). Although there are some morphological overlaps in the measurements of the species reported in this study, PCA supported the discrimination of these two species (Fig. 3). Although the 95% ellipses overlapped, the strong separation is consistent with the presence of two species.
PCA analyses of subsets of host/parasite/location combinations of isolates identified as K. whippsi suggest a complex structure. In all analyses there is significant morphological overlap between the two isolates of K. whippsi taken from Lizard Island hosts reported in this study (Figs. 3, 4). Due to the low intensity of infection and limited material, only two mature spores could be measured in apical view for K. whippsi from Ostorhinchus cyanosoma from off Ningaloo Reef and thus this isolate was not used in the principal component analyses for apical spore measurements. Minimal intra-specific variation in side view measurements was found between the isolates of K. whippsi reported in this study (figure not shown). Analyses were also performed between the isolates of K. whippsi from this study and three isolates from the original description (Burger & Adlard, 2010a) (Fig. 4). Minimal intra-specific morphological variation was found between all isolates of K. whippsi in the analysis comparing side view measurements (Figure not shown). However, in the analysis of measurements taken from spores in the apical view there is significant variation seen between the isolates taken from this study and those from the original description (Fig. 4). In particular, there is almost no morphological overlap between the isolates taken from the original description and those taken from Apogon doederleini in this study.
Phylogenetic analyses
Molecular analysis included 52 SSU and 62 LSU rDNA myxosporean sequences available from GenBank together with the sequences generated in this study (Figs. 6, 7). Trees of similar topology were produced for both SSU and LSU rDNA datasets using Bayesian, maximum likelihood, maximum parsimony and neighbour-joining analyses (Figs. 6, 7). In both SSU and LSU rDNA analyses, all isolates of Kudoa whippsi reported in this study fell within a well-supported clade comprising all muscle dwelling species with spores containing four polar capsules, one larger than the remaining three, with the exception of K. cheilodipteri n. sp. In SSU rDNA analyses this clade includes: K. minithyrsites, K. megacapsula Yokoyama & Itoh, 2005, K. thyrsites, K. lateolabracis and K. gunterae (Fig. 6). In LSU rDNA analyses this clade includes: K. thyrsites, K. whippsi and K. gunterae (Fig. 7). In both SSU and LSU rDNA analyses, the position of K. cheilodipteri varied due to low posterior probabilities and bootstrap support and, in view of this, a fixed position for this species was not generated. In both SSU and LSU rDNA analyses K. cookii n. sp. forms a well-supported monophyletic clade with K. ciliatae, the only other species with corresponding sequences available reported to solely infect the intestines.
Discussion
Host-specificity
Recent studies on myxozoan host-specificity have shown differing patterns between and also within different genera. Species of the bivalvulidan genera Ceratomyxa Thélohan, 1892, Auerbachia Meglitsch, 1968 and Coccomyxa Léger & Hesse, 1907 are thought to be highly host-specific and are generally restricted to a single host species (Heiniger et al., 2011; Gunter et al., 2009). Similarly, the majority of Kudoa species are apparently highly host-specific, having been reported from a single host species only (Moran et al., 1999). However, a significant proportion of species are reported to have much broader host ranges, infecting multiple host species from a number of families and occasionally different orders (Burger & Adlard, 2011; Egusa & Nakajima, 1980). One species, K. thyrsites, has been reported from 18 fish families representing nine fish orders (Whipps & Kent, 2006). With the availability and benefit of molecular data, however, it now appears that K. thyrsites represents a species complex (Burger & Adlard, 2011; Whipps & Kent, 2006). Other species for which wide host ranges have been reported [e.g. Kudoa nova Naidenova, 1975, K. iwatai and K. clupeidae (Hahn, 1917)] have not been subjected to such analysis. Thus, the extent to which host distribution can be taken as an indicator of likely identity or distinction remains unresolved for the Myxozoa as a whole.
Both muscle-dwelling species reported here were found in three fish species, whereas K. cookii n. sp. (found in the intestines) has, so far, been found in only a single host species. Interestingly, K. whippsi is reported from Ostorhinchus cyanosoma from two locations (off Lizard Island and Ningaloo Reef). Furthermore, both K. whippsi and K. cheilodipteri n. sp. were both found in O. aureus and O. cyanosoma from the same location, Ningaloo Reef. Although these two species have been found in the same host and geographical location there were no observed incidences of co-infection.
Kudoa cheilodipteri n. sp.
Molecular information was critical for discrimination of K. whippsi and K. cheilodipteri, two morphologically and biologically similar species. These two species are genetically distinct with inter-specific variation of 3.7–4% (55–59 nucleotides) in SSU rDNA. The genetic separation of the two species is supported by LSU rDNA, which has been shown to give higher resolution than SSU (Burger & Adlard, 2010a; Yurakhno et al., 2007), with inter-specific variation of 13.9–15.3% (83–90 nucleotides). K. cheilodipteri is genetically most similar to K. thalassomi (Tables 3A,B) and, although the genetic relatedness of these two species is supported in the Bayesian analysis using the LSU dataset, in all other analyses the phylogenetic position of this species remains unresolved.
Kudoa whippsi
The new records reported here for K. whippsi suggest the presence of complexity that requires further analysis. In summary, phylogenetic analyses suggests convincingly that samples from two species of the Apogonidae from off Ningaloo, four from apogonids from the Great Barrier Reef (GBR) and eight from other families from the GBR, all form a well-supported clade to the exclusion of all other recognised species. However, there is significant variation within this clade. LSU and SSU sequences differ between the various isolates by up to 12 nucleotides for both LSU and SSU, despite several replicates from individual host/locality combinations returning identical sequences. Within this clade, there is, however, no correlation between the genetic variation and host (both at the specific and familial levels) or geographical locality (Fig. 5). The value of molecular data in defining taxonomic boundaries is unequivocal; however, interpretation of these data still remains contentious. There is no set nucleotide or percentage difference of genetic variation for distinguishing species, and instead this needs to be assessed for each individual case. The levels of intra-specific genetic variation observed here for K. whippsi are similar to those previously reported for species of Kudoa. Burger & Adlard (2011) reported levels of intra-specific variation of up to 10 nucleotides (0.7% variation) in SSU and 14 nucleotides (2.1% variation) in LSU rDNA for K. thalassomi. Similarly, levels of intra-specific variation between isolates of K. thyrsites have been reported as high as 14 nucleotides (1% variation) in SSU and 59 nucleotides (9.2%) in LSU rDNA (Burger & Adlard, 2011; Whipps & Kent, 2006). In contrast, inter-specific genetic variation has previously been reported as low as 1 nucleotide (0.1% variation) in SSU and 25 nucleotides (3.6% variation) in LSU rDNA between K. quadricornis and K. paraquadricornis (see Burger & Adlard 2010a).
PCA analysis suggests that several of the host/locality combinations of K. whippsi are recognisably distinct from each other. Morphological plasticity is not uncommon for species of Kudoa, or indeed myxosporeans (Burger & Adlard, 2010b; Heiniger et al., 2008; Sitja-Bobadilla & Alvarez-Pellitero, 1993). Even the most obvious diagnostic characters have been shown to be plastic, and it is now recognised that a single species of Kudoa may contain spores with a variable number of spore valves and polar capsules (Burger et al., 2007; Burger & Adlard, 2010b; Gunter et al., 2006; Whipps et al., 2003a). Nevertheless, morphological characters at the light microscopic level provide a useful diagnostic tool when discriminating between Kudoa species of differing morphotypes. Finally, our host-specificity data are in themselves suspicious. If a single species is present, then it is evidently one capable of infecting a range of pomacentrids and a highly restricted range of apogonids relative to those that are present in tropical Australia. We can see no particular reason why this should be the case. It seems, therefore, at least possible that K. whippsi represents a species complex in the same way as has been suggested for K. thyrsites (see Whipps & Kent, 2006).
For the present, we feel unable to distinguish between what can reasonably be interpreted as inter- and intra-specific variation (using both morphological and molecular characters). On the basis of genetic, host and geographical (Lizard Island) similarities, and despite small morphological (especially in measurements taken from spores in apical view) and geographical (Ningaloo Reef) differences, we take a conservative approach and assign these new isolates to K. whippsi Burger & Adlard, 2010. In doing so we recognise that future evidence, e.g. more informative diagnostic characters and/or further isolates for comparison, may require a reassessment of this specific assignment.
Kudoa cookii n. sp.
Kudoa cookii is only the eighth species of Kudoa found that forms cysts in or around the intestines of its host. It is distinguished from K. valamugili Kalavati & Anuradha, 1993 and K. unicapsula Yurakhno, Ovcharenko, Holzer, Sarabeev & Balbuena, 2007 by the difference in spore shape, with the latter two species having spores with one larger polar capsule. K. intestinalis Maeno, Nagasawa & Sorimachi, 1993 from Mugil cephalus off Japan has overall much smaller spores than those of K. cookii. The remaining four species found infecting the intestines are all distinguished from the new species (Table 4, which also includes two morphologically similar species found infecting the musculature of their hosts). K. iwatai and K. ciliatae are the only species reported from the intestines to have corresponding rDNA sequences available. In both SSU and LSU rDNA analyses K. cookii formed a well-supported clade (>90% support in all analyses) with K. ciliatae. K. iwatai is primarily a parasite infecting the musculature of its hosts. Nevertheless, in the LSU rDNA analyses, the K. cookii and K. ciliatae clade displayed a close genetic relationship to K. iwatai and other muscle-dwelling species with quadrate spores and four equal polar capsules. To aid in the differentiation of morphologically similar species, previous studies have mapped several biological characters, such as host, geographical location and tissue site of infection, against phylogenies derived from genetic data in the hope of finding a uniting character that reflects the genetic relationships among the Myxosporea (see Burger et al., 2007; Eszterbauer, 2004). To date, tissue tropism has been shown to be the most significant uniting biological character (Burger et al., 2007; Burger & Adlard, 2010a; Miller & Adlard, 2012). The close genetic relationship between K. cookii and K. ciliatae, both intestine-dwelling species, further supports the value of tissue tropism when distinguishing between morphologically similar species.
Apogonids as hosts for kudoids
Only four species of Kudoa have been previously reported from apogonids, K. iwatai in the musculature of Apogon fleurieu from the Red Sea (Diamant et al., 2005), K. thalassomi in the musculature of Cheilodipterus macrodon from off the Great Barrier Reef (GBR), Australia (Burger & Adlard, 2011), and K. gunterae and K. whippsi in the musculature of Ostorhinchus properuptus also from off the GBR, Australia (Burger & Adlard, 2010a). This study examined over 700 individual apogonids and found only the two new species, K. cheilodipteri n. sp. and K. cookii n. sp., and three new host records for K. whippsi reported here. Interestingly, K. thalassomi and K. gunterae were not found. This was surprising given that both species have been previously reported from apogonids from off the GBR. Furthermore, both species are reported to have low host-specificity, K. gunterae being reported from 11 host species representing two families (dominated by Family Pomacentridae with 10 species) (Burger & Adlard, 2010a) and K. thalassomi from 18 host species representing six families (dominated by the Pomacentridae with eight species) (Burger & Adlard, 2011). Although the sample size of apogonid hosts previously reported for these species was small, we can conclude that certainly neither species is common in a wide range of GBR apogonids. Overall apogonids appear to be relatively poor hosts for kudoids given that perhaps five species are known from examinations of 700+ individuals of 24 species (including those examinations reported by Burger & Adlard, 2010a).
References
Adlard, R. D., Bryant, M. S., Whipps, C. M., & Kent, M. L. (2005). Multivalvulid myxozoans from Eastern Australia: three new species of Kudoa from scombrid and labrid fishes of the Great Barrier Reef, Queensland, Australia. Journal of Parasitology, 91, 1138–1142.
Aken’Ova, T. O. L., Cribb, T. H., & Bray, R. A. (2006). Helicometra Odhner, 1902 (Digenea: Opecoelidae) in Australian waters; problems of species identification and a description of H. sprenti n. sp. Systematic Parasitology, 63, 17–27.
Bott, N. J., & Cribb, T. H. (2005). First report of a bucephalid digenean from an apogonid teleost; Prosorhynchoides apogonis n. sp. from Cheilodipterus macrodon on the southern Great Barrier Reef. Australia. Systematic Parasitology, 30, 33–37.
Bray, R. A., & Cribb, T. H. (1989). Digeneans of the family Opecoelidae Ozaki, 1925 from the southern Great Barrier Reef, including a new genus and three new species. Journal of Natural History, 23, 429–473.
Bruce, N. L. (1986). Australian Pleopodias Richardson, 1910, and Anilocra Leach, 1818 (Isopoda: Cymothoidae), crustacean parasites of marine fishes. Records of the Australian Museum, 39, 85–130.
Bruce, N. L. (1987). Australian Renocila Miers, 1880 (Isopoda: Cymothoidae), crustacean parasites of marine fishes. Records of the Australian Museum, 39, 169–182.
Burger, M. A. A., & Adlard, R. D. (2010a). Four new species of Kudoa Meglitsch, 1947 (Myxosporea: Multivalvulida) from Australia with recommendations for species descriptions in the Kudoidae. Parasitology, 137, 793–814.
Burger, M. A. A., & Adlard, R. D. (2010b). Phenotypic variation in a significant spore character in Kudoa (Myxosporea: Multivalvulida) species infecting brain tissue. Parasitology, 137, 1759–1772.
Burger, M. A. A., & Adlard, R. D. (2011). Low host specificity in the Kudoidae (Myxosporea: Multivalvulida) including seventeen new host records for Kudoa thalassomi. Folia Parasitologica, 58, 1–16.
Burger, M. A. A., Cribb, T. H., & Adlard, R. D. (2007). Patterns of relatedness in the Kudoidae with descriptions of Kudoa chaetodoni n. sp. and K. lethrini n. sp. (Myxosporea: Multivalvulida). Parasitology, 134, 669–681.
Cribb, T. H. (1987). Studies on gorgoderid digeneans from Australian and Asian Freshwater fishes. Journal of Natural History, 21, 1129–1153.
Diamant, A., Ucko, M., Paperna, I., Colorni, A., & Lipshitz, A. (2005). Kudoa iwatai (Myxosporea: Multivalvulida) in wild and cultured fish in the red sea: redescription and molecular phylogeny. Journal of Parasitology, 91, 1175–1189.
Drummond, A. J., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Duran, C., et al. (2010). Geneious v5.4, Available from http://www.geneious.com.
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5, 113.
Egusa, S., & Nakajima, K. (1980). Kudoa amamiensis n. sp. (Myxosporea: Multivalvulida) found in cultured yellowtails and wild damselfishes from Amami-Ohshima and Okinawa, Japan. Bulletin of the Japanese Society of Scientific Fisheries, 46, 1193–1198.
Ernst, I., Whittington, I. D., & Jones, M. K. (2001). Diversity of gyrodactylids from some marine fishes in tropical and subtropical Queensland, Australia. Folia Parasitologica, 48, 165–168.
Eszterbauer, E. (2004). Genetic relationship among gill-infecting Myxobolus species (Myxosporea) of cyprinids: molecular evidence of importance of tissue specificity. Diseases of Aquatic Organisms, 58, 35–40.
Fogelman, R. M., & Grutter, A. S. (2008). Mancae of the parasitic cymothoid isopod, Anilocra apogonae: early life history, host-specificity, and effect on growth and survival of preferred young cardinal fishes. Coral Reefs, 27, 685–693.
Fogelman, R. M., Kuris, A. M., & Grutter, A. S. (2009). Parasitic castration of a vertebrate: effect of the cymothoid isopod, Anilcora apogonae, on the five-lined cardinalfish, Cheilodipterus quinquelineatus. International Journal for Parasitology, 39, 577–583.
Greenfield, D. W., & Johnson, R. K. (1990). Heterogeneity in habitat choice in Cardinalfish community structure. Copeia, 4, 1107–1114.
Grossel, G. W., Dyková, I., Handliinger, J., & Munday, B. L. (2003). Pentacapsula neurophila sp. n. (Multivalvulidae) from the central nervous system of striped trumpeter, Latris lineata (Forster). Journal of Fish Diseases, 26, 315–320.
Gunter, N. L., Cribb, T. H., Whipps, C. M., & Adlard, R. D. (2006). Characterisation of Kudoa monodactyli n. sp. (Myxosporea: Multivalvulida) from the muscle of Monodactylus argenteus (Teleostei: Monodactylae) from Moreton Bay, Queensland, Australia. Journal of Eukaryotic Microbiology, 53, 374–378.
Gunter, N. L., Whipps, C. M., & Adlard, R. D. (2009). Ceratomyxa (Myxozoa: Bivalvulida): Robust taxon or genus of convenience? International Journal for Parasitology, 39, 1395–1405.
Hammer, O., Harper, D. A. T., & Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4, 9 pp.
Heiniger, H., Gunter, N. L., & Adlard, R. D. (2008). Relationships between four novel ceratomyxid parasites from the gall bladders of labrid fishes from Heron Island, Australia. Parasitology International, 57, 158–165.
Heiniger, H., Gunter, N. L., & Adlard, R. D. (2011). Re-establishment of the family Coccomyxidae and description of five novel species of Auerbachia and Coccomyxa (Myxosporea: Bivalvulida) parasites from Australian fishes. Parasitology, 138, 501–515.
Hillis, D. M., & Dixon, M. T. (1991). Ribosomal DNA: molecular evolution and phylogenetic inference. The Quarterly Review of Biology, 66, 411–453.
Iversen, E. S., & Van-Meter, N. N. (1967). A new myxosporidian (Sporozoa) infecting the Spanish mackerel. Bulletin of Marine Science, 17, 268–273.
Jones, C. M., Nagel, L., Hughes, G. L., Cribb, T. H., & Grutter, A. S. (2007). Host specificity of two species of Gnathia (Isopoda) determined by DNA sequencing blood meals. International Journal for Parasitology, 37, 927–935.
Kent, M. L., Andree, K. B., Bartholomew, J. L., El-Matbouli, M., Desser, S. S., Devlin, R. H., et al. (2001). Recent advances in our knowledge of the Myxozoa. Journal of Eukaryotic Microbiology, 48, 395–413.
Kuiter, R. H., & Kozawa, T. (1999). Fishes of the Indo-West Pacific: Apogonidae (2nd Ed.). Seaford, Victoria, Australia: R. H. Kuiter & T. Kozawa, CD version.
Langdon, J. S. (1991). Myoliquefaction post-mortem (‘milky flesh’) due to Kudoa thyrsites (Gilchrist) (Myxosporea: Multivalvulida) in mahi mahi, Coryphaena hippurus L. Journal of Fish Diseases, 14, 45–54.
Lom, J., & Arthur, J. R. (1989). A guideline for the preparation of species descriptions in Myxosporea. Journal of Fish Diseases, 12, 151–156.
Lom, J., & Dykova, I. (2006). Myxozoan genera: definition and notes on taxonomy, life cycle terminology and pathogenic species. Folia Parasitologica, 53, 1–36.
Maddison, D. R., & Maddison, W. P. (2005). MacClade 4: Analysis of phylogeny and character evolution. Version 4.08. Sunderland, Massachusetts: Sinauer Associates.
Marnane, M. J. (2000). Site fidelity and homing behaviour in coral reef cardinalfishes. Journal of Fish Biology, 57, 1590–1600.
Matsumoto, K. (1954). On the two new Myxosporidia, Chloromyxum musculoliquefaciens sp. nov. and Neochloromyxum cruciformum gen. et sp. nov., from the jellied muscle of swordfish, Xiphias gladius Linné, and common Japanese sea-bass, Lateolabrax japonicus (Temmink et Schlegel). Bulletin of the Japanese Society of Scientific Fisheries, 20, 469–478.
Miller, M. A., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (CGE). San Diego Supercomputer Center, La Jolla, CA, USA, pp. 1–8.
Miller, T. L., & Adlard, R. D. (2012). Brain infecting kudoids of Australia’s coral reefs, including a description of Kudoa lemniscati n. sp. (Myxosporea: Kudoidae) from Lutjanus lemniscatus (Perciformes: Lutjanidae) off Ningaloo Reef, Western Australia. Parasitology International, 61, 333–342.
Moran, J. D. W., Whitaker, D. J., & Kent, M. L. (1999). Review of the myxosporean genus Kudoa Meglitsch, 1947, and its impact on the international aquaculture industry and commercial fisheries. Aquaculture, 172, 163–196.
Nagel, L., & Grutter, A. S. (2007). Host preference and specialization in Gnathia sp., a common parasitic isopod of coral reef fishes. Journal of Fish Biology, 70, 497–508.
Narasimhamurti, C. C., Calavati, C., Anuradha, I., & Dorothy, K. P. (1990). Studies on protozoan parasites of deep water fishes from the Bay of Bengal. In: Pillai, V.K., et al. (Eds) Proceedings of the First Workshop of Scientific Results of FORV Sagar Sampada, 5-7 June 1989. New Delhi: Department of Ocean Development, pp. 325–336.
Posada, D. (2008). jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution, 25, 1253–1256.
Ronquist, F., & Huelsenbeck, J. P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574.
Sitja-Bobadilla, A., & Alvarez-Pellitero, P. (1993). Light and electron-microscopic description of Ceratomyxa labracis n. sp. and a redescription of C. diplodae (Myxosporea, Bivalvulida) from wild and cultered Mediterranean sea bass Dicentrarchus labrax (L)(Teleostei, Serranidae). Systematic Parasitology, 26, 215–223.
Stamatakis, A., Hoover, P., & Rougemont, J. (2008). A rapid bootstrapping algorithm for the RAxML web-servers. Systematic Biology, 75, 758–771.
Swofford, D. L. (2002). PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates.
Thacker, C. E., & Roje, D. M. (2009). Phylogeny of cardinalfishes (Teleostei: Gobiiformes: Apogonidae) and the evolution of visceral bioluminescence. Molecular Phylogenetics and Evolution, 52, 735–745.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Vagelli, A., Parama, A., Sanmartin, M. L., & Leiro, J. (2005). Glugea vincentiae n. sp. (Microsporidia: Glugeidae) infecting the Australian marine fish Vincentia conspersa (Teleostei: Apogonidae). Journal of Parasitology, 91, 152–157.
Whipps, C. M., Adlard, R. D., Bryant, M. S., & Kent, M. L. (2003a). Two unusual myxozoans, Kudoa quadricornis n. sp. (Mulitvalvulida) from the muscle of goldspotted trevally (Carangoides fulvoguttatus) and Kudoa permulticapsula n. sp. (Multivalvulida) from the muscle of Spanish mackerel (Scomberomorus commerson) from the Great Barrier Reef, Australia. Journal of Parasitology, 89, 168–173.
Whipps, C. M., Adlard, R. D., Bryant, M. S., Lester, R. J. G., Findlay, V., & Kent, M. L. (2003b). The first report of three Kudoa species from Eastern Australia: Kudoa thyrsites from mahi mahi (Coryphaena hippurus), Kudoa amamiensis and Kudoa minithyrsites n. sp. from sweeper (Pempheris ypsilychnus). Journal of Eukaryotic Microbiology, 20, 215–219.
Whipps, C. M., Grossel, G., Adlard, R. D., Yokoyama, H., Bryant, M. S., Munday, B. L., et al. (2004). Phylogeny of the Multivalvulida (Myxozoa: Myxosporea) based on comparative ribosomal DNA sequence analysis. Journal of Parasitology, 90, 618–622.
Whipps, C. M., & Kent, M. L. (2006). Phylogeography of the cosmopolitan marine parasite Kudoa thyrsites (Myxozoa: Myxosporea). Journal of Eukaryotic Microbiology, 53, 364–373.
Yokoyama, H., Whipps, C. M., Kent, M. L., Mizuno, K., & Kawakami, H. (2004). Kudoa thyrsites from Japanese flounder and Kudoa lateolabracis n. sp. from Chinese sea bass: causative myxozoans of post-mortem myoliquefaction. Fish Pathology, 39, 79–85.
Yurakhno, V. M., Ovcharenko, M. O., Holzer, A. S., Sarabeev, V. L., & Balbuena, J. A. (2007). Kudoa unicapsula n. sp. (Myxosporea: Kudoidae) a parasite of the Mediterranean mullets Liza ramada and L. aurata (Teleostei: Mugilidae). Parasitology Research, 101, 1671–1680.
Acknowledgements
This study was funded by, and is a contribution from, the Australian node of the CReefs global research initiative (grant no. 209/29), a project generously sponsored by BHP Billiton in partnership with The Great Barrier Reef Foundation, the Australian Institute of Marine Science, the Australian Biological Resources Study and the Alfred P. Sloan Foundation. CReefs is a field programme of the Census of Marine Life. We thank Dr Terrence Miller for technical advice, phylogenetic discussions and assistance in field collections, Jeff Johnson for assistance with fish identification, Dr Mieke Burger for supplying raw morphological data taken from isolates described in the original description of Kudoa whippsi, Ricky Gleeson who assisted in field collection, and the staff at Lizard Island and Heron Island Research Stations for logistical support in the field.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Heiniger, H., Cribb, T.H. & Adlard, R.D. Intra-specific variation of Kudoa spp. (Myxosporea: Multivalvulida) from apogonid fishes (Perciformes), including the description of two new species, K. cheilodipteri n. sp. and K. cookii n. sp., from Australian waters. Syst Parasitol 84, 193–215 (2013). https://doi.org/10.1007/s11230-012-9400-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-012-9400-9