Abstract
Auxin response factor (ARF) and Auxin/INDOLE-3-ACETIC ACID (Aux/IAA) proteins are the foremost regulators of auxin action and play an essential role in the coordination of many aspects of plant growth and development. Though many members of both ARF and Aux/IAA gene families have been identified and characterized in tomato, they are less studied in other Solanaceae species. In the present study, we focused on gaining insights into their functional conservation as well as diversification during auxin-mediated responses in Solanaceae. First, we identified their full complement in tomato, potato, pepper, Nicotiana benthamiana, eggplant, and petunia and found that both the gene families have expanded in N. benthamiana. We also looked into the structural variations associated with all the members of these two classes of genes in tomato and showed that huge natural variation exists in their sequence in wild relatives. The comprehensive gene expression analysis provided evidence of high conservation in the expression of orthologous ARFs and Aux/IAAs during fruit development and ripening in tomato and pepper. Furthermore, the molecular changes caused by exogenous plant hormones and abiotic stress conditions on their transcript levels were investigated which showed that many members of both the gene families may participate in various hormone- and stress-mediated responses in tomato and potato. Some of these genes may play a role in linking the hormone-controlled plant growth and stress-related signaling pathways. Finally, we demonstrate that single tomato ARF can interact with multiple Aux/IAA proteins and vice versa. Overall, our study will be very helpful in establishing both conserved as well as non-conserved functions of these genes in Solanaceae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Combinatorial action of Auxin response factor (ARF) and Auxin/INDOLE-3-ACETIC ACIDacid (Aux/IAA) proteins is required for rapid regulation of auxin response genes during auxin signaling in plants. Additional players, such as the topless (TPL) co-repressor and transport inhibitor response 1 (TIR1) receptor families are also involved in execution of auxin responses. ARF proteins contain a conserved N-terminal DNA-binding domain (DBD), which is required for targeting TGTCTC and related auxin response elements (AuxREs), a non-conserved middle region (MR) that confers either activation (AD) or repression domain potential to the ARF, and a conserved C-terminal dimerization domain (CTD). The CTD of ARF resembles the domains III and IV of Aux/IAA proteins and mediates interaction among ARFs and Aux/IAAs (Ulmasov et al. 1997). Aux/IAA genes, along with the two other classes of auxin response genes viz. Gretchen Hagen3 (GH3) and Small AuxinUp RNA (SAUR) are the three early auxin response gene classes (Abel and Theologis 1996; Shen et al. 2010). Aux/IAAs encode short-lived nuclear proteins and generally share four highly conserved domains, referred to as domains I, II, III, and IV (Abel et al. 1994; Hagen and Guilfoyle 2002; Liscum and Reed 2002). Domain I acts as transcriptional repressor, and it is known to interact with TPLs (Tiwari et al. 2003; Szemenyei et al. 2008). Domain II is involved in rapid turnover of these proteins and ensures their degradation via ubiquitin-mediated pathway (Dharmasiri et al. 2005a, b; Gray et al. 2001; Kepinski and Leyser 2005).
The activity of ARFs during auxin action depends on auxin-dependent degradation of Aux/IAA repressors. When available, auxin binds to the intercellular auxin receptors TIR1 and auxin binding F-Box (AFB) proteins to facilitate ubiquitination of Aux/IAA repressors. Such interaction triggers the degradation of Aux/IAA proteins by the 26S proteasome pathway and releases the inhibition of ARF activity, which is required for transcriptional activation of ARF responsive genes (Dharmasiri et al. 2005a, b). Once free, ARF proteins directly bind to the AuxREs (TGTCTC/TGTC) present in the promoters of target genes and modulate auxin response (Chapman and Estelle 2009). Since both ARF and Aux/IAA genes are encoded by multigene families and an ARF can independently interact with several Aux/IAAs, the auxin signaling pathway in plants is very complex.
Full complement of both ARF and Aux/IAA gene families has already been identified for several plants, including Arabidopsis, rice, populous, tomato, etc. and expression profiling during vegetative and reproductive stages has revealed distinctive spatio-temporal pattern for expression of these genes (Audran-Delalande et al. 2012; Jain et al. 2006; Kalluri et al. 2007; Kumar et al. 2011, 2012a). The understanding of the diverse roles of ARFs and Aux/IAAs in planta has majorly been achieved by the characterization of gain-of-function mutants in Arabidopsis. Very few identifiable phenotypes associated with their loss-of-function mutants indicate about the functional redundancy among both ARF and Aux/IAA family members in Arabidopsis (Nagpal et al. 2005; Okushima et al. 2005; Wilmoth et al. 2005). In contrast, visible and distinct phenotypes have been observed with downregulation of various ARF and Aux/IAA genes in Solanaceae species. Transcript inhibition of SlARF4 causes abnormal ripening with modified fine pectin structure and tissue architecture in tomato fruit (Guillon et al. 2008; Jones et al. 2002). SlARF4 also regulates sugar metabolism during tomato fruit development (Sagar et al. 2013). SlARF7 acts as a negative regulator of fruit-set as inhibition of its transcription has been found to cause parthenocarpic fruit development in transgenic tomato (de Jong et al. 2009; Vriezen et al. 2008). SlARF6 and SlARF8 are also involved in floral development as their down regulation by microRNA 167 leads to female sterility in tomato (Liu et al. 2014). Likewise, SlIAA9 is an established regulator of leaf morphogenesis and fruit-set whereas SlIAA15 is implicated in trichome development (Deng et al. 2012; Wang et al. 2005, 2009). SlIAA3 is considered a connecting point between auxin and ethylene signaling pathways as its transcriptional inhibition resulted in phenotypes attributed to reduced auxin and enhanced ethylene levels in tomato transgenic plants (Chaabouni et al. 2009). Likewise, StIAA2 knockdown potato lines display increased plant height, curvature of growing leaf primordia in the shoot apex and petiole hyponasty (Kloosterman et al. 2006). Altogether, these reports suggest very limited functional redundancy among tomato ARFs and Aux/IAAs and clearly emphasize widening of scope of their functional characterization beyond Arabidopsis to uncover the novel roles played by these genes in Solanaceae species.
Solanaceae is one of the most important groups of plants in terms of economic importance. It broadly includes all the major vegetables grown all over the world which show high level of conservation at genome level of its members. In 2003, International Solanaceae Genome Project (SOL) was initiated with an aim to answer two of the most important questions; first, how can a common set of genes/proteins give rise to such widely diverse plants, as observed in Solanaceae, and second, how can a deeper understanding of this plant diversity be harnessed for meeting the needs of society in a sustainable manner. After over a decade, draft genomes of several species belonging to this taxon, including tomato, potato, N. benthamiana, and pepper have been reported (Bombarely et al. 2012; Kim et al. 2014; TGC 2012; Xu et al. 2011). Moreover, expression data of thousands of genes also became available which now provides an opportunity to look into conserved as well as non-conserved functional aspects underlying various developmental processes (for example, auxin-mediated responses) across Solanaceae species (Bombarely et al. 2012; Kim et al. 2014; Xu et al. 2011). Although members of both ARF and Aux/IAA gene families have been primarily characterized at least twice in tomato, the exact number of encoded Aux/IAAs still remains disputed. Moreover, to our knowledge, no comprehensive study has reported the analysis of ARF and Aux/IAA genes in other Solanaceae species, so far. In order to address the two questions, we did comprehensive identification, structural comparison, phylogenetic analysis of these genes in Solanaceae, including tomato, potato, eggplant, petunia, pepper, and N. benthamiana. To get clues about their functional conservation as well as diversification, we analyzed their expression patterns during developmental stages in tomato and pepper, especially fruit development and ripening (as fruits in both species undergo similar phase changes during fruit development) and under phytohormone and stress treatments in tomato and potato. Pepper is not included for phytohormone and stress analysis as to the best of our knowledge comprehensive expression data are not available for this species. Furthermore, sub-cellular localization and protein–protein interaction (PPI) analysis for select members of ARF and Aux/IAA gene families was done to get insights into their complex nature of interaction in tomato. In addition, we identified natural variations associated with both ARF and Aux/IAA genes in wild relatives of tomato. We envisage that the comprehensive information generated in this study will be very useful in future investigations to assign them new roles in auxin-mediated responses in Solanaceae.
Materials and Methods
Database Search and Sequence Analysis
Protein and coding sequences for the full complement of both tomato ARF (SlARF) and Aux/IAA (SlIAA) gene families were retrieved from ITAG Release 2.3 predicted proteins and CDS (Sl2.40), available in Solanaceae Genomics Network database (www.sgn.cornell.edu). Protein sequences of ARFs and Aux/IAA proteins from other model plant organisms, including Arabidopsis, rice (Oryza sativa ssp. Indica), Physcomitrella patens (a model moss plant), and Selaginella moellendorffii (a model lycophyte) were retrieved from their respective databases and Plant Transcription factor Database (http://planttfdb.cbi.edu.cn/). All these protein sequences were used as query sequences and TBALSTN search was performed to identify their homologs in other Solanaceae members, namely, potato (Solanum tuberosum; PGSC DM v3), eggplant (Solanum melanogena), Nicotiana benthamiana (Genome v0.4.4), pepper (Capsicum annum), and petunia (Petunia hybrida), using DFCI-Plant Gene Index Database (http://compbio.dfci.harvard.edu/tgi/plant.html) and SGN Database. The HMM (hidden Markov model) profiles of ARF (accession number PF06507) and Aux/IAA family (accession number PF02309) were extracted from the Pfam (http://pfam.sanger.ac.uk/). HMM searches were performed on translated protein sequences of potato (http://www.potatogenome.net/index.php/Data), pepper (www.sgn.cornell.edu), and N. benthamiana WGS scaffolds (http://www.sgn.cornell.edu/tools/blast/; The N. benthamiana Genome Sequencing Consortium.v0.4.4). In addition, name search was also performed to identify ARF and Aux/IAA genes. Retrieved sequences (score ≥100 and e value ≤ e − 10) were sorted for the unique sequences and used for further analysis. The protein sequences were further scanned through SMART (http://smart.embl-heidelberg.de/) and PFam (http://pfam.sanger.ac.uk/) tools to identify and verify characteristic and functionally important domains. For their promoter analysis, 2-kb upstream region from the translation start site was extracted for the identified ARF and Aux/IAA genes. Various cis-regulatory elements present in their promoters were subsequently analyzed in PLACE (Higo et al. 1999) and PlantCare (Lescot et al. 2002) databases.
Structural Analysis of SlARF and SlIAA Genes in Wild Relatives
Structural variation browser for tomato and its wild relatives available (http://www.tomatogenome.net/terms-of-service_vb.html) at SGN Webpage was used for this analysis. SGN-IDs of SlARF and SlIAA genes were used as queries against the available genomic sequences of the references, Solanum lycopersicum var. Heinz, two inbred lines namely Ailsa Craig and Moneymaker and 11 lines of wild relatives (one each of Solanum pimpinellifolium ‘LA1578’, Solanum peruvianum ‘LA1278’, Solanum chmielewski ‘LA2663’, Solanum galapagense ‘LA0483’, Solanum chilense ‘CGN15530’, Solanum habrochaites ‘LA1777’, Solanum pennellii ‘LA0716’, Solanum huaylense ‘LA1365’, Solanum corneliomuelleri ‘LA0118’, Solanum neorickii ‘LA2133’, and Solanum arcanum ‘LA2157’. The sequence generated and used by this browser included complete genomic sequence for each gene, including sequences of all the exons and introns. In addition, some portion of 5′ as well as 3′ UTR is also represented in the final sequence used in the analysis.
Multiple Sequence Alignment, Phylogenetic Analysis, and Gene Nomenclature
For phylogenetic analysis, amino acid sequences corresponding to conserved ‘aux_resp’ domain, in case of ARF members, and ‘Aux/IAA’ domain, in case of Aux/IAA candidates, were used for multiple sequence alignment, employing ClustalX v2.0 (Thompson et al. 2002). Subsequently, phylogenetic analysis was performed using MEGA v5 program. The unrooted phylogenetic tree was generated by neighbor-joining (NJ) algorithm with p-distance method and pairwise deletion of gaps, using default parameters and with a bootstrap statistical analysis for 1000 replicates to test the phylogeny (Tamura et al. 2011). In case of SlARFs and SlIAAs, the nomenclature systems adopted by Zouine et al. (2014) and Wu et al. (2012), respectively, were retained. Nomenclature of orthologous ARF and Aux/IAA genes in five other Solanaceae species was performed based on their relative homology with the SlARFs and SlIAAs, respectively.
Plant Growth and Chemical Treatment
Wild type tomato plants (Solanum lycopersicon cv Pusa Ruby) were grown in a green house at 28 ± 2 °C with a daily photoperiodic cycle of 16 h light/8 h dark. Three-week soil grown seedlings were harvested for root, shoot, and leaf tissues. Mature plants were used for harvesting flower buds and fully blossomed flowers. Fruits were harvested from various stages, including 8, 30, 35, 40, 43, 45, and 60 DAP as described previously (Kumar et al. 2011). The collected tissues were instantly frozen in liquid nitrogen.
For various chemical treatments, tomato seeds were sterilized with 4 % sodium hypochlorite and grown in culture room maintained at 25 ± 2 °C with a daily photoperiodic cycle of 16 h light/8 h dark. For treatment of abscisic acid (ABA; Sigma-Aldrich), 6-benzylamonopurine (BAP; Sigma-Aldrich), brassinosteroids (BL; Sigma-Aldrich), jasmonic acid (JA; Sigma-Aldrich), gibberellic acid (GA; Sigma-Aldrich), salicylic acid (SA; Sigma-Aldrich), and ethylene (ethrel, Sisco Research Laboratories Pvt. Ltd), 10-day-old tomato seedlings were immersed in their respective 50-μM solutions for two time points, i.e., 1 and 3 h, as described earlier (Kumar et al. 2012a). Auxin treatment was performed as described earlier (Kumar et al. 2012a). Water-treated seedlings for the same time periods served as controls. Ten-day-old seedlings were subjected to various stress treatments, including cold, desiccation, heat, and salt as described earlier (Kumar et al. 2012a; Sharma et al. 2010). To study the effect of light on transcript accumulation of SlARF and SlIAA genes, 3-day-old dark-grown etiolated tomato seedlings were harvested while light-grown seedlings served as their controls. Harvested tissues were flash frozen in liquid nitrogen and stored at −80 °C.
Gene Expression Analysis
Besides the in-house microarray data, generated during the study of fruit transcriptomes of wild type and rin mutant fruits during ripening, the publically available microarray data, submitted at the Gene Expression Omnibus (GEO) database, under the series accession numbers GSE19326 (expression data of various tomato tissues), GSE22304 (expression data of heat and drought stresses), GSE16401 (expression study of salt tolerance), GSE14637 (expression data for virulent infection of Botrytis cinarea), and GSE21999 (Colletotrichum coccodes) were used to study the expression analysis of Aux/IAA genes in tomato (Kumar et al. 2012b). Affymetrix GeneChip® tomato genome arrays were used to study fruit transcriptomes of wild type during ripening, as described earlier (Kumar et al. 2012b). Furthermore, RNA-seq expression data for ten stages/tissues each of tomato (TGC 2012) and pepper (Kim et al. 2014) were analyzed to study the differential gene expression at different developmental stages of fruits in two Solanaceae species and final heatmaps were developed using ‘gplots’ package in programming language ‘R’. The gene expression of tomato Aux/IAA genes in various tissues/organs was validated by quantitative real-time PCR (QPCR) analysis, as described earlier (Kumar et al. 2011). Total RNA from the tissues included in this study was isolated and processed following the protocol described earlier (Kumar et al. 2012a). Additionally, we employed QPCR analysis to study gene expression of tomato ARF and Aux/IAA genes under various phytohormone treatments, abiotic stress conditions, and in etiolated seedlings. Briefly, complementary DNA (cDNA) was synthesized from at least two biological RNA samples. Three technical replicates for each biological replicate were analyzed for QPCR analysis. The reactions were set in 96-well optical reaction plates (Applied Biosystems, USA) using SYBR® Green PCR Master Mix (Applied Biosystems, USA) for detection of gene expression in the ABI Prism 7000 Sequence Detection System (PE Applied Biosystems, USA). Sample variance for each gene in different tissues/stages during QPCR analysis was normalized by using GAPDH gene as the endogenous control. The expression level for each SlIAA gene in different tissues/stages was calculated by using ∆∆CT method. Microsoft excel was employed for plotting the bar charts.
Plasmid Construction for Sub-cellular Localization and Protein–protein Interaction Study
Protein coding regions (without stop codon) of respective SlARFs and SlIAAs were amplified from cDNA pools (cDNA samples, prepared separately from root, shoot, and fruit tissues and later mixed in equimolar ratio) of tomato (Pusa Ruby) with gene-specific primers (Supplementary Table S1), using Phusion high-fidelity DNA polymerase (New England BioLabs Inc.). All the YFP-IAA and CFP-ARF constructs were prepared by cloning the amplified IAA and ARF CDSs in Gateway H entry vector pENTR-D/TOPO (Invitrogen) and subsequent mobilization to Gateway compatible binary vector pSITE3CA and pSITE1CA (Invitrogen), respectively. In pSITE vectors, the 2XCaMV35S promoter regulates the expression of cloned gene. All the final constructs were subsequently confirmed by sequencing and only successful preparations were used for further experiments.
Particle Bombardment in Onion Peel and Confocal Microscopy
Epidermal peels were sliced from the surface of the spring onion bulb leaves and placed on 1/2 MS-agar plates, supplemented with 1 % sucrose. Approximately 2.5 μg of ARF and IAA plasmid constructs were coated onto 1 μm gold particle (Bio-Rad). These plasmids were used for bombardment. Manufacturer's instructions were followed for microprojectile bombardment, and the coated DNA plasmids were introduced into onion epidermal cells using Bio-Rad PDS/1000 helium-driven particle accelerator. All the samples were incubated in dark conditions at 28 °C and microscopic analysis was conducted after 16–24 h. Transiently transformed epidermal peels were analyzed in TCS SP5 laser scanning confocal microscope (Leica, Germany) for fluorescence detection. CFP signals were detected between 505–550 nm after exciting with 488 nm laser. Similarly, YFP signals were detected at 600–630 nm after exciting with 543 nm laser. Sequential scanning of CFP and YFP was performed for both the channels in PPI and the co-localization experiments. Thereafter, the signals were merged together to show the overlap. All the images were further processed using LAS-AF software.
Results
Identification and Sequence Analysis of the ARF and Aux/IAA Complement in Solanaceae
Based on BLAST and HMM profile searches and subsequent domain analysis of identified proteins, 22 and 26 ARF and Aux/IAA members, respectively, were identified in tomato (Supplementary Tables S2 and S3). Similar approach fetched a total of 22, 2, 21, 2 and 38 putative orthologous ARF genes and 26, 16, 28, 16, and 45 putative orthologous Aux/IAA genes in potato, eggplant, pepper, petunia, and N. benthamiana, respectively. All Solanaceae orthologous genes identified in this study were named based on their relative homology with the SlARF and SlIAA proteins(Supplementary Tables S2, S3, S4, and S5). For species where genome sequence is now available, gene structure analysis showed that most of tomato, potato, pepper, and N. benthamiana ARF and Aux/IAA genes are made up of multiple intron-exons. However, StIAA20 lacked any intron whereas NbARF2A-2 sequence included 19 introns; the highest among all the genes studied here (Supplementary Tables S2, S3, S4, and S5). While most of the ARF genes include more than ten introns in their genomic sequence, the number of introns present in Aux/IAA genes is always less than that.
Evolutionary Relationship and Synteny of the Tomato ARF and Aux/IAA Genes
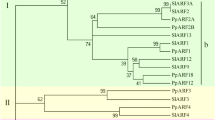
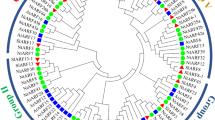
We studied evolutionary relatedness of ARF and Aux/IAA gene families, across flowering and non-flowering plant species. Phylogenetic analysis was performed among the gene family members of tomato vis-à-vis other model plant species, including Arabidopsis and rice, two model plants representing dicots and monocots, respectively, two non-flowering model plant species, including Phiscomitrella patens (a model moss plant) and S. moellendorffii (a model lycophyte) and six Solanaceae species. This analysis highlighted expansion of both ARF and Aux/IAA families during plant evolution. The phylogenetic study corroborated with the family based classification of both ARF and Aux/IAA proteins as Solanaceae members were always placed close to each other in comparison with that of Arabidopsis or other members (Figs. 1 and 2). Orthologs of non-flowering plants exhibited low sequence homology to their flowering counterparts and fell apart in a separate clade (Figs. 1 and 2). Among flowering plants, rice complement (monocot) showed distant phylogenetic relationship with Solanaceae members in comparison with that of Arabidopsis. We found that within Solanaceae, tomato ARF and Aux/IAA proteins exhibited highest phylogenetically relatedness to the potato complement, followed by that of eggplant and pepper (Figs. 1 and 2). Though N. benthamiana Aux/IAAs shared least similarity to their tomato orthologs as compared with the remaining solanaceous orthologs, we clearly identified at least two N. benthamiana orthologs for most of tomato Aux/IAAs and for a few ARF proteins (Figs. 1 and 2). Evolution and synteny of the Solanaceae ARFs and Aux/IAAs were studied using an online tool, SynFind at CoGe (http://genomevolution.org/CoGe/SynFind). In total, five and three pairs of paralogous SlARF and SlIAA genes, respectively, were identified in tomato. Two tomato ARFs (SlARF6A and SlARF8B) and eight Aux/IAAs (SlIAA1, SlIAA8, SlIAA12-13, SlIAA16-17, SlIAA21, and SlIAA23) lacked any syntelog in potato; whereas no syntelog in pepper and N. benthamiana could be retrieved (Supplementary Tables S2 and S3).
Phylogenetic relationship of Solanaceae ARF proteins encoded by members of ARF gene families with other flowering and non-flowering plants. This analysis included aux_resp domain sequences, from Arabidopsis (AtARF), rice (OsARF), six Solanaceae species, including tomato (SlARF), potato (StARF), eggplant (SmARF), pepper (CaARF), petunia (PhARF), and N. benthamiana (NbARF), and two non-flowering plants, including P. patens (PpARF) and S. moellendorffii (SmoARF). The unrooted tree was generated using MEGA5 program by neighbor-joining method. Significant bootstrap supports (>50 %) are indicated at each branch
Phylogenetic relationship of Solanaceae Aux/IAA proteins encoded by members of Aux/IAA gene families with other flowering and non-flowering plants. This analysis included Aux/IAA domain sequences, from Arabidopsis (AtIAA), rice (OsIAA), six Solanaceae species, including tomato (SlIAA), potato (StIAA), eggplant (SmIAA), pepper (CaIAA), petunia (PhIAA), and N. benthamiana (NbIAA), and two non-flowering plants, including P. patens (PpIAA) and S. moellendorffii (SmoIAA). Multiple sequence alignment was performed with ClustalX2.0.8 and MEGA5 was used to generate unrooted phylogenetic tree. Significant bootstrap supports (>50 %) are indicated at each branch
Expression Profiles of ARF and Aux/IAA Genes Under Dark and Auxin Treatment in Tomato and Potato
Light and auxin are two critical regulators of plant growth and development. In order to establish the role of these two important factors in controlling expression of entire tomato ARF and Aux/IAA complement, we analyzed their expression profiling by QPCR under dark and auxin treatment regimes. Most of the SlIAA genes were either upregulated or showed no substantial reduction in their expression in etiolated seedlings. On the contrary, out of six deferentially expressed SlARF genes, SlARF2A, SlARF6B, SlARF7B, and SlARF17 showed reduction in their transcript accumulation in etiolated seedlings (Tables 1 and 2). Furthermore, expression of many SlARF and SlIAA genes was found to be perturbed under both the physiological regimes, mostly in an antagonistic manner. Genes such as SlARF6B, SlARF7B, SlIAA3, and SlIAA8 that showed auxin-induced expression, were downregulated in etiolated seedlings whereas genes, including SlARF4, SlARF19, SlIAA7, and SlIAA26, inhibited by auxin, exhibited dark-induced expression (Tables 1 and 2). Interestingly, some of the SlIAA genes such as SlIAA13, SlIAA15, and SlIAA21 were found to be upregulated in both auxin-treated and etiolated seedlings; however, they differed in the magnitude of their upregulation. The remaining genes did not exhibit any change in their transcript accumulation in both the conditions.
The paralogous ARF genes belonging to their respective pairs viz. SlARF2A/SlARF2B (expression level is inhibited at the late stage of auxin treatment), SlARF8A/SlARF8B, and SlARF10A/SlARF10B (expression level remains unaltered) demonstrated similar expression profiles in the QPCR analysis in both auxin-treated and etiolated seedlings. The transcript levels of the remaining two paralogous pairs, SlARF9A/SlARF9B and SlARF16A/SlARF16B though demonstrated different expression profiles under auxin treatment but remained unaltered in etiolated seedlings (Table 1). Similarly, SlIAA paralogous gene pairs, including SlIAA7/SlIAA24 (unaltered expression in auxin-treated seedlings whereas their expression is induced in etiolated seedlings) and SlIAA5/SlIAA25 (their expression is induced in auxin-treated seedlings whereas unaltered expression in etiolated seedlings) also demonstrated almost similar profiles in the two studied conditions. Expression of the SlIAA genes of the third paralogous pair, SlIAA2/SlIAA3 was induced by auxin; however, both genes displayed different profiles in etiolated seedlings (Table 2). Furthermore, in silico analysis of 2-kb upstream regulatory sequence of three SlARF and two SlIAA paralogous gene pairs revealed that all these genes, expect SlARF10A and SlIAA5, lacked a typical AuxRE (TGTCTC); however, their promoters were found to be enriched by cis-acting elements involved in light signaling (Supplementary Table S6). In order to establish any conservation in the auxin responses mediated by these genes across Solanaceae species, we also analyzed the RNA-seq expression data available for potato (Xu et al. 2011). However, we could identify only two ARF (StARF4 and StARF6B) and four Aux/IAA (StIAA6, StIAA12, SlIAA17, and StIAA18) genes induced by auxin treatment (Fig. 3).
Expression profiles of potato ARF and Aux/IAA genes under various phytohormones and abiotic stresses. Heatmap showing expression of members of these two gene families under various phytohormone and abiotic stress treatments (labeled on top) in potato. Color scale represents log2 (FPKM) values of the publically available RNA-seq data (Xu et al. 2011). C.1 control for salt and mannitol treatments, C.2 control for BAP, ABA, IAA, and GA treatments, C.3 control for heat stress
We further studied the promoter architecture of other Solanaceae ARF and Aux/IAA genes and found significant conservation in the cis-acting elements among different Solanaceae species (Supplementary Table S6). In tomato, upregulation in expression of several ARF and Aux/IAA genes such as SlARF4, SlARF24, SlIAA2, SlIAA5, SlIAA6, and SlIAA8 was perfectly corroborated with the presence of typical AuxRE in their promoters (Tables 1 and 2; Supplementary Table S6). Interestingly, promoters of SlIAA14 and SlIAA26 though harbored at least one typical AuxRE, yet their expression was found to be inhibited by auxin. In case of potato orthologs, their promoters were found to be conserved for various cis-acting elements; however, comparatively fewer genes were induced by auxin treatment (Fig. 3; Supplementary Table S6).
Differential Gene Expression of ARF and Aux/IAA Genes Under Different Phytohormone Treatments
We further examined expression profiles of both ARF and Aux/IAA genes in 10-day-old tomato seedlings which were subjected to 50 μM solutions of various growth regulators viz. ABA, BAP, BL, C2H4, GA, JA, and SA for two time points by QPCR analysis. Most of the members were found to be differentially expressed under more than one phytohormone treatments. We noted that expression of SlARF1, SlARF18, and SlIAA9 remained majorly unaffected under most of the treatments. Transcript levels of three SlARFs, including SlARF7A, SlARF7B, and SlARF9A and seven SlIAA genes, including SlIAA6, SlIAA8, SlIAA10, SlIAA15, SlIAA19, SlIAA21, and SlIAA24 were induced while that of eight SlARFs, including SlARF2B, SlARF3, SlARF4, SlARF5, SlARF6A, SlARF10B, SlARF16A, and SlARF17 and six SlIAAs, including SlIAA2, SlIAA7, SlIAA9, SlIAA11, SlIAA14, and SlIAA17 were repressed by most of the treatments (Tables 1 and 2). Six SlIAAs, including SlIAA1, SlIAA3-5, SlIAA20, and SlIAA25 seemed to resist any change in their transcript accumulation under majority of the treatments. ABA and SA treatment resulted mainly in repression whereas BAP and JA treatments mainly resulted in induction in their expression (Tables 1 and 2). We further examined expression patterns of tomato ARF and Aux/IAA paralogs and observed that expression of SlARF2A/SlARF2B, SlARF8A/SlARF8B, and SlARF10A/SlARF10B paralogous genes was majorly in sync with each other and showed similar type of transcriptional changes under most of the treatment. The remaining SlARF and SlIAA paralogous gene pairs demonstrated distinct hormone-specific changes in their expression between the paralogous genes (Tables 1 and 2).
Expression profiling of potato ARF (StARF) and Aux/IAA (StIAA) genes under ABA, BAP, and GA treatments using RNA-seq data (Xu et al. 2011) further revealed that transcript accumulation of these genes is regulated by several cues. We observed that expression of six StARFs and seven StIAAs was inhibited (≥2-fold) by BAP and while ABA and GA had very limited or no influence on transcription of StARFs (Fig. 3). However, ABA treatment caused differential expression of seven StIAA genes. StIAA6 recorded an upregulation under all these treatment whereas the remaining StIAA genes, induced by GA, also exhibited increased expression levels under ABA and this trend was found to be in accordance with trend exhibited by their tomato orthologs (Fig. 3; Tables 1 and 2). Furthermore, we noticed good correlation between the presence of phytohormone related various cis-acting elements in the promoters of tomato and potato ARF and Aux/IAA genes and their differential expression in response to specific phytohormone treatments; however, this relation was not found to be strictly followed in case of all these genes (Fig. 3; Tables 1 and 2; Supplementary Table S6).
Expression Profiles of ARF and Aux/IAA Genes Under and Abiotic and Biotic Stresses
Furthermore, we investigated expression patterns of these genes under different abiotic stress conditions (cold, desiccation, heat, and salt) by QPCR analysis. We observed that 14 SlARF and 19 SlIAA genes were deferentially expressed in at least one or multiple stress treatments as compared with their unstressed control (Tables 1 and 2). Overall, cold stress resulted in differential expression of eight SlARF and 13 SlIAA genes whereas desiccation stress was found to be the least effective. Interestingly, cold stress had a negative influence on transcript accumulation of majority of differentially regulated SlARFs; however, SlIAA genes were found to be positively regulated by it. Transcript levels of SlARF4, SlARF5, SlIAA8, SlIAA11, SlIAA13, and SlIAA18 were induced whereas those of SlARF20, SlIAA10, SlIAA12, SlIAA15, and SlIAA25 were repressed in multiple stresses. Among the SlARF and SlIAA genes differentially regulated in heat and cold stresses, response of most of these genes, except for SlARF4, SlARF10B, SlIAA11, and SlIAA13, was found to be either antagonistic or very distinct. We noted mostly similar expression patterns of the two members of SlARF2A/SlARF2B, SlARF8A/SlARF8B, SlARF9A/SlARF9B, SlIAA2/SlIAA3, and SlIAA7/SlIAA24 pairs under at least three stresses. Additionally, exploration of publicly available Affymetrix GeneChip® tomato microarray data available at GEO database, including salt (GSE16401), drought, or heat stress (GSE22304), infections of tomato fruits with necrotrophic fungus Botrytis cineria (GSE14637) or C. coccodes (GSE21999) also showed modulation in the expression of a few SlIAA genes under at least one abiotic stress condition (Supplementary Figs. S1 and S2).
Furthermore, effect of stresses on potato StARF and StIAA genes was investigated. For this analysis, we used the publically available RNA-seq data (Xu et al. 2011). Besides many StIAAs, only single potato ARF gene, StARF9A, showed upregulation (>2-fold) in their expression under salt stress. Among all StARF and StIAA genes, StARF19 was the only member that was inhibited by salt stress (Fig. 3). Heat stress had a profound effect on transcript levels of most of the StARFs and StIAAs. Four StARFs, including StARF4, StARF5, StARF9B, and StARF24 and six StIAAs, including StIAA5, StIAA6, StIAA22, StIAA24, and StIAA25 were induced whereas seven SlARFs and five SlIAAs were inhibited by heat stress (Fig. 3). Analysis of the architecture of all the tomato and potato ARF and Aux/IAA genes further revealed the presence of various stress-related cis-acting elements; however, we observed that their presence/absence was not always correlated strictly to their response to these stresses (Fig. 3; Tables 1 and 2; Supplementary Table S6).
Expression Profiles of ARF and Aux/IAA Genes During Plant Development
Members of both ARF and Aux/IAA gene family play important role in plant development. The earlier investigations on SlARF and SlIAA genes have already covered their expression profiles at various developmental stages in tomato (Audran-Delalande et al. 2012; Wu et al. 2011, 2012). Earlier, we studied expression profiles of 17 SlARFs at various stages of fruit development and ripening in an Indian cultivar ‘Pusa Ruby’ and identified ARF genes with ripening-associated expression (Kumar et al. 2011). We further analyzed expression patterns of all SlIAA genes in the same genetic background in our in-house microarray and public available data (GSE19326) as well as by employing QPCR analysis and observed high correlation between the two datasets (Supplementary Figs. S3 and S4). It was observed that most of these genes are expressed at comparatively higher levels during vegetative development than in reproductive development (Fig. 4). Tomato (climacteric fruit) and pepper (non-climacteric fruit) are well-established models for comparisons of fruit ripening processes. Furthermore, to gain insights into the conservation of their functions during fruit development and ripening in Solanaceae, we looked into the expression profiles of members of both ARF and Aux/IAA orthologous genes. Based on the clustering, their expression profiles were mainly divided into three major clusters in both tomato and pepper. Cluster I included genes which showed high expression levels at most of the developmental stages, especially at the stages of early fruit development. Expression of six orthologous ARFs (ARF1, ARF2A, ARF2B, ARF3, ARF6A, and ARF8A) and three Aux/IAAs (IAA3, IAA4, and IAA8) was found to be conserved in both pepper and tomato. Cluster II included genes with intermediate level of expression during various developmental stages. Genes present in this group showed diverse expression profiles as only five orthologous genes, including four ARFs (ARF7B, ARF16A, ARF17, and ARF19) and one Aux/IAA (IAA22) exhibited similar expression in both the species. Cluster III included mainly those genes which showed a decline in their expression at ripening stages; however, we observed a high conservation in the expression of most of the orthologous genes. Such genes included four ARFs (ARF9A, ARF9B, ARF10B, and ARF18) and ten Aux/IAAs (IAA2, IAA10, IAA11, IAA14, IAA17-19, and IAA23-25). The remaining orthologous ARF and Aux/IAA genes showed distinct expression profiles in tomato and pepper (Supplementary Fig. S5). Overall, this analysis showed high conservation in the expression of orthologous ARFs and Aux/IAAs during fruit development and ripening in both tomato and pepper.
QPCR analysis of SlIAA genes in a total of 12 vegetative/reproductive tissues/stages. x-axis represents the different tissues/stages of development. y-axis represents the relative mRNA levels, detected by quantitative real-time PCR analysis, of genes. Expression of GAPDH was used as internal control and to normalize the expression of SlIAA genes. L leaf, S shoot, R root, FB flower bud, OF open flower. Ft is used to indicate fruit tissue while number represents days after pollination. Error bars show the standard error between three replicates performed
We further investigated expression profiles of SlARF and SlIAA paralogous genes during developmental stages. During investigation of their expression patterns using RNA-seq expression data of S. lycopersicum var. ‘Heinz’, we observed similar expression patterns between the two members of SlARF2A/SlARF2B and SlARF8A/SlARF8B gene pairs. On the contrary, members of SlARF9A/SlARF9B pair showed distinct expression profiles. Average RPKM value for all the samples has been presented as an area diagram (Fig. 5). Paired partners of the remaining ARF and Aux/IAA paralogous gene pairs showed distinct profiles and could lead to neo-functionalization of the paralogous genes. One of the paired partners of SlARF16A/SlARF16B gene pair seems to have lost its expression, as transcripts of SlARF16B could not be detected in RNA-seq as well as our real-time PCR expression data. Altogether, the expression data indicate a case of pseudo-functionalization for SlARF16B gene (Fig. 5).
Expression profile of tomato ARF and Aux/IAAsyntelogs. Expression pattern of paralogous genes was analyzed during various developmental stages in two genotypes using available RNA-seq data (TGC 2012). The syntelogs exhibited variable expression patterns and revealed retention of expression, neo-functionalization, and pseudo-functionalization. Each area graph depicts mean normalized RPKM values on y-axis and different developmental stages at x-axis. L leaf, R root, FB flower bud, F flower, 1-, 2-, and 3-cm fruit stages at these values of the diameter, MG mature green, B breaker, B + 10 10 days post-breaker stage of fruit ripening
Sub-cellular Localization and Protein–Protein Interaction Analysis of SlARFs and SlIAAs
Several members of ARF and Aux/IAA proteins are known to be localized in nucleus and also interact with each other. We also observed presence of nuclear localized signals in many of these proteins. To substantiate our observation, we studied sub-cellular localization of 18 SlIAAs, including, SlIAA1-3, SlIAA5, SlIAA7-13, SlIAA15-17, and SlIAA21-24 along with empty yellow fluorescent protein (YFP) vectors as their control. Among 18 SlIAA proteins, only eight, including SlIAA2-3, SlIAA8, SlIAA12, SlIAA15, SlIAA17, and SlIAA22-23 were found to be nucleus localized whereas the remaining SlIAA genes were detected throughout cell, including nucleus and cytoplasm (Fig. 6a; Supplementary Fig. S6). Three SlARFs, including one putative repressor, namely SlARF2A and two putative activators viz. SlARF6A and SlARF7A were selected to test PPI potential between tomato ARF and Aux/IAA proteins. The other reason for selecting these SlARF genes was their distinct expression profiles during fruit development stages. For PPI study, both SlARF-CFP and SlIAA-YFP constructs were co-bombarded in various combinations in the onion peel cells and using confocal microscopy their co-localization pattern and flourescence resonance energy transfer (FRET) scores were analyzed in at least three independent cells. We fixed 9 % FRET to be lower threshold and any value above that was considered to be an indication of positive interaction. Based on their co-localization and FRET scores, we found that SlARF2A interacts with at least five SlIAA proteins namely, SlIAA2, SlIAA3, SlIAA7 and SlIAA15-16 (Fig. 6b). SlARF7A also interacted with five SlIAAs, including SlIAA8, SlIAA15-16, SlIAA21 and SlIAA24. It was found that SlARF6A showed the highest interactive potential and interacted with at least 11 SlIAAs, including SlIAA2-3, SlIAA5, SlIAA9-11, SlIAA13, SlIAA15, SlIAA17, SlIAA21 and SlIAA24. Interestingly, SlIAA15 was found to interact with all the SlARFs while SlIAA16 interacted with only SlARF2A and SlARF7A. We also made three interesting observations in protein-protein interaction experiments. First, we used only a partial sequence of SlARF6A (truncated at 5' end and lacking the NLS motif), however, in few cases it was found to be nuclear localized. Second, even though both SlARF6A-SlIAA8 and SlARF6A-SlIAA12 were found to be co-localized in nucleus, they did not show any interaction between them. Third, in many cases where a positive interaction was observed, the SlARF-CFP was found to be localized outside nucleus with the SlIAA-YFP construct (Fig. 6a, b).
Sub-cellular localization and protein–protein interaction (PPI) between tomato ARF and Aux/IAA proteins in onion peel epidermal cells. a SlARF-CFP and SlIAA-YFP fusion protein constructs were bombarded in onion peel cells. Some extra nuclear interactions between some ARF and IAA proteins; as depicted for SlARF7A in ARF7A-IAA10 and ARF6A in ARF6A-IAA9 panels, were also observed. Surprisingly, ARF6A (which lacks the NLS as only a partial sequence was used in this study) and IAA8 and ARF6A and IAA12 (lowermost two panels) were found to be co-localized in nucleus; however, these proteins do not interact with each other. Both ARF and Aux/IAA proteins were found to be present in granular bodies. Cells transformed with CaMV35S-YFP were used as a control. Florescence was detected under a confocal laser-scanning microscope (wavelength: CFP, 488 nm; YFP, 543 nm). For PPI study, FRET analysis was performed at least in three independent cells. b Current status of PPI between three SlARFs and 18 SlIAAs. Green color represents positive interaction, red color depicts no interaction, and yellow color represents that PPI could not be ascertained between the ARF and Aux/IAA pair even after multiple attempts
Analysis for Sequence Variants of SlARF and SlIAA Genes in Other Tomato Genotypes
In this analysis, we identified 29,064 and 17,845 sequence variants, including both indels and SNPs, for 22 SlARF (covering ~166 kb genomic region) and 26 SlIAA (covering ~79 kb region of tomato genome) genes, respectively, in a total of 13 lines including, two tomato inbred lines, AC and Moneymaker (both red-fruited) and 11 lines of wild relatives (Table 3). These wild relatives included five red-fruited lines, including one line each of S. pimpinellifolium ‘LA1578’, S. galapagense ‘LA0483’, S. huaylense ‘LA1365’, S. corneliomuelleri ‘LA0118’, and S. arcanum ‘LA2157’ and the six green-fruited lines, including S. peruvianum ‘LA1278’, S. chmielewski ‘LA2663’, S. chilense ‘CGN15530’, S. habrochaites ‘LA1777’, S. pennellii ‘LA0716’, and S. neorickii ‘LA2133’. Although the number of identified structural variants was higher in case of SlARFs than SlIAAs, we observed a higher combined frequency (226.2 variants/kb in comparison with 175.6 variants/kb) for 26 SlIAA genes (Table 3). As expected, the inbred lines harbored very less sequence variations (<1 variants/kb) followed by a modest sequence divergence (<5 variants/kb) in the two closest red-fruited wild relatives, S. pimpinellifolium and S. galapagense. All the wild relatives, except S. galapagense, showed presence of a very high number of sequence variations for both SlARFs and SlIAAs (Table 3). Collectively, S. peruvianum (green-fruited) with 6290 sequence variations became the most diverged whereas S. galapagense (red-fruited) with only 870 sequence variants was the least diverged among wild relatives (Table 3). We observed that while almost equal representation of both indels and SNVs contributed to the sequence variations observed in AC and Moneymaker inbred lines, SNV was the over-represented category in the sequence variations observed in wild relatives.
Discussion
Solanaceae represents one of the most important groups of plant that includes most of the vegetable crop species. Members of this group show high level conservation at genome level within and across species which make them the best candidates amenable for comparative studies. With the availability of genome information for four important Solanaceae species, including tomato (TGC 2012), potato (Xu et al. 2011), pepper (Kim et al. 2014), and N. benthamiana (Bombarely et al. 2012), it becomes important to understand the degree of homology between genome sequences and phenotypes of evolutionarily divergent species. It is well established that the genetic changes associated with most of the traits involved in adaptation and diversification are not “loss of function” mutations; rather genetic variants that changed the function of protein. Therefore, in the present study we set to identify and characterize the genetic variants associated with the foremost regulators of auxin responses viz. ARFs and Aux/IAA in tomato and across Solanaceae species and report putative conserved as well as distinct functional roles played by these genes during similar developmental aspects in tomato, potato and pepper. We primarily focused on tomato ARF and Aux/IAA complement; however, to identify the conserved and non-conserved roles in Solanaceae, the publically available expression data on potato and pepper were also included in the final analysis.
We identified a total of 22 ARF and 26 SlIAA genes encoded by tomato and potato genomes and support the earlier findings (Wu et al. 2011; Zouine et al. 2014). In pepper, presence of similar number of ARF and Aux/IAA complement further supports the earlier observation that genomes of both tomato and pepper are highly conserved (Kim et al. 2014). However, both ARF and Aux/IAA gene families have significantly expanded in N. benthamiana (Figs. 1 and 2) (Kalluri et al. 2007; Liscum and Reed 2002). Two homologs per most tomato ARFs and Aux/IAAs encoded by N. benthamiana genome can be explained by the fact that it is an allotetraploid with a genome size of ~3 Gb (gigabases) and its genome is not completely sequenced. Therefore number of NbARFs and NbIAAs can also increase in future (Bombarely et al. 2012).
The Evolution of ARF and Aux/IAA Genes in Solanaceae
Phylogenetic analysis of different ARF and Aux/IAA members from non-flowering (P. patens and S. moellendorffii) and flowering plants, including Solanaceae members, rice and Arabidopsis revealed that family members of two non-flowering plant species are rightly placed in a distinct group as these plants have been proposed to be separated from flowering plants by more than 400 million years on evolutionary time scale (Figs. 1 and 2) (Nickrent et al. 2000). The phylogenetic trend revealed high degree of sequence similarity among dicot members followed by similarity between monocots and dicots for the conserved evolution of these groups of genes, through a common origin and ancestors. Expectedly, Arabidopsis orthologs were phylogenetically closer to solanaceous members than rice orthologs. Monocots and eudicots have been estimated to be diverged at least 150 million years ago (Mya) (Chaw et al. 2004; Salinas et al. 2012). Furthermore, members of potato and tomato are rightly placed closest to each other as analysis of the age of their most recent common ancestor (MRCA) suggested that age could be 7.3 million years (My), whereas it is 15.5 My for MRCA of tomato, potato, and eggplant (Wu and Tanksley 2010). Moreover, 19.6 My was found to be the age of MRCA of tomato, potato, eggplant, and pepper, which supports the phylogenetic relatedness among various ARF and Aux/IAA orthologs analyzed in the present study. Members of N. benthiamana were found to be most diverged and are rightly placed distantly in the phylogram as age of the MRCA for tomato, potato, eggplant, pepper, and Nicotiana is suggested to be 23.7 My (Wu and Tanksley 2010). Overall, phylogenetic analysis of ARFs and Aux/IAAs among the Solanaceae taxa reveals that members of these two gene families have followed the similar evolutionary trend as observed in case of species-level changes during evolution and it further supports the earlier findings about the evolutionary trends in Solanaceae (Clarkson et al. 2005; Kumar et al. 2011, 2012a; Salinas et al. 2012; Sharma et al. 2010; Wang et al. 2008; Wu et al. 2011, 2012; Zouine et al. 2014).
Study of their evolution and synteny further reveals high conservation among ARF and Aux/IAA genes of Solanaceae. Five ARF and three Aux/IAA genes emerged in homeologous pairs in tomato whereas 20 ARFs and 16 Aux/IAAs evolved in parallel manner in both tomato and potato (Supplementary Tables S2 and S3). In addition to the similar gene structures, the two members of homeologous and homologous gene pairs also shared close evolutionary relationships. While the homeologous gene pairs might have originated from a duplication event, the homologous gene pairs might have originated from same ancestor. Indeed, a whole-genome triplication event, followed by widespread gene loss ~71 My in the Solanum lineage; much before the tomato–potato divergence has been reported (TGC 2012). Moreover, presence of at least 18 tomato orthologous ARFs and 20 orthologous Aux/IAAs in pepper also indicates parallel evolution of these genes in tomato and pepper; similar to the evolutionary trends associated with tomato–potato divergence discussed in this paper.
ARF and Aux/IAA Genes Play Diverse Roles in Solanaceae
The expression profile of full complement of a gene family can provide clues for the functional divergence among the members. In the etiolated seedlings, altered expression of many tomato ARF and Aux/IAA genes further supports the earlier findings in other plants and suggests that these genes actively participate in the regulation of light-controlled aspects of seedling development across plant species (Jain et al. 2006; Halliday et al. 2009; Song et al. 2009). Furthermore, opposite expression patterns of many SlARF and SlIAA genes in auxin-treated and etiolated seedlings is in agreement with previous report where etiolated seedlings were found to be largely devoid of auxin (Bhalerao et al. 2002). Activation of most of the SlIAA genes by auxin is in accordance with the earlier studies and well correlated with the presence of AuxREs in their promoter region (Abel et al. 1995; Audran-Delalande et al. 2012). However, induction of only a few potato Aux/IAAs by auxin is surprising, even though we found high conservation of cis-regulatory elements in these homologous genes (Xu et al. 2011).
It is well known that different classes of genes involved in auxin signaling, including ARF, Aux/IAA, GH3, and SAUR are affected by various external as well as internal cues at transcript levels (Kumar et al. 2012a; Ghanashyam and Jain 2009; Song et al. 2009). Our present findings on tomato and potato regarding altered expression of most of the auxin-responsive ARF and Aux/IAA genes by at least two other phytohormone/growth regulator/biotic/abiotic stress treatments, are in agreement with previous reports and suggest that expression of these genes in Solanaceae members is also intricately controlled by many phytohormones and stresses, both biotic and abiotic (Fig. 3; Tables 1 and 2). Our observation that ABA and SA mostly inhibit whereas JA and BAP mostly activate their expression supports findings of a very recently published study where auxin-mediated expression of DR5::GUS was reported to be modulated in similar manner (Yuan et al. 2013). Enhanced expression of SlIAA genes under ethylene treatment supports the earlier observation made by Audran-Delalande et al (2012). Altogether, data on tomato and potato presented here as well as published earlier on rice suggest that modulation in the expression of ARF and Aux/IAA genes in response to different phytohormones/stresses is a conserved and important mechanism involved in auxin responses across plant species (Kumar et al. 2012a; Rahman 2013; Song et al. 2009; Yuan et al. 2013).
Moreover, differential regulation of several tomato Aux/IAA genes such as SlIAA1, SlIAA4, SlIAA8, SlIAA21, and SlIAA26 under biotic stress condition, including infection of C. coccodes (GSE21999) in tomato fruits or in response to necrotrophic fungus B. cineria (GSE14637) highlights their role in controlling auxin-mediated responses under biotic or similar stresses (Bari and Jones 2009; Ghanashyam and Jain 2009). Auxins interact with other hormones to mediate plant defense responses and SA has been reported to repress auxin signaling pathway by stabilizing Aux/IAA proteins as a part of disease resistance mechanism (Wang et al. 2007). These evidences suggest that the tomato and potato genes which were affected by majority of these stresses and phytohormones could play an active role in auxin-mediated cross talk between phytohormone pathways and stress signals in Solanaceae (Kumar et al. 2014).
Function of Several Paralogous and Orthologous ARFs and Aux/IAAs Appears To Be Conserved During Growth and Development in Solanaceae
High transcript accumulation of most of tomato and pepper orthologs in vegetative tissues and similar expression profiles of many ARF and Aux/IAA genes reveals that both classes of proteins can perform specific as well as redundant functions during vegetative and reproductive developmental stages and their function could be conserved across Solanaceae species (Audran-Delalande et al. 2012; Kumar et al. 2012a). Furthermore, identification of 14 orthologous ARFs and Aux/IAAs each with similar expression profiling in tomato and pepper during fruit development and ripening revealed that functions of these genes are conserved during fruit development and ripening; irrespective of the different ripening physiologies in tomato and pepper whereas the remaining members have diversified for their functions (Kim et al. 2014). Elevated transcript levels of SlIAA3 at the initiation of ripening supports the earlier findings (Chaabouni et al. 2009). Moreover, its induction, along with other SlARF and SlIAA genes, by both auxin and ethylene supports the hypothesis that not only Aux/IAAs but other auxin-related genes such as ARFs and GH3s can represent a molecular link between the two signaling pathways in Solanaceae members (Audran-Delalande et al. 2012; Chaabouni et al. 2009; Jones et al. 2002; Kumar et al. 2012a). Recently, inhibited expression of SlIAA27 (named SlIAA6 in this study), a gene that has comparatively higher expression in vegetative tissues and green fruits, correctly revealed multiple altered phenotypes related to vegetative and reproductive growth, including higher auxin sensitivity, altered root development, reduced chlorophyll content in leaves, altered flower morphology and smaller fruits. This study indicated its limited role in fruit ripening (Bassa et al. 2012). Similarly, SlARF4, SlARF6, and SlARF8 have also been implicated in various aspects of tomato development and fruit ripening (Guillon et al. 2008; Liu et al 2014; Sagar et al. 2013). Furthermore, similar expression pattern exhibited by many tomato and potato ARFs (such as ARF8B and ARF18) and Aux/IAA orthologous genes (such as IAA6) under various phytohormones and stresses further supports the retention of function theory whereas the other genes with dissimilar profiles would have undergone neo-functionalization.
Expression profiling of tomato paralogous ARF and Aux/IAA genes in a spectrum of developmental stages, phytohormone treatments and abiotic stresses showed similar expression pattern of SlARF2A/SlARF2B and SlARF8A/SlARF8B paralogous partners and suggested about retention of same function by the duplicated genes. Whereas similar expression pattern of paired partners in SlARF10A/SlARF10B, SlIAA5/SlIAA25, and SlIAA7/SlIAA4 in at least two conditions indicates that both duplicated genes can play both conserved and non-conserved functions (Fig. 5). Distinct expression patterns of the two members of SlARF16A/SlARF16B and SlIAA2/SlIAA3 pairs suggest about pseudo-functionalization and neo-functionalization, respectively, for the paralogous genes. It has been proposed that the variable expression pattern of the duplicated partners might have resulted due to lack of intensive selection pressure and is required for the functional diversification of various ARF and Aux/IAA genes in tomato. Moreover, it is well known that duplicated genes as well as even very closely placed sister pairs of a phylogenetic tree clade often display functional divergence (Kalluri et al. 2007; Paponov et al. 2009).
A Complex Matrix of Interaction Between ARFs and Aux/IAAs Regulates Auxin Responses
Auxin responses are mediated by interaction between ARF and Aux/IAA proteins (Liscum and Reed 2002; Shen et al. 2010). We further tested PPI potential, by performing co-localization and FRET assays, between a few ARF and Aux/IAA proteins and demonstrated that single ARF protein can interact with multiple Aux/IAAs and vice versa (Kenworthy 2001; Lleres et al. 2007). While SlARF2A and SlARF7A interacted with at least five SlIAA proteins each, SlARF16 displayed interaction with 11 SlIAAs. Since SlARF5 is a putative transcriptional repressor, with maximum transcript accumulation at fruit ripening stages, its interaction with a ripening-associated Aux/IAA (SlIAA3) indicates that physical interaction between these two proteins would be important in regulation of some aspects of ripening (Chaabouni et al. 2009). Further positive interaction between SlARF7A and SlIAA8 indicates that these two proteins are required in regulation of important fruit developmental aspects just before ripening, as both these genes display moderate expression at mature green stages in fruits. Similarly, interaction of SlARF6A with both ripening-associated SlIAA3 and other SlIAAs, with high expression in vegetative tissues, further supports our hypothesis that both ARF and Aux/IAA proteins are involved in the regulation of various aspects of tomato development. Moreover, higher interacting potential of an activator (SlARF6A) in comparison with a repressor (SlARF2A) is in agreement with the previous report published by Shen et al (2010). The poor affinity of ARFs, which function as repressors, to interact with Aux/IAAs has been highlighted in Arabidopsis (Piya et al. 2014). However, the interaction between these two class of proteins appear to be complex as one repressor (AtARF4) was reported to interact with almost all Arabidopsis Aux/IAAs. Interaction of AtARF17, which lacks the CTD, with at least nine Aux/IAA proteins in Arabidopsis further indicates that protein structure or regions other than CTD of ARFs may influence ARF-Aux/IAA interactions. Shen et al. (2010) have reported that the capacity of ARFs to interact with Aux/IAA proteins can differ even between full length and truncated ARF protein. Altogether, these observations suggest that ARF-Aux/IAA interactions are complex and can also vary in different plant species. No interaction between SlARF6A and SlIAA8 and SlARF6A and SlIAA12 even though they are co-localized and presence of SlARF6A in such cases in nucleus is surprising; however, these results provide support to our PPI results and indicate that every pair of co-localized proteins do not necessarily interact whereas it is very likely that the proteins which are co-localized and have shown ≥9 % FRET would be interacting to each other. The other interesting findings were interaction of SlIAA15 and SlIAA16 with multiple ARF proteins and in some cases extranuclear localization of ARF proteins along with their interacting Aux/IAA partners. However, the reason behind such observations remains unclear and needs further experimentation to identify the underlying mechanism.
Genomic Sequences of SlARF and SlIAA Genes Show High Structural Variations in Most Wild Relatives
Huge diversity in the genomic sequences of all SlARF and SlIAA loci present in wild relatives was observed. Our results demonstrate that cultivated accessions harbor very less structural variants (<1 variant/kb) while closest ancestors of the cultivated tomato, S. pimpinellifolium and S. galapagense, harbor modest number of structural variations (<5 variant/kb). The fact that both S. lycopersicum and S. galapagense inhabit Galapagos islands and have experienced strong genetic bottlenecks during domestication and island colonization, respectively, our observation that S. galapagense has lesser structural variation than S. pimpinellifolium, provides support to the previous findings and reveals that S. pimpinellifolium is a more distant relative of cultivated tomato (Table 3) (Koenig et al. 2013; Nuez et al. 2004). Collectively, S. peruvianum, a green-fruited species, exhibited highest structural variation and was found to be the most diverged in our analysis which is in concordance with the earlier observations where this species has been suggested to be ancestral to all other wild tomatoes (Peralta et al. 2005). Though still unexplored, the natural allelic variations identified in different wild relatives for SlARF and SlIAA genes might be responsible for either change in their expression levels/patterns or protein sequence, which could have contributed to their successful in-habitation in different environments. Several lines of recent evidence have demonstrated that hundreds of candidate genes have either evolved to cause new protein structure or are deferentially expressed in tomato wild relatives in response to natural selection (Chitwood et al. 2013; Koenig et al. 2013; TGC 2012). We also observed altered expression levels in SlARF and SlIAA genes in S. pimpinellifolium at fruit ripening stages in comparison with that of cultivated tomato. Based on this information, we speculate that since tomato wild relatives have differences in morphological attributes such as plant height, leaf shape, fruit size, fruit weight etc. and auxin are known to regulate such attributes across plant species, the existing natural alleles of SlARF and SlIAA genes in different genetic background could have performed important specialized/modified functions which might have helped survival of plants during evolution
Conclusion
In conclusion, we identified members of ARF and Aux/IAA gene families and highlighted both conserved and distinct spatio-temporal transcript accumulation patterns existing for many ARF and Aux/IAAhomeologs and orthologs in Solanaceae. Identification of many ARF and Aux/IAA genes with similar/dissimilar expression profiles in tomato and pepper fruits is especially interesting as the conservation and divergence of the transcription of these genes and their interactions may contribute to quantitative and qualitative differences associated with the specific ripening program in climacteric and non-climacteric fruits, respectively. Our study provides evidence for the role of ARFs and Aux/IAAs during environmental stress conditions in Solanaceae. Furthermore, the distinct morphological features exhibited by tomato wild relatives, their capability to tolerate unfavorable environmental conditions, presence of a number of natural structural alleles in the ARF and Aux/IAA genomic sequences and altered expression of many of these genes in one of the tomato wild relative suggests that these genes might have actively participated in the survival and evolution of plants. The results herein clearly suggest that in addition to their structural variation and multilevel transcriptional and post-transcriptional regulation, these genes achieve functional diversity by changing their interacting partners as well. Altogether, this work further bridges the knowledge of ARFs and Aux/IAAs in Solanaceae and contributes towards improved understanding of their plausible role during plant development.
References
Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111:9–17
Abel S, Oeller PW, Theologis A (1994) Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci U S A 91:326–330
Abel S, Nguyen MD, Chow W, Theologis A (1995) ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin. J Biol Chem 270:19093–19099
Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M (2012) Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53:659–672
Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488. doi:10.1007/s11103-008-9435-0
Bassa C, Mila I, Bouzayen M, Audran-Delalande C (2012) Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol 53:1583–1595. doi:10.1093/pcp/pcs101
Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29:325–332
Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25:1523–1530. doi:10.1094/MPMI-06-12-0148-TA
Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, Frasse P, Latche A, Pech JC, Bouzayen M (2009) Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot 60:1349–1362. doi:10.1093/jxb/erp009
Chapman EJ, Estelle M (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43:265–285. doi:10.1146/annurev-genet-102108-134148
Chaw SM, Chang CC, Chen HL, Li WH (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58:424–441. doi:10.1007/s00239-003-2564-9
Chitwood DH, Maloof JN, Sinha NR (2013) Dynamic transcriptomic profiles between tomato and a wild relative reflect distinct developmental architectures. Plant Physiol 162:537–552. doi:10.1104/pp. 112.213546
Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol 168:241–252. doi:10.1111/j.1469-8137.2005.01480.x
de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57:160–170. doi:10.1111/j.1365-313X.2008.03671.x
Deng W, Yang Y, Ren Z, Audran-Delalande C, Mila I, Wang X, Song H, Hu Y, Bouzayen M, Li Z (2012) The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytol 194:379–390. doi:10.1111/j.1469-8137.2012.04053.x
Dharmasiri N, Dharmasiri S, Estelle M (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445. doi:10.1038/nature03543
Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9:109–119. doi:10.1016/j.devcel.2005.05.014
Ghanashyam C, Jain M (2009) Role of auxin-responsive genes in biotic stress responses. Plant Signal Behav 4:846–848
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414:271–276. doi:10.1038/3510450035104500
Guillon F, Philippe S, Bouchet B, Devaux M-F, Frasse P, Jones B, Bouzayen M, Lahaye M (2008) Down-regulation of an Auxin Response Factor in the tomato induces modification of fine pectin structure and tissue architecture. J Exp Bot 59:273–288
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385
Halliday KJ, Martinez-Garcia JF, Josse EM (2009) Integration of light and auxin signaling. Cold Spring Harbor Perspect Biol 1:a001586. doi:10.1101/cshperspect.a001586
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6:47–59. doi:10.1007/s10142-005-0005-0
Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latche A, Pech JC, Bouzayen M (2002) Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J 32:603–613
Kalluri UC, Difazio SP, Brunner AM, Tuskan GA (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7:59
Kenworthy AK (2001) Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24:289–296. doi:10.1006/meth.2001.1189
Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435:446–451
Kim S, Park M, Yeom SI et al (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46:270–278. doi:10.1038/ng.2877
Kloosterman B, Visser RG, Bachem CW (2006) Isolation and characterization of a novel potato Auxin/Indole-3-Acetic Acid family member (StIAA2) that is involved in petiole hyponasty and shoot morphogenesis. Plant Physiol Biochem 44:766–775. doi:10.1016/j.plaphy.2006.10.026
Koenig D, Jimenez-Gomez JM, Kimura S, Fulop D, Chitwood DH, Headland LR, Kumar R, Covington MF, Devisetty UK, Tat AV, Tohge T, Bolger A, Schneeberger K, Ossowski S, Lanz C, Xiong G, Taylor-Teeples M, Brady SM, Pauly M, Weigel D, Usadel B, Fernie AR, Peng J, Sinha NR, Maloof JN (2013) Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci U S A 110:2655–2662. doi:10.1073/pnas.1309606110
Kumar R, Tyagi AK, Sharma AK (2011) Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol Genet Genomics 285:245–260. doi:10.1007/s00438-011-0602-7
Kumar R, Agarwal P, Tyagi AK, Sharma AK (2012a) Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol Genet Genomics 287:221–235
Kumar R, Sharma MK, Kapoor S, Tyagi AK, Sharma AK (2012b) Transcriptome analysis of rin mutant fruit and in silico analysis of promoters of differentially regulated genes provides insight into LeMADS-RIN-regulated ethylene-dependent as well as ethylene-independent aspects of ripening in tomato. Mol Genet Genomics 287:189–203. doi:10.1007/s00438-011-0671-7
Kumar R, Khurana A, Sharma AK (2014) Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 65(16):4561–4575
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49:387–400
Liu N, Wu S, Van Houten J, Wang Y, Ding B, Fei Z, Clarke TH, Reed JW, van der Knaap E (2014) Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J Exp Bot 65(9):2507–2520
Lleres D, Swift S, Lamond AI (2007) Detecting protein-protein interactions in vivo with FRET using multiphoton fluorescence lifetime imaging microscopy (FLIM). Current protocols in cytometry / editorial board, J Paul Robinson, managing editor Chapter 12:Unit12 10. doi:10.1002/0471142956.cy1210s42
Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, Ecker JR, Reed JW (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132:4107–4118
Nickrent DL, Parkinson CL, Palmer JD, Duff RJ (2000) Multigene phylogeny of land plants with special reference to bryophytes and the earliest land plants. Mol Biol Evol 17:1885–1895
Nuez F, Prohens J, Blanca JM (2004) Relationships, origin, and diversity of Galapagos tomatoes: implications for the conservation of natural populations. Am J Bot 91:86–99. doi:10.3732/ajb.91.1.86
Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463
Paponov IA, Teale W, Lang D, Paponov M, Reski R, Rensing SA, Palme K (2009) The evolution of nuclear auxin signalling. BMC Evol Biol 9:126. doi:10.1186/1471-2148-9-126
Peralta IE, Knapp S, Spooner DM (2005) New species of wild tomatoes (Solanum Section Lycopersicon: Solanaceae) from Northern Peru. Syst Bot 30:424–434
Piya S, Shrestha SK, Binder B, Stewart CN Jr, Hewezi T (2014) Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5:744
Rahman A (2013) Auxin: a regulator of cold stress response. Physiol Plant 147:28–35. doi:10.1111/j.1399-3054.2012.01617.x
Sagar M, Chervin C, Mila I, Hao Y, Roustan JP, Benichou M, Gibon Y, Biais B, Maury P, Latche A, Pech JC, Bouzayen M, Zouine M (2013) SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol 161:1362–1374. doi:10.1104/pp. 113.213843
Salinas M, Xing S, Hohmann S, Berndtgen R, Huijser P (2012) Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 235:1171–1184. doi:10.1007/s00425-011-1565-y
Sharma MK, Kumar R, Solanke AU, Sharma R, Tyagi AK, Sharma AK (2010) Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol Genet Genomics 284:455–475. doi:10.1007/s00438-010-0580-1
Shen C, Bai Y, Wang S, Zhang S, Wu Y, Chen M, Jiang D, Qi Y (2010) Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J 277:2954–2969. doi:10.1111/j.1742-4658.2010.07706.x
Song Y, Wang L, Xiong L (2009) Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229:577–591. doi:10.1007/s00425-008-0853-7
Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384–1386. doi:10.1126/science.1151461
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
TGC (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641. doi:10.1038/nature11119
Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics/editoral board, Andreas D Baxevanis Chapter 2:Unit 2 3. doi:10.1002/0471250953.bi0203s00
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543
Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276:1865–1868
Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C (2008) Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 177:60–76
Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech JC, Bouzayen M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17:2676–2692
Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17:1784–1790. doi:10.1016/j.cub.2007.09.025
Wang Y, Diehl A, Wu F, Vrebalov J, Giovannoni J, Siepel A, Tanksley SD (2008) Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae. Genetics 180:391–408
Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latche A, Pech JC, Fernie AR, Bouzayen M (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21:1428–1452
Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43:118–130. doi:10.1111/j.1365-313X.2005.02432.x
Wu F, Tanksley SD (2010) Chromosomal evolution in the plant family Solanaceae. BMC Genomics 11:182
Wu J, Wang F, Cheng L, Kong F, Peng Z, Liu S, Yu X, Lu G (2011) Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep 30:2059–2073. doi:10.1007/s00299-011-1113-z
Wu J, Peng Z, Liu S, He Y, Cheng L, Kong F, Wang J, Lu G (2012) Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol Genet Genomics 287:295–211
Xu X, Pan S, Cheng S et al (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195. doi:10.1038/nature10158
Yuan H, Zhao K, Lei H, Shen X, Liu Y, Liao X, Li T (2013) Genome-wide analysis of the GH3 family in apple (Malus x domestica). BMC Genomics 14:297. doi:10.1186/1471-2164-14-297
Zouine M, Fu Y, Chateigner-Boutin AL, Mila I, Frasse P, Wang H, Audran C, Roustan JP, Bouzayen M (2014) Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS One 9:e84203. doi:10.1371/journal.pone.0084203
Acknowledgments
This work was financially supported by grants received from the Department of Biotechnology, Government of India. RK, AP, and PA acknowledge DST, CSIR, and UGC, respectively, for the fellowship granted during their tenure as research fellows. RK is grateful to DST for INSPIRE-Faculty Award (grant number IFA-LSPA-15) and to CSIR for Travel Grant (TG/8137/HRD). Authors also acknowledge the Solanaceae Genomics Network for the use of Tomato BAC Sequence Database to retrieve putative promoter sequences. We are grateful to 150 tomato genome consortium for sequence data of wild relatives.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, R., Agarwal, P., Pareek, A. et al. Genomic Survey, Gene Expression, and Interaction Analysis Suggest Diverse Roles of ARF and Aux/IAA Proteins in Solanaceae. Plant Mol Biol Rep 33, 1552–1572 (2015). https://doi.org/10.1007/s11105-015-0856-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0856-z