Abstract
The advent of large data repositories and the necessity of distributed skillsets have led to a need to study the scientific collaboration network emerging around cyber-infrastructure-enabled repositories. To explore the impact of scientific collaboration and large-scale repositories in the field of genomics, we analyze coauthorship patterns in NCBIs big data repository GenBank using trace metadata from coauthorship of traditional publications and coauthorship of datasets. We demonstrate that using complex network analysis to explore both networks independently and jointly provides a much richer description of the community, and addresses some of the methodological concerns discussed in previous literature regarding the use of coauthorship data to study scientific collaboration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The emergence of cyberinfrastructure (CI) enabled research are thought to affect the structure and scale of scientific collaboration (Szalay and Blakeley 2009) by allowing scientists to share data and computing resources between geographically disparate teams. Scientific data repositories (SDRs) are one example of cyberinfrastructure. We argue that the emergence of CI-enabled science, and SDRs in particular, should be of interest to any researcher studying scientific collaboration, especially if the researcher uses network analytic lenses to study the phenomenon for two reasons. First, from a theoretical point of view, quantitative analyses of the impact of CI on the scale and size of scientific collaboration are sparse. Second, SDRs may serve as an additional source of trace data for studying scientific collaboration, thus addressing some of the concerns expressed about using coauthorship data to study scientific collaboration (Glänzel and Schubert 2005; Laudel 2002). This observation leads to the general research question motivating our paper—Can trace data from scientific data repositories supplement trace data from publications to provide additional insight into the structure and evolution of scientific collaboration networks?
The term scientific data repository (SDR) is used frequently, often in broader discussions of cyberinfrastructure (CI) enabled science, but rarely explicitly defined. Having said that, there appears to be an implicit consensus on the functions and characteristics of SDRs; consequently, there is little controversy when exemplars are given in the literature. The implicit consensus suggests that an SDR is a system of technologies and policies that enable, at a minimum, the storage of data sets in a centralized location with respect to the community the SDR serves. SDRs may also provide access to additional data services, including importing, exporting, handling, archiving, and curating the data, as well as supporting usage tracking and linking to publications or external sites (Marcial and Hemminger 2010).
The primary impetus for the investment in SDRs is to facilitate data sharing and reuse (Advanced Cyberinfrastructure Division 2012; Hey et al. 2009, p. XV). Many SDRs are designed to support entire research fields, and thus are underpinned by relatively complex technical architectures in order to support a range of services that connect to various parts of the community’s typical research workflow. As a result, the design and maintenance of the system is costly, and continued funding is almost always required. Given the high cost of most SDRs, there is a strong incentive to see them used. Consequently, there is an increasing emphasis on the development of policies and regulations that promote the use of SDRs, and more attention is being paid to the socio-cultural factors that influence scientists’ data sharing behaviors (Faniel and Jacobsen 2010; Faniel and Zimmerman 2011).
Examples of well-established SDRs that are integrated into the communities they serve include the LIGO data grid, which supports the Laser Interferometer Gravitational Wave Observatory (LIGO) experiments, the Long Term Ecological Research Network (LTER), which allows ecologists to gather and analyze data over spatial and temporal dimensions, the Worldwide Protein Data Bank (wwPDB), and GenBank, the international nucleotide sequencing databank. The extent to which the use of these repositories affects the work practices and collaborative interactions of the scientists in the surrounding communities is largely unknown. Before we attempt to answer the question of how the emergence of these repositories impacts the structure and scale of scientists’ collaborative interactions we need to answer the more basic question—What are the structural characteristics of the collaborative interactions of scientists who use these repositories?
The spatial metaphors of structure and scale refer to the presence, composition, and interconnectedness (structure) of sub-communities within research fields and the size (scale) of the teams and sub-communities within the larger research communities. Studying the structure and scale of scientific collaboration is often best done through network analytic lenses, made popular by the work of Albert, Barabási, and Newman (Barabási and Albert 1999; Newman 2001b, 2003). Conceptualizing scientists as a set of points on a plane (or less often, a three dimensional volume), connected by lines representing social relationships, has proven to be a useful approach for studying the social nature of science. The resultant network is amenable to a host of advanced statistical analyses, which provide a much richer description of the overall organization of scientific fields (macroscopic analysis), the prominence of groups within fields (mesoscopic analysis), as well as the role individuals play in those fields (microscopic analysis).
For this paper we focused on four major network measures, the clustering coefficient, mean degree and degree distribution coefficient (or power law fit), and size of the giant component as a percentage of all scientists active in the network.
Clustering coefficient measures the number of completed transitive triples, or the fraction of scientists who are connected to a common neighbor and are connected themselves. Small networks exhibit much higher clustering coefficients than random graphs, but will decrease over time or as the network gets larger (Barabási et al. 2002). The clustering coefficient varies between fields, and can be attributed to the tendency of friends to become friends, or to work in groups of 3 or more (Newman 2001a). Low values may indicate a hierarchically organized community, with few actors having opportunities to independently build relationships, even with colleagues of colleagues.
Mean degree provides a general sense of how many connections actors in a network have. When the mean degree is analyzed in conjunction with the degree distribution coefficient, the two measures can be used to discern the hierarchical nature of a community. The mean degree in a horizontally organized community will be higher than that of a comparable, vertically organized network. However, the degree distribution coefficient will provide a much better indication of the extent to which connectivity is centralized in a few prominent actors (Newman 2001a).
Giant Component is the largest component in a network in which there is at least one path between all nodes in that component. In disconnected networks, components, better thought of as isolated islands, are relatively small in terms of the overall number of nodes in the network. The absence of a giant component may indicate that the network being studied is not a true community, but instead many smaller isolated communities. The presence of a giant component may indicates the presence of a community, with larger giant components being indicative of a connected community.
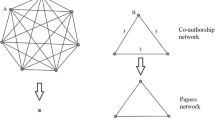
In this paper, we use the measures listed above to provide a more accurate description of the structural characteristics of scientific community emerging and evolving around a scientific data repository, as well as compare the collaboration networks that can be reconstructed using metadata from the repository. Specifically, we identify two types of coauthorship within a scientific data repository—coauthorship of datasets and coauthorship of publications. Each form of coauthorship can be used to construct an independent network, as well as combined to construct a more comprehensive network. By comparing and contrasting the three networks we answer our original research question—Can trace data from scientific data repositories supplement trace data from publications to provide additional insight into the structure and evolution of scientific collaboration networks?
By answering these questions, we can demonstrate that the large body of literature looking at scientific collaboration could be missing a significant portion of the collaborative process, as noted by others including (Laudel 2002). As far as we know, this is the first large scale study of scientific collaboration that includes and integrates both publication data and dataset generation. This paper also builds on past research by demonstrating the effect policy decisions have on the structure and activity rates of scientific networks.
Materials and methods
The most common approach to studying scientific collaboration is to use metadata from publications to extract coauthorship relationships. It is also possible to gather data using surveys, qualitative interviews, or some mixture of the three. There are known limitations to each approach; of direct interest to our research are the limitations of operationalizing collaboration as coauthorship. Both (Glänzel and Schubert 2005; Laudel 2002) go into greater detail on these limitations, so we will only cover them briefly. Specifically, using coauthorship as the basis for studying collaboration is known to both undercount and overcount interactions. Undercounting comes from the fact that scientists often informally collaborate on papers, receiving feedback from peers. Scientists have taken to using acknowledgements to give credit for this less than coauthorship level of collaboration (Cronin et al. 2003). Coauthorship can overstate collaboration as well, particularly when coauthorship is given freely to colleagues or more senior scientists in recognition of their support, or in attempts to use their name recognition to draw attention to the publication. Understanding exactly what collaboration is, and the relationship between any two scientists who coauthored a paper together, becomes even more difficult in the case of hyper-authored papers. In general, the number of authors per paper has increased steadily over the years, including the number of papers with over 1000 authors (King 2012).
One question that came to mind regarding capturing the full breadth of scientific collaboration was whether extracting metadata from a scientific repository could provide a more detailed picture of collaboration than just using metadata from publications alone. Additionally, results from a pilot study conducted by the authors of this paper suggested that scientists were beginning to behave as if the datasets submitted to the GenBank repository were intellectual contributions, (i.e., laying intellectual claim to their production). These observations prompted us to ask whether using the metadata from a scientific data repository would capture additional collaboration not present in a collaboration network based on publications, as well as if there are differences in the structure of dataset submission networks and publication networks. In order to answer these questions we chose to further explore the GenBank community.
Data source
GenBank overview
GenBank is the National Institute of Health (NIH) genetic sequence database of publicly available nucleotide sequences for almost 260,000 formally described species submitted by researchers from around the world (Benson et al. 2013). GenBank was formed in 1982 and is run by the National Center for Biotechnical Information. GenBank is now part of the International Nucleotide Sequencing Consortium in which each member exchanges information daily. Complete bimonthly releases and daily updates of the GenBank database are available by FTP. NCBI makes the GenBank data available at no cost over the Internet, through FTP and a wide range of web-based retrieval and analysis services.
From its inception, GenBank has doubled in size roughly every 18 months. In addition to storing genetic information, GenBank provides a number of tools for discovery and analysis. The repository has been well adopted by its respective community, which now requires all scientists to submit genetic data to the repository prior to publication, verified by an accession number. Additionally, genetic information associated with patents are now stored in the repository.
As a large cyberinfrastructure (CI)-enabled repository, GenBank facilitates the management, preservation, and access to a bibliographically and biologically annotated collection of DNA sequences, and connects the data with related publications (genome.gov). The interdisciplinarity of collaboration (Qin et al. 1997; Porter and Rafols 2009) calls for CI-enabled research to identify fruitful collaborative partners. The tool “BLAST” provides sequence similarity searches of GenBank and other sequence databases, enabling collaboration between scientists and building from the work of previous submissions to develop new or combine sequencing work.
Data entry into GenBank’s database
The primary units of submission into GenBank are References and Annotations. We define the terms and provide examples in Table 1. The sequences are obtained primarily through submissions from individual laboratories and batch submissions from large-scale sequencing projects, including whole-genome shotgun (WGS) and environmental sampling projects. Most submissions are made using the web-based BankIt or standalone Sequin programs. Daily data exchange with the European Nucleotide Archive (ENA) and the DNA Data Bank of Japan (DDBJ) as partners in the International Nucleotide Sequence Database Collaboration (INSDC) ensures that a uniform and comprehensive collection of sequence information is available worldwide.
NCBI builds GenBank primarily from the submission of sequence data from authors and from the bulk submission of expressed sequence tag (EST), genome survey sequence (GSS), whole-genome shotgun (WGS) and other high-throughput data from sequencing centers. The U.S. Patent and Trademark Office also contributes sequences from issued patents. In a recent development, NCBI is in the process of creating a unified submission portal that will provide a single access point for data submitters (submit.ncbi.nlm.nih.gov).
Metadata standardization continues to be a problem (Costa et al. 2014; Qin et al. 2014). The submitting institution indicated in a record may or may not be equivalent to the institutional affiliation of all the authors associated with the submission. For journal articles, the fields contain the expected citation information, although there is no indication that the journal abbreviations are standardized across the database. This is due to the fact that GenBank is not intended to be a literature repository; yet, analysis indicates that the literature referenced in the repository leaves traces of a large, very well connected community.
The Bermuda Principles and changes to data submission practices
There are a number of policy related decisions impacting the data sharing practices of scientists working in the genetic sequencing community. Of particular interest to our research are the Bermuda Principles, which were adopted in 1996. The principles altered the way in which DNA sequence data were to be uploaded to public repositories. Specifically, prior to the Principles, data were uploaded and made available after publication. However, the accord suggested a change in the practice, establishing that all sequence of human genome data above 1 kb be uploaded within 24 h (Arias et al. 2015; Collins et al. 2003; Rodriguez et al. 2009).
The Bermuda Principles were reaffirmed and extended in a 2003 meeting sponsored by the Wellcome Trust (Rodriguez et al. 2009), extending the push to submit data from all sequencing techniques and sources. A number of other policy changes have been affected over the years, most notably, the 2003 Fort Lauderdale Policy made data sharing as a prerequisite for funding. As stated in NHGRI’s 2003 “Reaffirmation and Extension of NHGRI Rapid Data Release Policies: Large-Scale Sequencing and Other Community Resource Projects: “users have responsibilities consistent with scientific norms” (Arias et al. 2015).
These policies continue to evolve, influenced by ongoing tension between the actors in the system, including data producers and users, and funding agencies, and social concerns including confidentiality and data access. Members of the community recognize data producers’ wish to maximize their opportunities to analyze the data, while data consumers require access to the data for their own research purposes. Furthermore, the economic impact of genomic research cannot be underestimated—our data extraction has identified over 26 million patent entries in the database (see below). The overall trajectory of the changing policy landscape is to make the data more freely available to the community, which has resulted in a substantial increase in data stored in the repository.
Data collection
GenBank provides an entire copy of its database in compressed semi-structured text format via an ftp site. Data were downloaded from the ftp site in August 2013. A parsing script was written to extract the metadata from records into a relational database while dropping the genomic data. The initial parsing process extracted approximately 150 million annotation records, plus an additional 26 million patents associated with publications. The remainder of the results reported here do not include patent analysis. We further identified 599,318 authors associated with 1.35 million references. After the metadata set was parsed, we normalized author names by stemming last names, reducing the number of unique scientists to 531,019.
Based on our knowledge of the differences in direct submissions and publication citations in GenBank, we can revise and elaborate on the general research questions of (1) whether using coauthorship metadata from a data repository would capture additional collaborative interactions not present in a standard publication network, and (2) whether there would be differences in the structure of the collaboration networks, as operationalized by dataset coauthorship and publication coauthorship. These two issues can be addressed by answering the following three questions:
-
RQ1 What are the differences and similarities between the structural characteristics of collaboration networks based on dataset coauthorship, publication coauthorship, and a combination of the two forms of coauthorship? Researchers have studied the macroscopic structure of scientific fields, using measures such as clustering coefficient, power law coefficients of degree distribution, and degree correlation coefficient to describe the extent to which (a) scientists in a particular network tend to collaborate with collaborators of collaborators (clustering coefficient); (b) current status impacts future opportunities, or cumulative advantage (power law coefficient of degree distribution); (c) scientists of similar status choose to collaborate with other scientists of similar status (degree correlation coefficient).
-
RQ2 To what extent does the dataset coauthorship network capture collaborative interactions not present in a network assembled from traditional publication coauthorship metadata and vice versa? Our initial hypothesis was that looking at the coauthorship of datasets would provide additional insight into the collaboration patterns of scientists. Answering these two related questions helped us determine whether using the dataset coauthorship provided the additional information as we hypothesized, as well whether it is beneficial to use one or both data sources. Does using the coauthorship metadata associated with datasets provide the same information using coauthorship metadata from publication provides, the same information plus some, or no additional information? In other words, can we get by using one or the other sources of data, or does the use of both result in a more detailed picture of the structure of the community?
-
RQ3 Are collaborations on the production of a dataset leading or lagging indicators of collaborations on publications? A commonly held and often implicit belief is that the generation of data is part of a research process that should culminate in a publication. If this is true, the generation and datasets should precede a publication. However, research into scientists’ data sharing behaviors indicate that scientists often hold off sharing their data in the hopes of maximizing their ability to publish from the data.
The answers to our research questions will lead to a better understanding of the utility of using trace data from scientific data repositories to study scientific collaboration, as well as the phenomenon of scientific collaboration and how it is impacted by the adoption of advanced information technology. The operationalization of concepts in our research questions, as well as the approaches to answering those questions, are outlined in the following section.
Operationalization of concepts
Scientific collaboration can be defined as the “the system of research activities by several actors related in a functional way and coordinated to attain a research goal corresponding with these actors’ interests” (Laudel 2002). Here we operationalize collaboration as coauthorship on either a directly submitted dataset with no associated publication (dataset coauthorship), or coauthorship on a publication (publication coauthorship). We recognize the limitations of using coauthorship as a source of data for studying scientific collaboration articulated by Laudel (2002), but agree with Glänzel and Schubert (2005), that coauthorship continues to be a useful approach to operationalizing the concept. Furthermore, our research asks whether using metadata from dataset publications provides additional information to metadata extracted from formal publications only.
A scientific collaboration network is a set of nodes and edges depicting actors (nodes) and the presence of a relationship (edge) between those actors. We constructed three unimodal, undirected networks from the metadata extracted from GenBank—a publication network, dataset submission network, and combined network. Networks were constructed from records, and do not include records that have no date attached to them (except for one explicitly stated instance, where the undated records are used to explore the potential range of differences between networks).
The dataset submission network was constructed by identifying references in the database whose title field was empty or the phrase “direct submission”, or with the journal field containing the phrase “unpublished”. Publications were extracted by eliminating all other possible alternatives (i.e., direct submissions, dissertations, theses). Once the datasets and publications were identified, related authors were extracted to form edgelists, and unique authors were then extracted from these edgelists.
With respect to the construction of the networks, graphs were simplified because of the gross disparity in the submission rates of datasets and publications. Including edge weights or duplicate edges for multiple submissions dramatically skewed the results, making comparisons between the two networks impossible. For example, one scientist had over 1.2 million edges (some redundant) from 13,000 dataset submissions and 50 publications. That same scientist had 2350 edges from the 50 publications, suggesting that there were over 45 authors per paper on average.
The time dynamics were analyzed using a series of cumulative snapshots to better represent the growth of the network and not the immediate activity. This choice is in line with our research question, which is more focused on determining what additional historical information using trace data from dataset coauthorship can provide, and less on mapping the current state of the field. For a review of different approaches to constructing temporally evolving networks, see (Holme and Saramäki 2012). Structural characteristics of the networks were calculated using functions in R’s igraph package.
In addition to comparing the macroscopic structural characteristics of the two networks, we also focused on determining the difference between the dataset and publication networks with respect to scientist membership and relationship presence. This involved a series of set operations, looking for individuals present in one, but not the other, or present in both, as well as relationships between scientists present in one, but not the other, or present in both.
Our final research question focused on whether collaboration on the production of a dataset preceded or lagged collaboration on publications. For this question, we focused on the years 1990 through 2012, creating a logical cumulative adjacency matrix for each year, as well as logical adjacency matrices for individual years 1991–2013 (Table 2). Next, we create a cumulative logical adjacency matrix of scientists that have collaborated on both publications and submissions (MB). The sum of MB gives us the total number of scientists who have collaborated on both publications and datasets.
We can identify who collaborated on a dataset first, by year, by subtracting the logical publication matrix for the year of interest from the cumulative logical dataset matrix up to, but not including the year of interest, then counting all instances in the Boolean intersect of the resulting matrix that equal one (i.e., a relationship that is present in the dataset submission network, but not the publication network) and the publication adjacency list for that year. The same can be done in inverse for publications.Footnote 1 Dividing by the sum of all collaborative relationships that have coauthored a paper and dataset together gives us the fraction of relationships attributable to that year for that order of precedence.
Equation 1: P Adp = the percentage of coauthorship relationships that have published both publications and datasets together where publication of a dataset precedes publication of an article
Equation 2: P Apd = the percentage of coauthorship relationships that have published both publications and datasets together where publication of an article precedes publication of a dataset
If \({\text{CP}}_{{{\text{YR}} - 1}} - {\text{CP}}_{\text{YR}} = = - 1\;{\text{for}}\;A_{ij}\) then two scientists (i, j) have first collaborated on a publication in year YR. Similarly, if \({\text{CD}}_{{{\text{YR}} - 1}} - {\text{CD}}_{\text{YR}} = = - 1\;{\text{for}}\;A_{ij}\) then two scientists (i,j) have first collaborated on a dataset in year YR. If \(A_{ij} = - 1\) in both matrices, then scientists i and j have collaborated for the first time on both an article and a dataset in year YR.
Results and discussion
Descriptive analysis
In total, the GenBank community includes 545,354 unique scientists, with the number of scientists publishing per year rising from a total 2563 scientists in 1982, to 31,554 in 1995. The 545,354 scientists includes 404,465 scientists in the publication network and 386,139 scientists in the dataset submission network. Up through 1995, the number of scientists active only in the dataset submission network was relatively marginal. However, after the Bermuda Principles were adopted, the number of scientists active in the dataset submission network grew rapidly (Fig. 1). It wasn’t until 2007 that the number of scientists submitting datasets surpassed the number of scientists contributing publications. However, the number of scientists active in only one or the other network grew quickly, indicating a portion of the population was not laying intellectual claims to both activities.
In terms of the number of scientists entering the network, scientists would first be noted entering the network as a coauthor on a paper. However, after 1996 the pattern changed; the number of scientists who first entered the network by submitting a dataset increase 550 % in five years, rising from 1825 in 1995 to 10,034 in 2000. To put that in perspective, the number of scientists entering the network with a publication declined by 10 % in the same period, from 11,566 in 1995 to 10,400 in 2000. Note that the majority of the uncertainty with respect to how scientists first make a contribution to the network lies in the dataset submission population, with 26,439 scientists coauthoring datasets with no date attached and only 242 scientists coauthoring publications with no date attached. Of the 26,439 scientists entering the network without a date attached to their entry, 21,744 only had one submission and 0 were associated with a publication (Table 3).
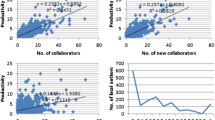
A lower percentage of scientists had a single dataset entry than a single publication entry, with 47.0 % of scientists having only one publication and 31.1 % of scientists having only one dataset submission (Fig. 2). However, comparing the productivity of dataset submissions to publications is not trivial; scientists seem to frequently break up dataset submissions into subsequences of base pairs, with no immediately apparent relationship between the two types of submissions. Looking at the general question of transiency rates in the network, 50.4 % of scientists who submitted a publication were only active for 1 year, while 48.9 % of scientists who submitted datasets first were active for only 1 year. Furthermore, 45.5 % of scientists who submitted both a publication and submission in their first year were active for only 1 year. Globally, the transiency rate (Braun et al. 2001; Price and Gürsey 2001) was 49.3 %.
The Bermuda Principles had a clear impact on the submission practices of scientists in the GenBank community. The number and overall percentage of scientists who first contributed to the data repository via a dataset submission were both negligible prior to the policy shift. After 1995, the number of scientists making their first knowledge contribution via a dataset submission increased dramatically, perhaps at the expense of getting their name on a publication. An alternative explanation for the observed patterns is that a greater percentage of scientists and graduate students were able to make some contribution before leaving the network, versus never publishing at all in the submission regime prior to the Bermuda Principles.
Not only did a greater percentage of scientists begin to enter the network submitting datasets after the adoption of the Bermuda Principles, but the number of dataset submissions grew rapidly as well. Publication submission rates increased approximately linearly from 1996 to 2012, while dataset submissions increased steeply linearly upward (Fig. 3).
Productivity disparities in terms of datasets to publications continues to affect analysis. The actual number of submissions to publications should not be taken at face value, but instead the growth curves should be compared. Similarly, when looking at the number of authors per dataset submission versus authors per publication submission, the large number of submissions skews the results. In the years where the mean number of authors per submission increased from approximately 5 to almost 13, there were over 5000 dataset submissions with more than 100 authors. A better indicator of general authors per paper continues be the number of authors per publication, where no group of scientists completely skew the results (Fig. 4).
Structural changes
Another way to look at the evolution of this community is to analyze the structural changes of the collaboration network over time. Looking at, the number of relationships between members of the network increased dramatically between the 1990–1993 and 1994–1997 time slices. Although the number of edges nearly doubled, there was still significant overlap between the edges present in the dataset submission and publication networks, as evidenced by the marginal increase the dataset network contributed to the combined network (Table 4).
More interestingly, the giant component of both networks exceeds 90 % by the second time window, remains above 95 % for the combine network, and above 90 % for the dataset submission network only for the last 3 time windows. Although the submission network grew more rapidly over the past 20 years, the publication network still remains larger and better connected.
Scientists in the publication network are also, on average, slightly more connected than the scientists in the dataset submission network. The mean degree (unique edges, not weighted edges or counts of repeat relationships) is 62 % higher in the publication network. The overall centralization of the network decreased over time as well, with the alpha coefficient on the power law fit of the degree distribution decreasing near monotonically over time (except for the final time window) as the network grew (see Table 5). There was some instability in the alpha values for the power law fits for the initial time windows, which was not significant for any of the networks until the 1990–1993 time window, and not significant for the dataset network until the 1998–2001. This was due to the fact that the networks had not reached a critical mass in the early years, particularly for the submission network. One other point to note is that the power law fits have relatively high minimum thresholds which is due to the fact that most scientists will start off with many connections because there are many authors on papers and datasets. This reveals the bipartite nature of the network. Overall, once the distribution takes on a power law form, the numbers indicate a slightly hierarchical organizational structure, although increasingly less so.
The clustering coefficient values further support the arguments that the growth in the network has made it more diffuse. Instead of the clustering coefficient increasing over time as active members consolidate their relationships and take advantage of weak ties, the growth in the network drives separation, reducing the clustering coefficient over time. The high clustering coefficient in the early years of the dataset network suggest that there was a relatively small group of well-connected individuals submitting datasets prior to 1996. However, after the Bermuda Principles, many scientists began submitting datasets, rapidly decreasing the density of the network, which decreased from 5.1 × 10−4 in 1982–1985 to 4.23 × 10−5 in 2010–2013.
The combination of the lower mean degree, lower clustering coefficient, and higher power law coefficient in the dataset network suggests that it is more hierarchically organized, with a few more prominent scientists coordinating the sequencing of genomic data of disparate teams comprised of members who remain relatively ultra-peripheral, having fewer opportunities to collaborate outside of their core group of collaborators. This could mean that more students are getting credit for work, or it could mean that the general social structure is changing. The data on the cumulative and publication only networks seems to suggest that the network is not getting more centrally organized, but local variations within the entire network may be different.
Added insight of the submission data
Although the dataset submission network appears at first glance to add only marginal value to our understanding of the collaboration network, delving deeper into the data suggest otherwise. Specifically, out of the cumulative network, 28.1 % of scientists contributed to the publication network but not the data submission network, 24.75 % of scientists who have contributed datasets but not publications, and 46.05 % that are in both. This indicates that large portions of the community do not overlap with both submission and publication networks. Looking at the relationships between the scientists, there are 4,393,748 unique edges in the publication network and 2,842,271 edges in the submission network. 55.6 % of the edges present in the submission network are not present in the publication network, 71.2 % are present in the publication network that are not present in the submission network, and 28.75 % edges are present in both.Footnote 2
We suggested that the ability to submit datasets only allows lesser experienced researchers to get some credit for a knowledge contribution. This suggestion was based on the number of scientists entering the network via a dataset submission, and the relatively hierarchical structure of the dataset submission network. Furthermore, the disparity in dataset submissions to publications further supports the argument that a dataset submission is “less than” a publication in terms of productivity. This leads to the general question of whether collaboration on a dataset submission precedes collaboration on a publication, or vice versa.
Analysis of the temporal sequencing of submissions supports the idea that sequencing and submission of genomic datasets precedes the submission of a publication. There are two numbers provided in an upper and lower bound on our estimates of precedence in Table 6. The lower bound was calculated by eliminating all dataset submissions that did not have a date associated with them, the upper bound included all submissions, regardless of whether or not there was a date associated with the dataset. The latter choice was done under the assumption that the lack of metadata is homogenously distributed over the years analyzed.
Results in Table 6 seem to suggest that submission collaboration proceeded to publication coauthorship more frequently than the opposite, approximately one-third of the collaborations occurred concurrently, with a fraction of collaborations that published paper before submitting data sets. This prompted us to speculate that the ability of an author to garner a large number of submission nodes as the support, or “collaboration capacity”, is vital for him/her to rise to the rank in the network and maintain the status. In other words, we may assume submission nodes that cluster around a node or a hub are an indication of the collaboration capacity of that node or hub. A closer examination of the annotations and name tracking revealed that concurrent submission-publication collaborations involved a certain level of intellectual maturity, which is necessary for translating into higher level of collaboration (concurrent collaboration) in which the primary author is also deeply involved in laboratory work (direct submission). It is likely this happened to most postdoc researchers. Dataset-first collaboration reflects a hierarchy of division of labor in laboratory work, with junior researchers (graduate students) performing the lab work and carrying out to its end (submission) and principal investigators (as represented by publication author nodes primarily) steering the research and less involved in lab work. It is possible for a junior node at the beginning of time to be primarily in submission network and later disappear from the submission network to either evolve into a publication-network-only node or never show up again in the whole network. How long it took a dataset-first node to become a concurrent node (or disappear altogether) and eventually in a publication-only network will be another issue worth exploring.
Conclusions and future work
The growth in cyberinfrastructure enabled science is affecting the scale and structure of scientific collaboration. The extent to which this is true is not fully known, yet early research is demonstrating clear increases in large scale projects and associated publications, as well as growth in use of cyberinfrastructure to support multinational collaborations. In this paper we argued that it was not possible to understand the effect CI has on the scale and structure of scientific collaboration until we understand the general structural characteristics of communities that have emerged around large sale CI investments.
We chose to study one example of CI supported scientific community, the Genomics community. The community uses GenBank, a large nucleotide sequence repository integrated into an international consortium of repositories, to archive and make available DNA sequence data to the international research community. Because GenBank allows scientists to upload and share datasets, we hypothesized that using trace metadata from the datasets would provide additional information regarding the collaborative behaviors of scientists than if we were to just use metadata from formal publications. A number of factors specific to the genomics community contributed to our belief this was possible. First, the scientific community voluntarily accepted a standard where all data needed to be submitted to the database prior to the related article being published. Therefore, scientists have a strong incentive to share their data. Furthermore, repository has developed tools to support batch uploading, batch record importing from the patent office, and bulk downloading of data.
An important point to consider is that the results from the research in this paper may not generalize directly to other communities that use repositories. The overview of GenBank provided in the Data Source section suggests that the repository is fully integrated into the practice and culture of its community. Accepted modes of practice ensure that the database gets used; this level of integration is not something that should be assumed to be present in other fields.
Within the context of this community, analysis shows that constructing collaboration networks using coauthorship of datasets and publications provides much more information regarding the collaborative behaviors of scientists than using traditional publication data alone. Viewing the two networks independently, we see structural differences, with the submission network being slightly more hierarchical; scientists within the dataset network have lower clustering coefficients, suggesting they are not as likely to work with collaborators of collaborators, they have fewer formal connections than scientists in the publication network, and exhibiting lower tendencies for triadic closure. Our analysis suggests that less experienced researchers are more likely to work on and publish a dataset than a formal publication, thus providing a better indication of the true transiency rate of the community.
Furthermore, scientists were much more likely to collaborate on a dataset prior to collaborating on a formal publication, or nearly as likely to submit both data and publications simultaneously, than collaborate on a publication first. There are several potential reasons for this observation. First, researchers are more likely to use graduate students to sequence the data, which would precede or at least be concurrent with, a publication. This supports our intuition that generating data is still only part of the process of research, with publication being the end goal (according to the norms of science). Also, because such a large portion of researchers collaborate on a dataset submission and not a publication, we can argue that the ability to submit datasets unattached to formal publications allows researchers to rapidly contribute datasets without having to wait for formal publications. Our observations are limited by the fact that metadata standards for the community are relatively low, with many dataset submission records missing important information. We are also unable to determine the differences in metadata entry practices for the various researchers and institutions that contribute data, and have not included collaborations on patents, which may constitute an increasingly important component of this field given the economic value of genomics research.
At this point, we have assumed that all scientists place equal emphasis on claiming intellectual ownership of data submitted to the repository, yet this may not be true. Nevertheless, our research provides evidence to support the argument that trace data from a scientific data repository can be used to construct a more accurate representation of scientific collaboration than using publication data alone. Some of the methodological concerns raised by Laudel (2002), and Glänzel and Schubert (2005) are being addressed by advances in computational science, which are facilitating the extraction of unstructured data from multiple sources to provide a more detailed picture of scientific interactions than publication data alone.
There are also several policy related implications for our research as well. First, social mechanisms play a clear role in facilitating the integration of cyberinfrastructure into the research practice of the community. The genomics community functionally mandates use of the repository, thus providing significant incentive for practitioners to contribute. Several community driven policy decisions have also impacted submission behaviors. Specifically, the Bermuda Accords set the stage for scientists to being directly submitting data without that data being attached to a formal publication. The result of the Accords, and subsequent agreements, was the rapid expansion of contributions to the repository.
Our next project will focus on individual scientists within the network and their collaboration patterns, exploring in greater detail the networks of scientists across both dataset and publication submission networks. This includes looking at the role scientists play in those networks, whether common measures of status (i.e., centrality) are stable across networks, and with whom they collaborate in the different networks. Further in the future, our work will focus on integrating patent metadata into the analysis, as well as reconciling the differences in scale between publications and datasets in order to better estimate strengths of relationships between scientists in the community. Once the patent metadata is integrated, we can begin to explore the rate of international collaboration and the differences in collaborative behaviors by country of affiliation. Exploring the international aspect of collaboration will facilitate the exploration of the relationships between funding practices and collaborative behaviors, and thus give a better insight into the results of countries’ investments in the building of scientific capacity in this research field.
Notes
In an undirected network, these calculations should only be done for the upper or lower triangle, not both.
These totals differ slightly from 5 because datasets and publications with no dates are included in the figures.
References
Advanced Cyberinfrastructure Division, Cyberinfrastructure framework for 21st century science and engineering: Vision. http://www.nsf.gov/cise/aci/cif21/CIF21Vision2012current.pdf.
Arias, J. J., Pham-Kanter, G., & Campbell, E. G. (2015). The growth and gaps of genetic data sharing policies in the United States. Journal of Law and the Biosciences, 2, 56–68.
Barabási, A. L., & Albert, R. (1999). Emergence of scaling in random networks. Science, 286, 509–512.
Barabási, A.-L., Jeong, H., Néda, Z., Ravasz, E., Schubert, A., & Vicsek, T. (2002). Evolution of the social network of scientific collaborations. Physica A: Statistical Mechanics and its Applications, 311, 590–614.
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., & Sayers, E. W. (2013). GenBank. Nucleic Acids Research, 41(Database issue), D36–D42.
Braun, T., Glänzel, W., & Schubert, A. (2001). Publication and cooperation patterns of the authors of neuroscience journals. Scientometrics, 51, 499–510.
Collins, F. S., Morgan, M., & Patrinois, A. (2003). The Human Genome Project: Lessons from large-scale biology. Science, 300, 286–290.
Costa, M., Qin, J., & Wang, J. (2014). Research networks in data repositories. In Joint conference of digital libraries (JCDL) London, UK, September 8–10, 2014.
Cronin, B., Shaw, D., & La Barre, K. (2003). A cast of thousands: Co-authorship and sub-authorship collaboration in the twentieth century as manifested in the scholarly literature of psychology and philosophy. Journal of the American Society for Information Science and Technology, 54, 855–871.
Faniel, I. M., & Jacobsen, T. E. (2010). Reusing scientific data: How earthquake engineering researchers assess the reusability of colleagues’ data. Computer Supported Cooperative Work (CSCW), 19, 355–375.
Faniel, I. M., & Zimmerman, A. (2011). Beyond the data deluge: A research agenda for large-scale data sharing and reuse. International Journal of Digital Curation, 6, 58–69.
Glänzel, W., & Schubert, A. (2005). Analysing scientific networks through co-authorship. In Handbook of quantitative science and technology research (pp. 257–276). http://springerlink.bibliotecabuap.elogim.com/chapter/10.1007%2F1-4020-2755-9_12.
Hey, T., Tansley, S., & Tolle, K. (Eds.). (2009). “The fourth paradigm” data-intensive scientific discovery. Redmond, WA: Microsoft.
Holme, P., & Saramäki, J. (2012). Temporal networks. Physics Reports, 519, 97–125.
King, C. (2012). Multiauthor papers: Onward and upward. Science Watch Newsletter, July 2012. http://archive.sciencewatch.com/newsletter/2012/201207/multiauthor_papers/.
Laudel, G. (2002). What do we measure by co-authorships? Research Evaluation, 11, 3–15.
Marcial, L. H., & Hemminger, B. M. (2010). Scientific data repositories on the Web: An initial survey. Journal of the Association for Information Science and Technology, 61, 2029–2048.
Newman, M. E. J. (2001a). Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality. Physical Review E, 64, 016132.
Newman, M. E. J. (2001b). Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality. Physics Review E, 64(1). http://pre.aps.org/pdf/PRE/v64/i1/e016132.
Newman, M. E. J. (2003). The structure and function of complex networks. SIAM Review, 45, 167–256.
Porter, A. L., & Rafols, I. (2009). Is science becoming more interdisciplinary? Measuring and mapping six research fields over time. Scientometrics, 81, 719–745.
Price, D. J. S., & Gürsey, S. (2001). Studies in scientometrics I transience and continuance in scientific authorship. Ciência da Informação, 4. http://revista.ibict.br/cienciadainformacao/index.php/ciin-f/article/view/1611.
Qin, J., Costa, M., & Wang, J. (2014). Attributions from data authors to publications: Implications for data curation. In The 9th international digital curation conference, 24–27 February 2014, San Francisco.
Qin, J., Lancaster, F. W., & Allen, B. (1997). Levels and types of collaboration in interdisciplinary research. Journal of the American Society for Information Science, 48, 893–916.
Rodriguez, H., Snyder, M., Uhlén, M., et al. (2009). Recommendations from the 2008 International Summit on Proteomics Data Release and Sharing Policy: The Amsterdam Principles. Journal of Proteome Research, 8, 3689–3692.
Szalay, A. S., & Blakeley, J. A. (2009). Grey’s laws: Database-centric computing in science. In T. Hey & S. Tansley (Eds.), The fourth paragim: Data-intensive scientific discovery (pp. 5–11). Redmond, WA: Microsoft Research.
Acknowledgments
This research is sponsored by the NSF’s Science of Science Policy Program, Grant Number 1262535. The authors thank Jun Wang, Qianqian Chen for their technical assistance in data processing and analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Costa, M.R., Qin, J. & Bratt, S. Emergence of collaboration networks around large scale data repositories: a study of the genomics community using GenBank. Scientometrics 108, 21–40 (2016). https://doi.org/10.1007/s11192-016-1954-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11192-016-1954-x