Abstract
Accurate maize plant counting plays an essential role in prediction of leaf area index (LAI), aboveground biomass (AGB) and yield. Plant counting of maize inbred lines at early growth stage will result in counting bias caused by death and growth of small seedlings. Therefore, the estimation of LAI and AGB might be negatively affected by plant counting bias at early growth stage. In this study, morphologic discrimination model (MDM) and interpolation discriminant model (IDM) were proposed for plant counting of maize inbred lines at second to fourth (V2–V4) leaf and fourth to sixth (V4–V6) leaf stages with different uncrewed aerial vehicles (UAV) flight heights. Automatic optimum angle calculation of each row, location-based plant cluster segmentation and mosaic method were presented to improve the estimation accuracy of plant counting. Then, the impact of accurate plant counting was evaluated in LAI and AGB prediction at the two growth stages. The results indicated that germination rate difference of some inbred lines could reach up to 38% between V2–V4 and V4–V6 leaf stages. The proposed method accurately estimated the plant counting in the UAV images during V2–V4 leaf stage (R2 = 0.98, RMSE = 7.7, rRMSE = 2.6%) and V4–V6 leaf stage (R2 = 0.86, RMSE = 2.0, rRMSE = 5.5%). The estimated LAI and AGB with plant numbers calculated at V4–V6 leaf stage correlated better with the field measurements (R2 = 0.85 and R2 = 0.9, respectively) compared with those estimated at V2–V4 leaf stage (R2 = 0.8 and R2 = 0.86, respectively). This research indicates that better estimation of LAI and AGB in the field were obtained by accurate plant counting in the late growth stage using UAV images and provides valuable insight for more accurate prediction of yield and crop management and breeding.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.) is one of the main grain crops widely cultivated around the world. Effective calculation of maize seedling numbers can not only assess the quality of the machine sowing, but also evaluate the seedling emergence rate and distribution characteristics of row and plant spacing (Findura et al., 2018; Font et al., 2014; Ormond et al., 2018). Maize plant counting by visual detection is labour-intensive and time-consuming. To address this issue, recent researches have investigated the potential of plant counting using machine learning approach implemented to images obtained from different sensors of crop phenotyping platform (Aharon et al., 2020; Bendig et al., 2015; Herrmann et al., 2019; Virlet et al., 2017; Varela et al., 2018). Ground fixed and moving platform were found effectively at high-resolution and high-quality data collection, but were destructive to the experimental sites and limited to the sampled areas. Increased costs were also needed for large-scale experiments.
Recent technological advances have led to a boom in high-throughput phenotypic studies using uncrewed aerial vehicles (UAV) mounted on multiple low-cost portable sensors (Maimaitijiang et al., 2020). UAV has the advantages of high-resolution, real-time image acquisition and easy operation. Images obtained from UAV were used to calculate the seedling number (Feng et al., 2020; Osco et al., 2020), seedling distance (Gnädinger & Schmidhalter, 2017), plant density (Thorp et al., 2008), plants detection and classification (Guo et al., 2021), plant height (Hu et al., 2018), tassel number (Liu et al., 2020), biomass (Togeiro de Alckmin et al., 2021; Yu et al., 2016), LAI (Che et al., 2020; Lei et al., 2019) and yield (Herrmann et al., 2020). Different machine vision-learning approaches like random forest classifier (Li, Li, et al., 2019; Li, Xu, et al., 2019), support vector machine (Jin et al., 2017) and deep learning (Fan et al., 2018; Feng et al., 2020) were applied for plant counting. Deep learning can obtain higher estimation accuracy, but needs a larger quantity of sampled data, longer training time and higher cost in order to classify all of the plants (Finizola et al., 2019; Kamath et al., 2018; Shinde & Shah, 2018).
Current studies on the plant counting have been focused on the second to fourth (V2–V4) leaf stage with two or four fully developed leaves (Feng et al., 2020; Li, Li, et al., 2019; Li, Xu, et al., 2019). Maize inbred lines have different germination rates, different weak seedling ratios and the survival of the seedlings varies between them (Hu et al., 2017; Zhang et al., 2020). Ground-measured LAI and AGB per square meter were calculated using the plant numbers per square meter multiplied by the average leaf area and average biomass of plant samples, respectively (Bendig et al., 2015; Gong et al., 2021; Li et al., 2020). Bias will occur for LAI and AGB per square meter calculations when the plant counting was inaccurate. LAI and AGB per square meter from UAV were extracted using UAV-estimated leaf area and biomass of the plant sample divided by the area of each plot. Leaf area and biomass estimation model were built using spectral (Dong et al., 2019; Li et al., 2020), texture (Maimaitijiang et al., 2020; Zheng et al., 2019) and structure (Che et al., 2020; Jimenez-Berni et al., 2018; Lei et al., 2019) information extracted from UAV images. The quantitative analysis of LAI and AGB during the growth stage is of great significance for crop growth monitoring, yield estimation and crop breeding (Dong et al., 2019; Houborg & McCabe, 2018; Xie et al., 2018). Therefore, the effect of late-stage accurate maize plant counting on LAI and AGB estimation will be evaluated in this study.
A key question in this respect is whether plant counting of maize inbred lines with extensive genetic diversity in the later growth stage might avoid bias caused by death and growth of small seedlings. And whether plant counting of maize inbred lines in the later growth stage as compared with early growth stage might contribute to higher estimation accuracy in LAI and AGB. The aims of this study were to (1) identify the better growth stage for plant counting of maize inbred lines with extensive genetic diversity; (2) propose the optimal plant discrimination models for more accurate plant counting of maize inbred lines in the better growth stage; (3) evaluate the importance of accurate plant counting in predicting LAI and AGB.
Materials and methods
Field experiments and images acquisition by UAV
Field experiments were conducted at four experimental farms in Jilin (43° 16′ 45″ N, 124° 26′ 10″ E), Hebei (39° 27′ 37″ N, 115° 50′ 50″ E), Henan (36° 04′ 32″ N, 114° 31′ 41″ E) and Hainan (18° 23′ 06″ N, 109° 10′ 57″ E) in 2018 (Fig. 1). The soil types in Henan and Hebei are similar, composed of tidal soil and brown soil. The main soil types of Hainan and Jilin are black land and yellowish red soil, respectively. Before sowing, basal chemical fertilizers were applied in all plots at a rate of 60 kg P ha−1 and 80 kg K ha−1. Water management was conducted according to natural rainfall.
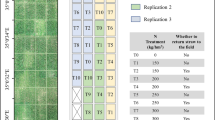
The experiments were set with three replicates. There were 30 maize inbred lines with extensive genetic diversity in each replicate. The plot size was 5 m × 1.2 m with two rows and 42 evenly sown maize seeds (Fig. 2). The distance between rows was 60 cm and the distance between plants was 25 cm. Destructive sampling area was 1.2 m × 1.2 m distributed on the left side of each plot. The total plant number were 90 for destructive sampling with 30 plants from each replicate. Three plants from ten inbred lines (Sample 10) were sampled to measure leaf area and dry biomass on 50 and 65 days after emergence. The ten inbred lines with different genetic backgrounds were screened from four subpopulations by breeders to ensure there were a range of LAI and AGB. Leaf area was measured with a LI-3000C Portable Leaf Area Meter (LI-COR Biosciences, Lincoln, U.S.A.). Dry biomass was obtained after the fresh samples without roots and dried at 60 °C until their weights stabilized. The LAI per square meter and AGB per square meter were calculated using the plant numbers per square meter multiplied by the average LAI and average biomass of plant samples, respectively. Plant counting of all 180 plots were completed by visual recognition with the image of 35 and 50 days after emergence corresponding to V2–V4 and V4–V6 leaf stage. The rest of the other 20 inbred lines (Rest 20) were not sampled for LAI and AGB measurement.
PHANTOM 4 (DJI, Shenzhen, China) was used for image acquisition. The imaging sensor was a 1-inch CMOS with 20 million pixels. DJI GS Pro (DJI, Shenzhen, China) was used to determine the target aerial photography area on a satellite map and to plan the aerial photography route by inputting relative flight and camera parameters. Autonomous flight plans were used to have an 80% overlap between adjacent images with a 2 s interval. RGB images were collected at two to six leaf stages with flight heights of 10 m, 15 m and 30 m at Jilin and Henan and 30 m at Hebei and Hainan. The ground sample distances were 2.73 mm for 10 m flight height, 4.10 mm for 15 m flight height and 8.21 mm for 30 m flight height. UAV and ground sampling were synchronized and completed under low wind speed and clear sky conditions on 36, 48 and 60 days after emergence (Table 1).
Point cloud and digital orthophoto map generation
UAV images were processed by the Agisoft Metashape (Agisoft, Russia) to generate point clouds and Digital Orthophoto Map (DOM) of the flight area. Scale Invariant Feature Transform algorithm (Lowe, 2004) was used to extract and describe the information of feature points. Structure From Motion (SFM) algorithm was used to perform a bundle adjustment based on matching features between the images. Ground control points were evenly distributed in the field, recorded by RTK (CHCNAV-T8, Shanghai, China) and then used for geometry correction. Finally, the DOM is exported in.tiff image format and point clouds data in.txt file format. Images randomly clipped from DOM with a size of 1500 pixel × 1500 pixel were used to verify model accuracy for maize plant counting. Images and point clouds of each individual plots were segmented for LAI and biomass estimation according to their positions in the experimental design layout and assigned with a unique ID based on their geographical location.
Row detection and plant identification
Excess green index (EXG; Meyer and Neto, 2008), Lab colour space (Liu, Baret, et al., 2017), excess green minus excess red index (EXGR; Upendar et al., 2021), green leaf index (GLI; Blancon et al., 2019) and support vector machine (SVM; Jin et al., 2017) were proposed in previous research for well identification of the green pixels. EXG (Eq. 1), EXGR (Eq. 2), GLI (Eq. 3) was calculated for each pixel using the three colours [R, G, B] of the RGB image. Lab colour space were transformed from RGB, to enhance its sensitivity to variations in greenness. Otsu automatic thresholding method (Otsu, 1979) was used to separate the green plants from the background pixels. Binary images were thus transformed using threshold value and assigning 1 to the green pixels and 0 to the background. For SVM, the image classification model was trained on image fragments (vegetation or background) from five UAV images and applied to all other UAV images to obtain binary images. Binary image was then processed using morphological opening operation to remove the weeds in smaller area, to break narrow strips and to smooth the contour objects (Abid Hasan & Ko, 2016).
where R, G and B are three colour components of the RGB image.
The skeleton of the plant obtained using parallel thinning algorithm (Zhang & Suen, 1984) had numerous isolated points. The neighbourhood threshold method was used to determine the outliers of the skeleton of the plants with a threshold (N; Eq. 4). The isolated points were then removed. The value of threshold was set to 1 to determine the abnormal skeletons, as there were no pixels of the plant around the isolated points. The final skeleton diagram is obtained after several iterations of the above process.
where A is a binary image. The image size is c*r. sn is sliding neighbourhood.
After removing the abnormal skeletons, the plant row was detected with Hough transform (Hough, 1962) and the angle of plant row was calculated with swing adjustment method. Then, row positions can be calculated with regular and uniformly distributed plant pixel peaks. The number of plant pixels covered by these lines with a slope of K1 and intercepts from zero to (r*tanK1 + c) was calculated (Fig. 3a, bottom). The x coordinates of the pixel number distribution curve in the row orientation were separately extracted from y = 0 to the maximum of peaks (blue points in Fig. 3a). Major peak spacing was calculated by the distance between P points (red points in Fig. 3a) obtained by the medium of adjacent blue points. The crop row positions can be identified by the minimum standard deviation of the major peak spacing.
The process of row detection and plant identification. a An example of determining the position of evenly spaced plant pixel peaks slicing along the Y axis. b Pixel number distribution in the row direction. c The process of angle adjustment by swing adjustment method. d The difference in angle of each row. e The position of the row layout, and f flow of separating the individual plants from each other by the location-based plant cluster segmentation and mosaic (LSM) method
Finally, the optimum slope (K2) of different rows was automatically calculated by swing adjustment method. The plant pixels were counted by these lines with the angle varied from (\({\text{tan}}^{-1}{K}_{1}-\Delta\)) to (\({\text{tan}}^{-1}{K}_{1}+\Delta\)) and the sowing angle (\(\Delta\)) was set to 2°. The slope (K2) of the maximum peak was the optimum angle of each row (Fig. 3b; Eq. 5). The width of the row layout (L’) was calculated according to the size of plants (Fig. 3c; Eq. 6). Only the regions in these row layouts were analysed, and any weeds between the rows were effectively excluded.
where Δ is 2°. \({(x}_{m},{y}_{m}\)) are the coordinates of a known point. A is a binary image. The image size is c*r. K1 is the slope of the skeleton detected by the Hough transform.
where L is the position of each row and w is mean of the plants size.
The overlapping of plant seedlings is inevitable in the obtained images. A location-based plant cluster segmentation and mosaic (LSM) method were proposed to separate the individual plants from each other (Fig. 3f). Plant clusters (P) were firstly separated into several regions based on plant spacing. When there were two connected domains in one region, marked as large area part P(t) and small area part P(t′). P(t′) was used as a custom sliding neighbourhood and connected to P(t+1). The minimum difference between the last line of P(t’) and the first line of P(t+1) was calculated to identify the optimum mosaic position (P′; Eq. 7). The inaccurately segmented leaves and plants were then mosaiced together to obtain a more complete plant morphology.
where P is the binary image of a plant cluster. Total number of parts in the image was represented with n, and t denotes one part of n (\(t\in \left[1,n\right]\)). The image size of P(t+1) is c(t+1)*r(t+1). K1 is the slope of the skeleton detected by the Hough transform.
Plant characterization and classification
Morphological features were divided into four categories: area (A), roundness (R), skeleton (S) and position (P). The details of the morphological features were described in Table 2. Area and roundness characteristics were obtained using the ‘regionprops’ function of MATLAB (version R2019b, MathWorks, USA). Range of ‘extent’, ‘solidity’ and ‘eccentricity’ were 0–1. Higher ‘extent’ and ‘solidity’ meant more spatial area of objects within bounding box and convex hull. ‘Eccentricity’ was calculated by ratio of focal length to length of major axis. A circle had ‘eccentricity’ equal to 0, and a longer ellipse had ‘eccentricity’ close to 1.
The contour point was the endpoint for the skeleton if it satisfied the following conditions: the centre point of the computed matrix was 1, and N = 2 (Eq. 2; Fig. 4b). Only condition N ≥ 4 was changed in the calculation of the cross-point for the skeleton, and the rest remained the same (Fig. 4c). Lengthskelet, Numend and Numbranch were both calculated using skeleton. Offset and distanceup represent position information of the target plant.
Plant discrimination models
Four types of morphological characteristics were combined and trained by SVM to relate to the number of plants and weeds inside each image using morphologic discrimination model (MDM). The total dataset consists of 1120 images, which were divided into three parts: training (60%), validation (20%) and test (20%) set. The training and the validation set were used to train the internal parameters of MDM, and adjust the hyperparameters for MDM with the best classification performance respectively. Test set without participating in any modelling or preparation was treated as new, unknown data. Commonly used indicators, such as precision, recall and F1-score were used to evaluate the degree to which the measured values conformed to the estimated values, and to evaluate the generalization ability of MDM. The detail information of precision, recall and F1-score will be outlined in the following section.
Classification model produces four types of outcomes: correctly recognized maize (true positives), correctly recognized weeds (true negatives), incorrectly recognized maize (false positives) and incorrectly recognized weeds (false negatives). Retrieved features are the total number of true positives and false positives. Relevant features are the total number of true maize and false recognized weed. |{relevant features} ∩ {retrieved features}| means instances of true positives. As proposed in Osco et al (2020), precision (Eq. 8) is defined as the fraction of correctly recognized maize among all retrieved features. Recall (Eq. 9) is the fraction of true positives among all relevant features and is the measure of a classifier effectively to identify true positives. F1-score (Eq. 10) is a way of combining the precision and recall of the model, and is defined as the harmonic mean of the model’s precision and recall. Precision, recall and F1-score of a perfect classifier equal to one.
Therefore, interpolation discriminant model (IDM) was further proposed to estimate the plant counting at V4–V6 leaf stage due to serious overlapping of plant seedlings. The steps were: (1) MDM was first used to obtain the plant positions in images of V2–V4 leaf stage. Plant positions in the images of V4–V6 leaf stage were first registered with that of V2–V4 leaf stage by perspective transformation. (2) Positions without seedling was calculated with IDM by equidistance interpolation. Region of interest (ROI) was established at the location without seedling. (3) The plant pixel ratio in ROI (Eq. 11) were calculated and compared with the threshold of plant discrimination. The presence of plants in ROI was judged according to whether the plant pixel ratio exceeds the threshold. The thresholds were range from 0 to 0.3, with an interval of 0.01.
The number of plants was estimated using different thresholds and compared with the measured values. The minimum RMSE proportion was selected as the best threshold of plant discrimination.
where PPR is plant pixel ratio. \({\mathrm{A}}_{plant}\) is the area of plant within ROI and \({A}_{ROI}\) is the area of ROI.
Pseudo code of interpolation discriminant model were shown below:

LAI and AGB estimations
A best plant classification model with the best classification performance was firstly selected from these models built by EXG (Eq. 1), Lab colour space, EXGR (Eq. 2), GLI (Eq. 3) and SVM classification model (Fig. 5a). In order to construct these classification models based different vegetation indices, EXG, Lab colour space, EXGR, GLI were calculated using the colour information of point clouds. Point clouds of maize inbred lines were then well identified by filtering out soil background based on Otsu automatic thresholding method (Fig. 5a). Another plant classification model was SVM classification model trained by image fragments (vegetation or background) from five UAV images. The components of background and noises of original point clouds were picked out using SVM classification model (Fig. 5a). And then point clouds structure information of maize inbred lines were down sampling based on voxel grid to improve computing efficiency as follows: (1) Point clouds of the canopy were summarized into voxels with equal dimensions of 0.2–20 cm, at intervals of 0.1 cm (Fig. 5b). (2) The point cloud numbers were calculated within each voxel. Voxels with point cloud numbers < 3 were regarded as abnormal voxels and removed accordingly (Fig. 5b). (3) Optimal voxel size was determined according to the comparisons between measured and calculated LAI and AGB estimation models. Finally, LAI and AGB estimation models were built using point clouds of maize inbred lines after voxelization (Fig. 5). LAI and AGB were estimated using the plant counting with MDM and IDM by Eqs. 12 and 15. Image processing was conducted using MATLAB (code available on request from the corresponding author).
where \({\mathrm{n}}_{i}\) is the number of 3D grids with point clouds in the ith layer and \({\mathrm{nt}}_{i}\) is the total number of 3D grids in the ith layer. \({N}_{MDM}\) and \({N}_{IDM}\) were the plant numbers obtained with MDM and IDM. Optimal voxel size was calculated with 0.09 m and 0.19 m for best estimation of LAI and AGB respectively. The unit of AGB is kg/m2.
Statistical analyses
The coefficients of determination (R2; Eq. 16), root mean square error (RMSE; Eq. 17) and relative root mean square error (rRMSE; Eq. 18) were used to assess the coincidence degree between the measured and calculated values. Significant analysis of germination rate (Eq. 19) difference between two different growth stages was evaluated using ANOVA with Student's t-Test (α = 0.05). The dispersion degree of data was calculated by InterQuartile Range (IQR). Range of germination rate was calculated subtracting the maximum number from the minimum number. 75% quartile was subtracted from 25% quartile for IQR calculation to ignore outliers. Quantitative statistics and graphical statistics were processed using tidyverse, stats and ggplot2 packages in R (R Development Core Team).
where \({N}_{plant}\) is the actual plant numbers, and \({N}_{seed}\) is the plant number sowed within each plot.
Results
Separations of the green plants from the background and the classification accuracies were compared and shown in Fig. 6. Green leaf index (GLI) and support vector machine provided a better identification of the green pixels as compared to other methods. The classifier training by support vector machine needs more time to build train dataset and optimize model hyperparameters than GLI. Therefore, GLI was selected to separate the green pixels from the background here.
Examples of phenotypic parameters of maize seedlings are shown in Fig. 7, including convex hulls (Fig. 7a), bounding boxes (Fig. 7c), the distance between adjacent plant centres (Fig. 7e; L1), deviation of the target plant relative to the position of the row (Fig. 7e; L2), projection areas (Fig. 7b and Fig. 7d), skeletons and their intersection points (Fig. 7f). L1 is the distance between adjacent plants (Fig. 7c).
Schematic diagram for calculating the phenotypic parameters of maize seedlings. a Convex hull. b The area of the plants. c A minimum bounding box. d The ‘filledarea’ of the plants. e Seedling position information. L1 is the distance between adjacent plant centres. L2 is the deviation of the target plant relative to the mid-row position. f Outline skeleton and cross-point of the skeleton
The prediction accuracy were compared between the measured and estimated plant counting at the V2–V4 leaf stage for different flight heights at two locations (Fig. 8). Morphologic discrimination model (MDM) exhibited higher accuracy for plant counting at the flight heights of 15 m with the R2 of 0.99 and averaged rRMSE of 1.95%. Performance of MDM built at the flight heights of 30 m was the averaged R2 value of 0.98 and averaged rRMSE of 2.55%. Plant counting at the flight heights of 10 m yielded slighter lower performance with R2 = 0.93 and rRMSE = 5.55% (Fig. 8).
The estimated and measured plant counting were compared using maize images of V2–V4 leaf stage at the flight heights of 10 m (a and b), 15 m (c and d) and 30 m (e and f) at two locations (Jilin and Henan). One point stand for maize plant counting from randomly clipped DOM within a size of 1500 pixel × 1500 pixel
MDM based on area information such as ‘originalarea’ and ‘convexarea’ presented the better performance in plant counting prediction with averaged precision = 0.95, recall = 0.95 and F1 = 0.95 than other models constructed separately using area, roundness and skeleton information. Classification accuracy of MDM built by combination of area (averaged precision = 0.93), roundness (averaged precision = 0.93) and skeleton (averaged precision = 0.92) information with position information was improved as compared to MDM built by position information with relatively low averaged precision of 0.90. Estimation accuracy of MDM based on the combination of individual plant area, roundness and position (A + R + P) was better than the other models, with the plant density estimation of precision > 0.94, recall > 0.98 and F1 > 0.96 (Table 3). Maize (Fig. 9, grey) and weeds (Fig. 9, white box) were separated accurately using MDM built by groups A, R and P.
The differences in the morphological characteristics of plants and weeds are presented in Fig. 10. The distribution of ‘originalarea’ and ‘convexarea’ of plants and weeds was positively right-skewed. The ‘extent’ (marked with a red quadrangle) and ‘solidity’ (marked with a yellow polygon) of weeds were larger than those of the plants. The ‘extent’ mode of the plants was 0.35 (Fig. 10c) and that of the weeds was 0.55 (Fig. 10g). The ‘solidity’ of the plant is similar to a normal distribution, but that of weeds is positively right-skewed.
Empirical histogram of different morphological characteristics of plants (marked with green circles) and weeds (marked with yellow triangles). These morphological characteristics belong to area, roundness and position. ‘Extent’, ‘solidity’ and ‘eccentricity’ of plant and weed were marked with a red quadrangle, a yellow polygon and a green ellipse
The area of weeds is smaller than that of maize (Fig. 10e). Narrow strips and thin leaf tips of weeds were eliminated after morphological opening operation (the pictures in Fig. 10p). The ellipse built by small weeds tend to be round as shown in the last group picture in Fig. 10p. The ellipse eccentricity of the plants and weeds had a negatively left-skewed distribution, and the other morphological characteristics of roundness were positively right-skewed distributions. The eccentricity of the weeds was closer to 1 than that of the plants indicating the longer axis of ellipse by weeds (Fig. 10l vs p).
The distance distribution between adjacent plants was a double wave crest. The second peak along X-axis (green arrow in Fig. 10s) was 50 cm and that of the first peak was 25 cm. The deviation distribution of the plants and weeds were positively right-skewed relative to mid-row. Maize was concentrated within 10 cm of plant row (Fig. 10q). The distribution of weeds was relatively scattered (Fig. 10r).
The distance distribution between adjacent plants (Fig. 10, Position) and plant area distribution (Fig. 10, Area) indicated the seedling deficiency and seedling weakness at V2–V4 leaf stages. The maize position of some inbred lines such as Sy1032, 647 and CML473 was only detected at V4–V6 leaf stages due to inconsistent germination time (Fig. 11). Germination rate of 30 inbred lines including Sample 10 and Rest 20 was then compared at V2–V4 leaf and V4–V6 leaf stages (Fig. 12). Germination rate difference (hollow bar in Fig. 12) between two leaf stages were highest for Shen5003 with the difference > 30%. Germination rate of GEMS9 and huangC at V2–V4 leaf stage was higher than that at V4–V6 leaf stage (Fig. 12a), which means seedlings of these materials died at V4–V6 leaf stage. No significant differences between V2–V4 leaf and V4–V6 leaf stages were found in germination rate for Sample 10, but germination rate for Rest 20 was significantly different (Fig. 12d). Sample 10 group (Mean = 5%, Range = 21%) had lower germination rate difference, dispersion and variability than Rest 20 group (Mean = 12%, Range = 38%; Table 4).
The construction process of the interpolation discriminant model (IDM). Plant position of V4–V6 leaf stage was detected based on plant position of V2–V4 leaf stage obtained by MDM and then calculated using IDM for isometric interpolation. Red points and blue points were the plant positions obtained using MDM and IDM
Comparisons of germination rate for different maize inbred lines at V2–V4 and V4–V6 leaf stages. a–c The plant germination rate differences of each material, d the plant germination rate differences of all materials with two groups. Sample 10 means ten materials were sampled to measure LAI and AGB. And the rest of the other 20 inbred lines were not sampled and named as Rest 20. Plant counting of Sample 10 and Rest 20 were visually identified at V2–V4 and V4–V6 leaf stages. Lines in boxplots means standard deviation of germination rate for different maize inbred lines. ANOVA was carried out, and ‘**’ on the bars indicates significant differences at the .01 level, ‘#’ means significant differences at the .05 level
IDM was constructed to address the unpredictable errors caused by the above germination differences in Fig. 11. The estimated plant number has a better correlation with the measured value than the other threshold when the plant discrimination threshold is 0.07, with R2 = 0.86, rRMSE = 5.5% (Fig. 13). The agreement between the measured and estimated plant counting using different models was presented in Fig. 14. The plant counting estimated using the images of V2–V4 leaf stage has lower correlation with R2 = 0.78 (Fig. 14a). There were still some materials that germinate or die in the late growth stage leading to inaccurate prediction of seedling number. The plant counting at V4–V6 leaf stage estimated by IDM was better than that by MDM, with R2 = 0.79 for MDM (Fig. 14b) and R2 = 0.86 for IDM (Fig. 14c).
Morphological discriminant model (MDM, a and b) and interpolation discriminant model (IDM, c) were used to compare the estimated and measured plant counting. The measured value was the plant counting at V4–V6 leaf stage. a Based on the maize images of V2–V4 leaf stage, b and c based on the maize images of V4–V6 leaf stage. One point represents the number of maize plants in each plot of 5 × 1.2 m
The plant counting estimated by IDM at V4–V6 leaf stage was better for predicting LAI and AGB than that estimated by MDM at V2–V4 leaf stage (Fig. 15). The agreements between calculated and measured LAI and AGB are presented in Fig. 15. R2 of LAI estimated by MDM was lower than that by IDM (0.8 vs. 0.85). R2 of AGB estimated by MDM was 0.86 lower than that by IDM (R2 = 0.9).
Comparison of estimated and measured leaf area index (LAI) and aboveground biomass (AGB) using Morphological Discriminant Model (MDM, a and b) at V2–V4 leaf stage and Interpolation Discriminant Model (IDM, c and d) at V4–V6 leaf stage. One point stand for leaf area index and aboveground biomass of each maize inbred line
Discussion
Application of UAV technologies in plant counting and plant identification
Plant counting was an important reference index for evaluating the quality of maize seed and machine planting, and also helpful for making up the yield losses by reseeding and transplanting in time (Assefa et al., 2018; Li et al., 2018; Ranđelović et al., 2020). Ground fixed and moving platform was found effective at high-resolution and high-quality data collection. Herrmann et al., (2019) evaluated soybean plant population with R2 of 0.51–0.69 at early developmental stages using a ground moving platform, which was destructive to the experimental sites and limited to the sampled areas. Increased costs were also needed for large-scale experiments. UAV have the advantages with the merits of fast image acquisition, automation and easy operation (Feng et al., 2020; Jin et al., 2017; Koh et al., 2019). Varela et al. (2018) used UAV to automatically scout fields with an overall crop classification accuracy of 0.96. Feng et al. (2020) captured high-resolution image frames by UAV to evaluate plant seedling number using a pretrained deep learning model with high accuracy (R2 = 0.95). Plant counting calculated with UAV images provides important application prospects in grain yield maximization with modern agricultural production.
A complex field environment presents many challenges. The robustness in crop rows estimation is a crucial precondition, especially for automatically performing weeding/fertilizing operations in future farming systems. The methods detected crop rows, including blob analysis (Ahmed et al., 2019), linear regression (Montalvo et al., 2012), and Hough transform (Ji & Qi, 2011; Liu, Zhou, et al., 2017). A pipeline for the automatic calculation of the optimum angle for each row was presented to avoid non-parallelism of rows caused by initial plantation (Fig. 3). Location-based plant cluster segmentation and mosaic method can effectively segment complete individual plants at different test sites (Figs. 3f & 8). However, the current application of the method is limited to plant counting under the condition of uniform planting in the field (Fig. 9).
Challenge and improvements of current plant discrimination models based on morphological characteristics
Liu, Baret, et al. (2017) proposed that there was redundancy between morphological features, which may confuse the training of the discriminative model. The morphological characteristics of maize seedlings were compared here with weeds and the contribution of various morphological characteristics were then assessed to improve the accuracy of the discriminative model at V2–V4 leaf stage (Table 2). The best model was obtained using the combinations of individual plant area, roundness and position with the plant density estimation of mean precision, recall and F1 above 0.95 (Table 3). Performance of the morphologic discrimination model was in accordance with the results of Varela et al., (2018), which provides a precision of 0.96–0.97 and recall of 0.95–0.97 between crop and weed objects detection. Therefore, the groups A, R and P were taken as the optimal training samples for morphologic discrimination model to separate maize and weed, which will improve the estimation accuracy and training efficiency. In some cases, the weeds distributed within maize rows were misidentified as maize because they had same shape and colour. This denotes the importance to the further usage of other deep morphological characteristics of maize for better classification results.
Currently, most studies suggest that the V2–V4 leaf stage was the best growth stage to estimate plant emergence in high estimation accuracy with UAV images (Feng et al., 2020; Jin et al., 2017; Li, Li, et al., 2019; Li, Xu, et al., 2019; Osco et al., 2021). However, germination time and growth status of maize inbred lines are different (Figs. 11, 12 and Table 4). It was necessary to use images of late growth stage for more accurate plant counting due to different germination rate and senescence of maize inbred lines (Fig. 14). The overlapping of plant seedlings at V4–V6 leaf stage is inevitable in the obtained images. IDM was therefore proposed for plant counting at V4–V6 leaf stage. The accuracy obtained with IDM (R2 = 0.86) was better than MDM (R2 = 0.79).
The impact of plant counting on the estimation accuracy of LAI and AGB
The effect of accurate plant counting were demonstrated on improving the model performance in LAI and AGB estimation. The results showed that LAI (R2 = 0.8 vs. R2 = 0.85) and AGB (R2 = 0.86 vs. R2 = 0.9) can be more accurately estimated only when precise plant counting is obtained (Fig. 15). There were significant differences between V2–V4 leaf and V4–V6 leaf stages in germination rate for Rest 20, but no significant differences were found for Sample10. Sample10 (Mean = 5%, Range = 21%) had lower germination rate difference, dispersion and variability than Rest 20 (Mean = 12%, Range = 38%). The inaccuracy for LAI and AGB prediction using the plant counting at V2–V4 leaf stage would be greater if all materials were sampled due to the greater variation of Rest 20. It should be emphasized that ten materials in Sample10 group were used to measure LAI and AGB. The rest of the other 20 inbred lines in Rest 20 were not sampled for LAI and AGB measurement due to the heavy workload. LAI and AGB of Rest 20 will be measured in a later study in order to further verify the results. This study focused on providing the application potential of precision agriculture on maize inbred lines with extensive genetic diversity, but lowered the contribution to LAI and biomass prediction as it is done in the late growth stage. In the future, the exact time of accurate plant counting from UAV images of maize inbred lines at different leaf stage will be detected to improve its contribution to effective fertilization management and crop yield prediction.
Conclusion
In this study, plant counting at V4–V6 leaf stage was better than that at V2–V4 leaf stage for maize inbred lines with extensive genetic diversity, as plant counting at V4–V6 leaf stage can efficiently prevent bias counting caused by death and growth of small seedlings. The designed IDM considered the time differences of germination in maize inbred lines, thus plant counting by IDM was closer to the field measurements compared with that of MDM. Performance of LAI and AGB estimation model based on the accurate maize plant counting of maize inbred lines in V4–V6 leaf stage was superior to that based on V2–V4 leaf stage. Therefore, accurate plant counting in later leaf stage can improve the estimation accuracy of LAI and AGB for maize inbred lines with extensive genetic diversity. In the future, the exact leaf stage for accurate plant counting will be further refined to improve the application potential of precision agriculture on maize management and breeding.
References
Abid Hasan, S. M., & Ko, K. (2016). Depth edge detection by image-based smoothing and morphological operations. Journal of Computational Design and Engineering, 3, 191–197. https://doi.org/10.1016/j.jcde.2016.02.002
Aharon, S., Peleg, Z., Argaman, E., Ben-David, R., & Lati, R. N. (2020). Image-based high-throughput phenotyping of cereals early vigour and weed-competitiveness traits. Remote Sensing, 12, 3877. https://doi.org/10.3390/rs12233877
Ahmed, I., Eramian, M., Ovsyannikov, I., Kamp, W. van der, Nielsen, K., Duddu, H.S., et al. (2019). Automatic detection and segmentation of lentil crop breeding plots from multi-spectral images captured by UAV-mounted camera. In 2019 IEEE Winter Conference on Applications of Computer Vision, Waikoloa Village, HI, USA (pp. 1673–1681). https://doi.org/10.1109/WACV.2019.00183
Assefa, Y., Carter, P., Hinds, M., Bhalla, G., Schon, R., Jeschke, M., et al. (2018). Analysis of long-term study indicates both agronomic optimal plant density and increase maize yield per plant contributed to yield gain. Scientific Reports, 8, 4937–4949. https://doi.org/10.1038/s41598-018-23362-x
Bendig, J., Yu, K., Aasen, H., Bolten, A., Bennertz, S., Broscheit, J., et al. (2015). Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. International Journal of Applied Earth Observation and Geoinformation, 39, 79–87. https://doi.org/10.1016/j.jag.2015.02.012
Blancon, J., Dutartre, D., Tixier, M.-H., Weiss, M., Comar, A., Praud, S., et al. (2019). A high-throughput model-assisted method for phenotyping maize green leaf area index dynamics using unmanned aerial vehicle imagery. Frontiers in Plant Science, 10, 685–700. https://doi.org/10.3389/fpls.2019.00685
Che, Y., Wang, Q., Xie, Z., Zhou, L., Li, S., Hui, F., et al. (2020). Estimation of maize plant height and leaf area index dynamics using an unmanned aerial vehicle with oblique and nadir photography. Annals of Botany, 126, 765–773. https://doi.org/10.1093/aob/mcaa097
Dong, T., Liu, J., Shang, J., Qian, B., Ma, B., Kovacs, J. M., et al. (2019). Assessment of red-edge vegetation indices for crop leaf area index estimation. Remote Sensing of Environment, 222, 133–143. https://doi.org/10.1016/j.rse.2018.12.032
Fan, Z., Lu, J., Gong, M., Xie, H., & Goodman, E. D. (2018). Automatic tobacco plant detection in UAV images via deep neural networks. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 11, 876–887. https://doi.org/10.1109/JSTARS.2018.2793849
Feng, A., Zhou, J., Vories, E., & Sudduth, K. A. (2020). Evaluation of cotton emergence using UAV-based imagery and deep learning. Computers and Electronics in Agriculture, 177, 105711–105726. https://doi.org/10.1016/j.compag.2020.105711
Findura, P., Krištof, K., Jobbágy, J., Bajus, P., & Malaga-Toboła, U. (2018). Physical properties of maize seed and its effect on sowing quality and variable distance of individual plants. Acta Universitatis Agriculturae Et Silviculturae Mendelianae Brunensis, 66, 35–42. https://doi.org/10.11118/actaun201866010035
Finizola, J. S., Targino, J. M., Teodoro, F. G. S., & Lima, C. A. M. (2019). Comparative study between deep face, autoencoder and traditional machine learning techniques aiming at biometric facial recognition. In 2019 International Joint Conference on Neural Networks, Budapest, Hungary (pp. 1–8). https://doi.org/10.1109/IJCNN.2019.8852273
Font, D., Pallejà, T., Tresanchez, M., Teixidó, M., Martinez, D., Moreno, J., et al. (2014). Counting red grapes in vineyards by detecting specular spherical reflection peaks in RGB images obtained at night with artificial illumination. Computers and Electronics in Agriculture, 108, 105–111. https://doi.org/10.1016/j.compag.2014.07.006
Gnädinger, F., & Schmidhalter, U. (2017). Digital counts of maize plants by unmanned aerial vehicles (UAVs). Remote Sensing, 9, 544–558. https://doi.org/10.3390/rs9060544
Gong, Y., Yang, K., Lin, Z., Fang, S., Wu, X., Zhu, R., et al. (2021). Remote estimation of leaf area index (LAI) with unmanned aerial vehicle (UAV) imaging for different rice cultivars throughout the entire growing season. Plant Methods, 17, 88–103. https://doi.org/10.1186/s13007-021-00789-4
Guo, Y., Du, C., Zhao, Y., Ting, T. F., & Rothfus, T. A. (2021). Two-level K-nearest neighbours approach for invasive plants detection and classification. Applied Soft Computing, 108, 107523. https://doi.org/10.1016/j.asoc.2021.107523
Herrmann, I., Bdolach, E., Montekyo, Y., Rachmilevitch, S., Townsend, P. A., & Karnieli, A. (2020). Assessment of maize yield and phenology by drone-mounted super spectral camera. Precision Agriculture, 21, 51–76. https://doi.org/10.1007/s11119-019-09659-5
Herrmann, I., Vosberg, S. K., Townsend, P. A., & Conley, S. P. (2019). Spectral data collection by dual field-of-view system under changing atmospheric conditions-a case study of estimating early season soybean populations. Sensors, 19, 457. https://doi.org/10.3390/s19030457
Houborg, R., & McCabe, M. F. (2018). A hybrid training approach for leaf area index estimation via Cubist and random forests machine-learning. ISPRS Journal of Photogrammetry and Remote Sensing, 135, 173–188. https://doi.org/10.1016/j.isprsjprs.2017.10.004
Hough, P. V. C. (1962). Method and means for recognizing complex patterns. US, US3069654A [P]. https://patents.google.com/patent/US3069654/en
Hu, G., Li, Z., Lu, Y., Li, C., Gong, S., Yan, S., et al. (2017). Genome-wide association study identified multiple genetic loci on chilling resistance during germination in maize. Scientific Reports, 7, 10840–10850. https://doi.org/10.1038/s41598-017-11318-6
Hu, P., Chapman, S. C., Wang, X., Potgieter, A., Duan, T., Jordan, D., et al. (2018). Estimation of plant height using a high throughput phenotyping platform based on unmanned aerial vehicle and self-calibration: Example for sorghum breeding. European Journal of Agronomy, 95, 24–32. https://doi.org/10.1016/j.eja.2018.02.004
Ji, R., & Qi, L. (2011). Crop-row detection algorithm based on random hough transformation. Mathematical and Computer Modelling, 54, 1016–1020. https://doi.org/10.1016/j.mcm.2010.11.030
Jimenez-Berni, J. A., Deery, D. M., Rozas-Larraondo, P., Condon, A. G., Rebetzke, G. J., James, R. A., et al. (2018). High throughput determination of plant height, ground cover, and above-ground biomass in wheat with LiDAR. Frontiers in Plant Science, 9, 237–254. https://doi.org/10.3389/fpls.2018.00237
Jin, X., Liu, S., Baret, F., Hemerlé, M., & Comar, A. (2017). Estimates of plant density of wheat crops at emergence from very low altitude UAV imagery. Remote Sensing of Environment, 198, 105–114. https://doi.org/10.1016/j.rse.2017.06.007
Kamath, C. N., Bukhari, S. S., & Dengel, A. (2018). Comparative study between traditional machine learning and deep learning approaches for text classification. In Proceedings of the ACM Symposium on Document Engineering 2018, New York, USA (pp. 1–11). https://doi.org/10.1145/3209280.3209526
Koh, J. C. O., Hayden, M., Daetwyler, H., & Kant, S. (2019). Estimation of crop plant density at early mixed growth stages using UAV imagery. Plant Methods, 15, 64–72. https://doi.org/10.1186/s13007-019-0449-1
Lei, L., Qiu, C., Li, Z., Han, D., Han, L., Zhu, Y., et al. (2019). Effect of leaf occlusion on leaf area index inversion of maize using UAV–LiDAR data. Remote Sensing, 11, 1067–1081. https://doi.org/10.3390/rs11091067
Li, B., Xu, X., Han, J., Zhang, L., Bian, C., Jin, L., et al. (2019). The estimation of crop emergence in potatoes by UAV RGB imagery. Plant Methods, 15, 15–27. https://doi.org/10.1186/s13007-019-0399-7
Li, B., Xu, X., Zhang, L., Han, J., Bian, C., Li, G., et al. (2020). Above-ground biomass estimation and yield prediction in potato by using UAV-based RGB and hyperspectral imaging. ISPRS Journal of Photogrammetry and Remote Sensing, 162, 161–172. https://doi.org/10.1016/j.isprsjprs.2020.02.013
Li, J., Xie, R. Z., Wang, K. R., Hou, P., Ming, B., Zhang, G. Q., et al. (2018). Response of canopy structure, light interception and grain yield to plant density in maize. The Journal of Agricultural Science, 156, 785–794. https://doi.org/10.1017/S0021859618000692
Li, Y., Li, C., Li, M., & Liu, Z. (2019). Influence of variable selection and forest type on forest aboveground biomass estimation using machine learning algorithms. Forests, 10, 1073–1096. https://doi.org/10.3390/f10121073
Liu, Q., Zhou, X., Li, J., & Xin, C. (2017). Effects of seedling age and cultivation density on agronomic characteristics and grain yield of mechanically transplanted rice. Scientific Reports, 7, 14072–14082. https://doi.org/10.1038/s41598-017-14672-7
Liu, S., Baret, F., Andrieu, B., Burger, P., & Hemmerlé, M. (2017). Estimation of wheat plant density at early stages using high resolution imagery. Frontiers in Plant Science, 8, 739–748. https://doi.org/10.3389/fpls.2017.00739
Liu, Y., Cen, C., Che, Y., Ke, R., Ma, Y., & Ma, Y. (2020). Detection of maize tassels from UAV RGB imagery with Faster R-CNN. Remote Sensing, 12, 338–354. https://doi.org/10.3390/rs12020338
Lowe, D. G. (2004). Distinctive image features from scale-invariant key points. International Journal of Computer Vision, 60, 91–110. https://doi.org/10.1023/B:VISI.0000029664.99615.94
Maimaitijiang, M., Sagan, V., Sidike, P., Hartling, S., Esposito, F., & Fritschi, F. B. (2020). Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sensing of Environment, 237, 111599. https://doi.org/10.1016/j.rse.2019.111599
Meyer George, E., & Camargo, N. J. (2008). Verification of colour vegetation indices for automated crop image application. Computers and Electronics in Agriculture, 63, 282–293. https://doi.org/10.1016/j.compag.2008.03.009
Montalvo, M., Pajares, G., Guerrero, J. M., Romeo, J., Guijarro, M., Ribeiro, A., et al. (2012). Automatic detection of crop rows in maize fields with high weeds pressure. Expert Systems with Applications, 39, 11889–11897. https://doi.org/10.1016/j.eswa.2012.02.117
Ormond, A., Furlani, C., Oliveira, M., Noronha, R., & de Tavares, T. (2018). Maize sowing speeds and seed-metering mechanisms. Journal of Agricultural Science, 10, 468–476. https://doi.org/10.5539/jas.v10n9p468
Osco, L. P., de Arruda, M. S., Gonçalves, D. N., Dias, A., Batistoti, J., de Souza, M., et al. (2021). A CNN approach to simultaneously count plants and detect plantation-rows from UAV imagery. ISPRS Journal of Photogrammetry and Remote Sensing, 174, 1–17. https://doi.org/10.1016/j.isprsjprs.2021.01.024
Osco, L. P., De Arruda, M. D. S., Junior, J. M., Da Silva, N. B., Ramos, A. P. M., Moryia, É. A. S., Imai, N. N., Pereira, D. R., Creste, J. E., Matsubara, E. T., & Li, J. (2020). A convolutional neural network approach for counting and geolocating citrus-trees in UAV multispectral imagery. ISPRS Journal of Photogrammetry and Remote Sensing, 160, 97–106. https://doi.org/10.1016/j.isprsjprs.2019.12.010
Otsu, N. (1979). A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics, 9, 62–66. https://doi.org/10.1109/TSMC.1979.4310076
Ranđelović, P., Đorđević, V., Milić, S., Balešević-Tubić, S., Petrović, K., Miladinović, J., et al. (2020). Prediction of soybean plant density using a machine learning model and vegetation indices extracted from RGB images taken with a UAV. Agronomy, 10, 1108–1117. https://doi.org/10.3390/agronomy10081108
Shinde, P. P., & Shah, S. (2018). A review of machine learning and deep learning applications. In 2018 Fourth International Conference on Computing Communication Control and Automation, Pune, India (pp. 1–6). https://doi.org/10.1109/ICCUBEA.2018.8697857
Thorp, K., Steward, B. L., Kaleita, A., & Batchelor, W. (2008). Using aerial hyperspectral remote sensing imagery to estimate corn plant stand density. Transactions of the Asabe, 51, 311–320. https://doi.org/10.13031/2013.20855
Togeiro de Alckmin, G., Kooistra, L., Rawnsley, R., & Lucieer, A. (2021). Comparing methods to estimate perennial ryegrass biomass: Canopy height and spectral vegetation indices. Precision Agriculture, 22, 205–225. https://doi.org/10.1007/s11119-020-09737-z
Upendar, K., Agrawal, K. N., Chandel, N. S., & Singh, K. (2021). Greenness identification using visible spectral colour indices for site specific weed management. Plant Physiology Reports, 26, 179–187. https://doi.org/10.1007/s40502-020-00562-0
Varela, S., Dhodda, P. R., Hsu, W. H., Prasad, P. V. V., Assefa, Y., Peralta, N. R., et al. (2018). Early-season stand count determination in corn via integration of imagery from unmanned aerial systems (UAS) and supervised learning techniques. Remote Sensing, 10, 343–356. https://doi.org/10.3390/rs10020343
Virlet, N., Sabermanesh, K., Sadeghi-Tehran, P., & Hawkesford, M. (2017). Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Functional Plant Biology, 44, 143–153. https://doi.org/10.1071/FP16163
Xie, Q., Dash, J., Huang, W., Peng, D., Qin, Q., Mortimer, H., et al. (2018). Vegetation indices combining the red and red-edge spectral information for leaf area index retrieval. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 11, 1482–1493. https://doi.org/10.1109/JSTARS.2018.2813281
Yu, N., Li, L., Schmitz, N., Tian, L. F., Greenberg, J. A., & Diers, B. W. (2016). Development of methods to improve soybean yield estimation and predict plant maturity with an unmanned aerial vehicle-based platform. Remote Sensing of Environment, 187, 91–101. https://doi.org/10.1016/j.rse.2016.10.005
Zhang, H., Zhang, J., Xu, Q., Wang, D., Di, H., Huang, J., et al. (2020). Identification of candidate tolerance genes to low-temperature during maize germination by GWAS and RNA-seq approaches. BMC Plant Biology, 20, 333–349. https://doi.org/10.1186/s12870-020-02543-9
Zhang, T. Y., & Suen, C. Y. (1984). A fast parallel algorithm for thinning digital patterns. Communications of the ACM, 27, 236–239. https://doi.org/10.1145/357994.358023
Zheng, H., Cheng, T., Zhou, M., Li, D., Yao, X., Tian, Y., et al. (2019). Improved estimation of rice aboveground biomass combining textural and spectral analysis of UAV imagery. Precision Agriculture, 20, 611–629. https://doi.org/10.1007/s11119-018-9600-7
Acknowledgements
We thank Shilin Li and Ziwen Xie for valuable help in image acquiring and processing. This work was supported by the National science foundation of China (Grant No. 31000671), Science and Technology projects Inner Mongolia (Grant Nos. 2019CG093; 2019ZD024).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Che, Y., Wang, Q., Zhou, L. et al. The effect of growth stage and plant counting accuracy of maize inbred lines on LAI and biomass prediction. Precision Agric 23, 2159–2185 (2022). https://doi.org/10.1007/s11119-022-09915-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11119-022-09915-1