Abstract
Southeast Asia is a major shark product trade hub, and many of the world’s largest shark product selling markets are located in the region. Malaysia is the second largest importer of shark fins in the world, approximately 2500 metric tonnes of fins were imported between 2000 and 2016. The country is also a major shark fisher—the eighth largest in the world in terms of yearly catch. Yet the composition of the species of shark involved in the Malaysian trade remains largely unknown. Not knowing the composition of the sharks involved in the fin trade in a major trade hub presents challenges when it comes to determining conservation goals, setting appropriate catch quotas and establishing new, or revisiting previous conservation listings. Using a fragment of the mtDNA COI gene we attempted to DNA barcode 147 dried shark fins. We identified a number of fins that belonged to species listed on either CITES appendix I, or II, and the composition of the sharks identified in samples collected from Peninsular Malaysia appears to be different from that of other trade hubs within the region. Given these differences, we suggest that further DNA barcoding studies be performed throughout the region at regularly repeated intervals to build a more comprehensive picture of the sharks involved in the trade within the region and globally, this information will be useful to policy makers and conservation planners.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shark populations have declined precipitously over the past 40 years, and many species are now considered at risk of extinction, a consequence of the demand for shark fin products in many Asian communities (Dulvy et al. 2014; Fields et al. 2020; Pacoureau et al. 2021). Furthermore, an increasing demand for shark meat and liver oil supplies a near USD 1 Billion industry (Dulvy et al. 2014; O’Bryhim et al. 2017). Even though many countries are obligated by international treaty, applied under the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), to monitor the species of shark that are traded within, and through their boarders (Dulvy et al. 2017; Cardeñosa et al. 2020; Prasetyo et al. 2020), products from endangered species of shark are readily available to members of the public (Sembiring et al. 2015; Wainwright et al. 2018; Cardeñosa 2019; Liu et al. 2021).
The monitoring of this trade is not a trivial matter, shipments of shark fins are frequently given generic labels (i.e., “seafood” or “dried marine product”) to avoid detection by the authorities. Further compounding this difficulty, once a fin is processed and dried it becomes nearly impossible to identify from visual characteristics alone (Wainwright et al. 2018). The common practice of shark finning, where the high value fin is removed and the low value carcass is disposed of at sea means that many of the diagnostic features used in identification are no longer present. This makes the task of regulatory bodies more challenging, and in some cases impossible (Shivji et al. 2002). DNA barcoding techniques have long been used to overcome these challenges, they allow positive identifications to be made from small samples of dried and processed fins that would otherwise be impossible to identify. These methods have already been applied throughout the world to identify the species of shark that a fin or meat came from (Shivji et al. 2002; Ward et al. 2008; Sembiring et al. 2015; O’Bryhim et al. 2017; Fields et al. 2018; Appleyard et al. 2018; Hobbs et al. 2019; Abdullah et al. 2020; Liu et al. 2021; French and Wainwright 2022; Neo et al. 2022).
Work examining the shark fin trade in East and Southeast Asia has tended to concentrate on Hong Kong (Fields et al., 2018), mainland China (Cardeñosa et al. 2020), Indonesia (Sembiring et al. 2015; Prasetyo et al. 2020) and Singapore (Wainwright et al. 2018), while each of these represents a major trade hub for shark products, there are other trade hubs in the region. It is important that conclusions and management decisions are based on representative data from all major shark markets and distribution centers. Each country is likely to have its own complex supply chain logistics, fisheries and customer specific nuances. For example, Singapore does not host a major shark fishery, rather, shark products are imported, processed and then exported to other markets (Boon 2017). The supply chain dynamics of the fin trade in Mainland China differ from those in Hong Kong and consequently the composition of the shark species landed differs between each market (Cardeñosa et al. 2020). These differences make it important that shark landings, and the species involved in the shark product trade are monitored wherever shark fishing takes place, or shark products are sold.
Hong Kong, Malaysia, China and Singapore account for 90% of all global imports of shark fin (Oakes and Sant 2019). Malaysia is the second largest importer of shark fins in the world importing approximately 2500 metric tonnes of fins between 2000 and 2016. In terms of metric tonnes of shark caught, Malaysia is the world’s eighth largest shark fisher, with a mean yearly catch of over 21,000 tonnes between 2007 and 2017 (Oakes and Sant 2019). Despite the scale of the trade in Malaysia and the implications this could have for shark conservation, very little is known about the species composition of the sharks traded within and through this country. With this study we aim to shed light on what species of shark are traded in Malaysia, and by doing so we provide data that can be used for future comparisons and policy decisions.
Methods
In total 147 dried shark fins were collected from stores in Dungun and Kuantan, Terengganu and Pahang States respectively, east coast Peninsular Malaysia. DNA was extracted from approximately 25 mg of tissue using a Qiagen DNeasy Blood & Tissue Kit following the manufactures instructions. Initially, we attempted to amplify an approximate 650 bp fragment of the mtDNA COI gene using the Fish-BCL-5′ TCAACYAATCAYAAAGATATYGGCAC-3′ and Fish-BCH-5′ ACTTCYGGGTGRCCRAARAATCA-3 primer pair (Appleyard et al. 2018) in 25 µl reaction volumes of 12.5 µl of GoTaq Master Mix Green (Promega), 1 µl Bovine Serum Albumin, 1.0 µl of each 10 µM primer, 7.5 µl water and 2 µl of template DNA. PCR cycling conditions were 94 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 50 °C for 1 min 30 s, 72 °C for 1 min and a final extension step of 72 °C for 10 min. Samples that failed to amplify were re-extracted and amplification was attempted again with the same primer set. If amplification failed again we targeted a shorter fragment (313 bp) of the mtDNA COI gene using the mlCOIintF & LoboR1 primers (Leray et al. 2013; Lobo et al. 2013), PCR cycling conditions were: 94 °C for 60 s, 5 cycles of 94 °C for 30 s, 48 °C for 120 s, 72 °C for 60 s, and 35 cycles of 94 °C for 30 s, 54 °C for 120 s, 72 °C for 60 s; and 72 °C for 5 min (Wainwright et al. 2018). Finally, in any sample that failed with either of the two previously described primer sets, we attempted to amplify a small 150 bp fragment of the mtCOI gene using the VF2_tl and Shark150R primer set with the following cycling conditions: 2 min at 94 °C, followed by 35 cycles at 94 °C for 1 min, 52 °C for 1 min, 72 °C for 1 min, and a final extension period of 10 min at 72 °C (Cardeñosa et al. 2017). Negative PCR controls containing no template DNA were included to identify any possible contamination issues, and successful PCR amplification was confirmed on a 1% TAE agarose gel. All PCR reactions were enzymatically cleaned (ExoSAP) and bidirectionally Sanger sequenced by Macrogen, Inc.
Sequences were viewed, and primer sites removed with Geneious v2020.2.4 (http://www.geneious.com/, Kearse et al. 2012). Consensus sequences were constructed from forward and reverse reads, and species identifications were made using the Barcode of Life Database (BOLD) (http://www.boldsystems.org/) and the BLASTn function in GenBank (http://www.ncbi.nlm.nih.gov/). We considered species identification positive if; (1) BOLD returned a 100% match for a single species, and indicated that no closely allied congeneric species currently exist, and (2) the same ID was made in GenBank with a 100% match.
Results
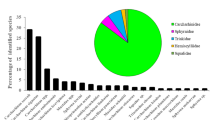
Using a combination of primers, we were able to positively identify 120 of the collected fins to the species level (n = 21), and a further 24 to genera (n = 3) (Table 1, Fig. 1). Ninety-nine samples were amplified with the Fish-BCL/BCH primer set, 31 amplified with the mlCOIintF & LoboR1 primer set, and amplification for 14 could only be achieved with the VF2_tl and Shark150R primers. We failed to produce usable sequences (low quality, or no amplification) in four samples.
Species and genus composition of the identified shark fins. Also indicated is the IUCN status of each. The category ‘not applicable’ is not an IUCN category, in this instance it indicates a genus that contains multiple species that have different IUCN designations, see Table 1 for additional details and CITES list status
Ten of the positively identified species are listed on the CITES appendix II, or 48% of all species identified in our work. We were unable to resolve samples identified as belonging to the genus Pristis (sawfish) to the species level, but, it should be noted that all members of this genus are listed under CITES appendix I. Similarly, we could not ID members of the Rhynchobatus (wedgefish) beyond genera, but all members of this genus are listed under CITES appendix II. Fourteen of the positively identified species (67%) are listed as threatened (vulnerable, endangered, or critically endangered) on the IUCN Red List, the remainder are listed as near threatened (n = 5), data deficient (n = 1), or least concern (n = 1) (Table 1). Eighteen (12%) of the products sold as shark fin were identified as belonging to rays.
Discussion
As with other work that has used DNA barcoding techniques to identify the species of shark a fin was removed from (Fields et al., 2020, 2018; Hobbs et al., 2019; Kuguru et al., 2018; Marchetti et al., 2020; O’Bryhim et al., 2017; Sembiring et al., 2015; Wainwright et al., 2018; Ward et al., 2008), we show numerous species of shark that have some degree of control over their trade are freely available to consumers. Most of these regulations are implemented under CITES appendix II which applies to species not necessarily threatened with extinction, but in which trade must be controlled in order to avoid utilization incompatible with their survival. Additionally, we identified one sample belonging to a genera of shark species (Pristis spp. or the sawfish) that is listed under CITES appendix I, meaning that trade in this species is permitted only in exceptional circumstances. Despite success in amplifying the longest fragment of the three primer pairs used, we were unable to identify the exact species of sawfish that this fin came from, this sequence matched 100% with all four current living species. In spite of this limitation, the presence of this genus should be of concern to regulatory bodies—all members of this genus are listed under CITES appendix I, and classified as Vulnerable or Endangered on the IUCN Red List.
The top five species most frequently occurring in this work (bull shark, spot-tail shark, great hammerhead shark, silky shark and scalloped hammerhead shark) accounted for 56% of the 21 species we were able to identify to species level, this possibly indicates a targeted fishery where these sharks are caught in preference to others, this suggestion is consistent with work from Indonesia that showed a bias in fishing effort towards silky, and hammerhead sharks (Sembiring et al. 2015). Similarly the silky shark was one of the top most frequently occurring sharks sold in Singapore (Wainwright et al. 2018). The frequent occurrence of fins from the silky shark in Southeast Asian markets is unfortunately not surprising, this shark is one of the most commonly caught shark species on the planet and is one of the top contributors of fins to the shark fin trade (Kraft et al., 2020; Oliver et al., 2015), and correspondingly this once widespread and abundant shark has seen populations decline by an estimated 85% over the past two decades (Kraft et al. 2020).
The two most frequently encountered sharks in this survey were the bull shark (Carcharhinus leucas) and spot-tail shark (Carcharhinus sorrah), these two sharks make up over a quarter of all the samples identified in our work. Bull sharks have been identified in other studies examining the shark fin trade in Southeast Asia (Sembiring et al. 2015; Wainwright et al. 2018), Hong Kong and Guangzhou (Cardeñosa et al. 2020), however, bull sharks tend to be much less frequently encountered in these regions—generally making up less than 2% of all the fins examined. In the samples we collected from Malaysia the bull shark accounted for 14% of all identifications. It is a similar situation with the spot-tail shark, in the Malaysian samples it makes up 14% of all identifications, in Indonesia it was 7%, Hong Kong 1% and Guangzhou 2%. It should be noted that the work performed in Indonesia dates from 2015 and the composition of the sharks involved in the fin trade could be different now, however, the work from Singapore, Hong Kong and Guangzhou is recent and provides a good comparison of the species composition in each market. These differences reinforce the need to monitor multiple trade hubs, if this is not done, it may not be possible to design effective conservation strategies for sharks. For example, bull sharks are not CITES listed, and are classified as near threatened on the IUCN red list. Considering the low occurrence of this species in the markets of Hong Kong, Guangzhou and Singapore, this could be an appropriate designation if only data from these markets is used, but, the high occurrence of bull shark fins in Malaysian markets suggest a targeted fishery. Bull sharks are known to aggregate in near-shore locations such as estuaries and the associated high human populations found in these areas typically results in elevated fishing pressure, these factors make bull sharks particularly vulnerable to local extirpation, which in turn elevates the potential for global extinction (Karl et al. 2011; Martin 2005).
The scalloped hammerhead (Sphyrna lewini), one of the most commonly encountered species in the samples collected from Malaysia, is also frequently observed in Singapore, Hong Kong, and Guangzhou. This is despite its listing on the CITES appendix II and designation as critically threatened on the IUCN Red List. Notable by its frequent occurrence in samples collected in Malaysia is the presence of fins from the great hammerhead shark (Sphyrna mokarran), this species made up more than 10% of all individuals identified here, yet it was absent in fins examined in Singapore, Indonesia and present at a frequency of < 1% in samples from Hong Kong and Guangzhou, this discrepancy further reiterates the need to monitor multiple trade hubs to build a comprehensive picture of the species of shark involved in the fin trade. This information is critical when setting appropriate catch quotas, determining the conservation status and appropriate CITES designation of shark species. Recent work suggests that the populations of scalloped hammerhead and great hammerhead sharks have declined by more than 80% (Pacoureau et al. 2021), and both of these are in the top five most frequently encountered species in our work.
To our knowledge, this is the first study to explicitly examine the composition of the shark fin trade in Malaysia, a country that is one of the largest importers, and fishers of shark in the world. We show distinct differences in terms of species composition to other markets in Southeast, and East Asia. We hope this information will be useful to policy makers and law enforcement agents, we provide baseline data and suggest that continued monitoring of the shark fin trade throughout Malaysia and Southeast Asia be implemented to assist in the management and conservation of sharks within the region, and throughout the world. The continuing advancement and development of increasingly sophisticated and cost effective molecular diagnostic techniques will provide additional support to these biodiversity monitoring activities. The evidence for the overexploitation of sharks is unequivocal, and many species of shark are at serious risk of extinction. If these extinctions are to be avoided, it is critical that national and international trade obligations are met, and enforcement is taken seriously to prevent the unsustainable trade of threatened species that continues unabated throughout the world.
References
Abdullah A, Nurilmala M, Muttaqin E, Yulianto I (2020) DNA-based analysis of shark products sold on the Indonesian market towards seafood labelling accuracy program. Biodivers J Biol Divers. https://doi.org/10.13057/biodiv/d210416
Appleyard SA, White WT, Vieira S, Sabub B (2018) Artisanal shark fishing in Milne Bay Province, Papua New Guinea: biomass estimation from genetically identified shark and ray fins. Sci Rep. https://doi.org/10.1038/s41598-018-25101-8
Boon (2017) http://www.trafficj.org/publication/17_The_Shark_and_Ray_Trade_in_Singapore.pdf
Cardeñosa D (2019) Genetic identification of threatened shark species in pet food and beauty care products. Conserv Genet. https://doi.org/10.1007/s10592-019-01221-0
Cardeñosa D, Fields A, Abercrombie D et al (2017) A multiplex PCR mini-barcode assay to identify processed shark products in the global trade. PLoS ONE 12:e0185368. https://doi.org/10.1371/journal.pone.0185368
Cardeñosa D, Fields AT, Babcock EA et al (2020) Species composition of the largest shark fin retail-market in mainland China. Sci Rep 10:12914. https://doi.org/10.1038/s41598-020-69555-1
Dulvy NK, Fowler SL, Musick JA et al (2014) Extinction risk and conservation of the world’s sharks and rays. eLife 3:e00590. https://doi.org/10.7554/eLife.00590
Dulvy NK, Simpfendorfer CA, Davidson LNK et al (2017) Challenges and priorities in shark and ray conservation. Curr Biol 27:R565–R572. https://doi.org/10.1016/j.cub.2017.04.038
Fields AT, Fischer GA, Shea SKH et al (2018) Species composition of the international shark fin trade assessed through a retail-market survey in Hong Kong: Shark Fin Trade. Conserv Biol 32:376–389. https://doi.org/10.1111/cobi.13043
Fields AT, Fischer GA, Shea SKH et al (2020) DNA zip-coding: identifying the source populations supplying the international trade of a critically endangered coastal shark. Anim Conserv. https://doi.org/10.1111/acv.12585
French I, Wainwright BJ (2022) DNA barcoding identifies endangered sharks in pet food sold in Singapore. Front Mar Sci 9:9
Hobbs CAD, Potts RWA, Walsh MB et al (2019) Using DNA barcoding to investigate patterns of species utilisation in UK shark products reveals threatened species on sale. Sci Rep 9:1–10. https://doi.org/10.1038/s41598-018-38270-3
Karl SA, Castro ALF, Lopez JA et al (2011) Phylogeography and conservation of the bull shark (Carcharhinus leucas) inferred from mitochondrial and microsatellite DNA. Conserv Genet 12:371–382. https://doi.org/10.1007/s10592-010-0145-1
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kraft DW, Conklin EE, Barba EW et al (2020) Genomics versus mtDNA for resolving stock structure in the silky shark (Carcharhinus falciformis). PeerJ 8:e10186. https://doi.org/10.7717/peerj.10186
Kuguru G, Maduna S, da Silva C et al (2018) DNA barcoding of chondrichthyans in South African fisheries. Fish Res 206:292–295. https://doi.org/10.1016/j.fishres.2018.05.023
Leray M, Yang JY, Meyer CP et al (2013) A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool 10:34. https://doi.org/10.1186/1742-9994-10-34
Liu CJN, Neo S, Rengifo NM et al (2021) Sharks in hot soup: DNA barcoding of shark species traded in Singapore. Fish Res 241:105994. https://doi.org/10.1016/j.fishres.2021.105994
Lobo J, Costa PM, Teixeira MA et al (2013) Enhanced primers for amplification of DNA barcodes from a broad range of marine metazoans. BMC Ecol 13:34. https://doi.org/10.1186/1472-6785-13-34
Marchetti P, Mottola A, Piredda R et al (2020) Determining the authenticity of shark meat products by DNA sequencing 9:1194
Martin R (2005) Conservation of freshwater and euryhaline elasmobranchs: a review. J Mar Biol Ass 85:1049–1073. https://doi.org/10.1017/S0025315405012105
Neo S, Kibat C, Wainwright BJ (2022) Seafood mislabelling in Singapore. Food Control 135:108821. https://doi.org/10.1016/j.foodcont.2022.108821
Oakes and Sant (2019) https://www.traffic.org/publications/reports/an-overview-of-major-shark-and-ray-catchers-traders-and-species/
O’Bryhim JR, Parsons ECM, Lance SL (2017) Forensic species identification of elasmobranch products sold in Costa Rican markets. Fish Res 186:144–150. https://doi.org/10.1016/j.fishres.2016.08.020
Oliver S, Braccini M, Newman SJ, Harvey ES (2015) Global patterns in the bycatch of sharks and rays. Mar Policy 54:86–97. https://doi.org/10.1016/j.marpol.2014.12.017
Pacoureau N, Rigby CL, Kyne PM et al (2021) Half a century of global decline in oceanic sharks and rays. Nature 589:567–571. https://doi.org/10.1038/s41586-020-03173-9
Prasetyo AP, McDevitt AD, Murray JM, et al (2020) Disentangling the shark and ray trade in Indonesia to reconcile conservation with food security. bioRxiv 2020.12.08.416214. https://doi.org/10.1101/2020.12.08.416214
Sembiring A, Pertiwi NPD, Mahardini A et al (2015) DNA barcoding reveals targeted fisheries for endangered sharks in Indonesia. Fish Res 164:130–134. https://doi.org/10.1016/j.fishres.2014.11.003
Shivji M, Clarke S, Pank M et al (2002) Genetic identification of pelagic shark body parts for conservation and trade monitoring. Conserv Biol 16:1036–1047
Wainwright BJ, Ip YCA, Neo ML et al (2018) DNA barcoding of traded shark fins, meat and mobulid gill plates in Singapore uncovers numerous threatened species. Conserv Genet 19:1393–1399. https://doi.org/10.1007/s10592-018-1108-1
Ward RD, Holmes BH, White WT, Last PR (2008) DNA barcoding Australasian chondrichthyans: results and potential uses in conservation. Mar Freshwater Res 59:57. https://doi.org/10.1071/MF07148
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Simon Hew is an independent researcher.
Rights and permissions
About this article
Cite this article
Seah, Y.G., Kibat, C., Hew, S. et al. DNA barcoding of traded shark fins in Peninsular Malaysia. Rev Fish Biol Fisheries 32, 993–999 (2022). https://doi.org/10.1007/s11160-022-09713-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11160-022-09713-y