Abstract
Auxin plays key roles in a wide variety of plant activities, including embryo development, leaf formation, phototropism, fruit development and root initiation and development. Auxin/indoleacetic acid (Aux/IAA) genes, encoding short-lived nuclear proteins, are key regulators in the auxin transduction pathway. But how they work is still unknown. In order to conduct a systematic analysis of this gene family in Solanaceae species, a genome-wide search for the homologues of auxin response genes was carried out. Here, 26 and 27 non redundant AUX/IAAs were identified in tomato and potato, respectively. Using tomato as a model, a comprehensive overview of SlIAA gene family is presented, including the gene structures, phylogeny, chromosome locations, conserved motifs and cis-elements in promoter sequences. A phylogenetic tree generated from alignments of the predicted protein sequences of 31 OsIAAs, 29 AtIAAs, 31 ZmIAAs, and 26 SlIAAs revealed that these IAAs were clustered into three major groups and ten subgroups. Among them, seven subgroups were present in both monocot and dicot species, which indicated that the major functional diversification within the IAA family predated the monocot/dicot divergence. In contrast, group C and some other subgroups seemed to be species-specific. Quantitative real-time PCR (qRT-PCR) analysis showed that 19 of the 26 SlIAA genes could be detected in all tomato organs/tissues, however, seven of them were specifically expressed in some of tomato tissues. The transcript abundance of 17 SlIAA genes were increased within a few hours when the seedlings were treated with exogenous IAA. However, those of other six SlIAAs were decreased. The results of stress treatments showed that most SIIAA family genes responded to at least one of the three stress treatments, however, they exhibited diverse expression levels under different abiotic stress conditions in tomato seedlings. SlIAA20, SlIAA21 and SlIAA22 were not significantly influenced by stress treatments even though at least one stress-related cis-element was identified in their promoter regions. In conclusion, our comparative analysis provides an insight into the evolution and expression patterns in various tissues and in response to auxin or stresses of the Aux/IAA family members in tomato, which will provide a very useful reference for cloning and functional analysis of each member of AUX/IAA gene family in Solanaceae crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Auxin, as one of the most important hormones, plays a key role in many processes of plant development, including embryo, root, flower and fruit development. AUX/IAA family genes are early auxin response genes that encode short-lived nuclear proteins (Abel and Theologis 1995; Quint and Gray 2006). Increasing evidences emerge that AUX/IAA genes act as transcription repressors in auxin signal transduction pathway by dimerizing with auxin response factor (ARF) (Leyser 2002). Most Aux/IAA proteins contain four highly conserved domains, called domain I, II, III and IV (Abel and Theologis 1995; Tiwari et al. 2001). Domain I, represented by an “LxLxL” motif (Tiwari et al. 2004), is a repression domain that can interact with the TOPLESS (TPL) co-repressor (Szemenyei et al. 2008). Domain II is required for auxin-regulated signaling by interacting with a component of the ubiquitin–proteasome protein (TIR1) degradation pathway (Dharmasiri et al. 2005), and this interaction is abolished by mutations within motif II (Gray et al. 2001). Auxin and Aux/IAA bind to the same TIR1 pocket. Auxin might act as a “molecular glue”, increasing the affinity of the two kinds of proteins (AUX/IAAs and TIR1) by simultaneously interacting in a cavity at the protein interface (Tan et al. 2007; Hayashi et al. 2008). Domains III and IV are responsible for the homo- and hetero-dimerization among Aux/IAA family members and between the Aux/IAA proteins, and auxin response factors (ARFs) (Kim et al. 1997; Ulmasov et al. 1997a; Ouellet et al. 2001; Hardtke et al. 2004). Domain III in Aux/IAAs might be sufficient for dimerization by itself, but domain IV which contains a dimerization region and a functional nuclear localization signal (NLS) sequence (Reed 2001) is also thought to be responsible for the dimerization. Under low auxin concentration, ARFs are thought to be inhibited by dimerize with the Aux/IAAs via domains III and IV that are conserved between the two protein families (Ulmasov et al. 1997b; Hagen and Guilfoyle 2002). Elevated auxin concentration releases ARFs from repressor heterodimer by promoting the degradation of Aux/IAA proteins through the ubiquitin–proteasome protein (TIR1) pathway (Tiwari et al. 2003; Berleth et al. 2004; Dharmasiri et al. 2005).
AUX/IAA family genes play important roles in many aspects of plant development. In Arabidopsis, an auxin-resistant mutant, iaa28-1, is insensitive to auxin, cytokinin, and ethylene. Rogg et al. (2001) proved that iaa28-1 might act as transcription repressor in promoting lateral root initiation in response to auxin signals. SLR/IAA14 was regarded as a fundamental regulator in lateral root formation because its mutant completely lacked lateral roots (Fukaki et al. 2002). In tomato, the down-regulation of Sl-IAA3 resulted in the alteration of several auxin-related vegetative growth phenotypes, such as apical dominance, apical hook curvature and petiole epinasty (Chaabouni et al. 2009). When tomato SlIAA9 was down-regulated, the compound leaves were replaced by the simple leaves, and the order of fruit development was also reversed and the ovary began to develop before fertilization and produce the parthenocarpic fruit (Wang et al. 2005, 2009). In potato, down-regulation of StIAA2 resulted in an increased plant height petiole hyponasty and extreme curvature of growing leaf primordia in the shoot apex (Kloosterman et al. 2006).
Aux/IAA gene was first isolated in soybean (Walker and Key 1982). Subsequently, many Aux/IAAs genes have been characterized based on the analysis of gain-of-function mutants in Arabidopsis (Park et al. 2002; Yang et al. 2004), mung bean (Yamamoto 1994), rice (Thakur et al. 2001; Nakamura et al. 2006) and Populus (Kalluri et al. 2007). In Arabidopsis, 29 Aux/IAA genes were distributed in 5 different chromosomes (Liscum and Reed 2002). In rice, a total of 31 AUX/IAAs distributed on 10 of the 12 rice chromosomes have been reported (Jain et al. 2006). In maize, a total of 31 AUX/IAAs genes were also identified, which are distributed in all the 10 chromosomes except chromosome 2 (Wang et al. 2010a, b). In tomato, only three complete SlIAAs sequences (SlIAA3, SlIAA4 and SlIAA9) and eight partial sequences (SlIAA1-2, SlIAA5-8, SlIAA10, and SlIAA11) shown to be homologous to AtIAAs were identified (Nebenführ et al. 2000; Wang et al. 2005; Chaabouni et al. 2009). Until now, 17 IAA genes in potato genome and several IAA genes in other Solanaceae species have been reported (Cle’ment et al. 2006; Kloosterman et al. 2006; Terrile et al. 2010; Zanetti et al. 2003). However, to our knowledge, no systematic investigations of IAA gene family have been conducted in Solanaceae species. Tomato, as the representation of Solanaceae plants, is not only one of the most important vegetables but also is considered one of the model dicot plants for fruit development. The Genome Sequencing Project for tomato genome has been completed lately (http://solgenomics.net/organism/Solanum_lycopersicum/genome). Recently, the potato genome was also released (Xu et al. 2011). In the present study, taking advantage of the available SGN database, we carry out a genome-wide search for the homologues of AUX/IAA family genes in Solanaceae crops. As a result, we identified 26 and 27 putative genes with IAA domains in tomato and potato genome, respectively. The detailed information on the genomic structures, chromosomal locations, sequence homologies and expression patterns of tomato IAA genes was presented. In addition, the phylogenetic relationships among IAA genes in Arabidopsis, tomato, rice and maize were also compared. Furthermore, the different expression patterns during flower and fruit development and in response to various abiotic stress conditions in tomato plants were determined for each SlIAA gene using quantitative real-time PCR (qRT-PCR) analysis. The data would facilitate future studies on elucidating the biological functions of AUX/IAA in Solanaceae crops.
Materials and methods
Searching for Aux/IAA family genes
To find all IAA genes in Arabidopsis (AtIAAs), rice (OsIAAs) and maize (ZmIAAs),“early auxin-responsive Aux/IAA” was used as a query to search the protein and nucleotide databases of NCBI (The National Center for Biotechnology Information) and the matching genes were confirmed by previous reports (Nebenführ et al. 2000; Wang et al. 2010a, b).
To find previously identified and potential IAA family genes in Solanaceae species, multiple database searches were performed. First, “early auxin-responsive Aux/IAA” was used as a query to search the SGN database (http://solgenomics.net). In tomato, three formerly known SlIAA family genes (SlIAA3, SlIAA4 and SlIAA9) with full-length cDNA sequences and eight other SlIAA genes with partial sequences (SlIAA1-2, SlIAA5-8, SlIAA10, and SlIAA11) were identified. Similarly, four formerly known IAA genes (StIAA1-4) in potato were found. To find other potential IAAs in tomato and potato, we initially surveyed the tomato and potato genome database of SGN by TBLASTN (Search translated nucleotide database using a protein query) using the whole amino acid sequences of the conserved IAA domains from all the known IAA families (including AtIAAs, OsIAAs, ZmIAAs, and PoptrIAA) as queries. Based on the combined results from all above searches, we finally identified all members of tomato and potato IAA family from the currently available genomic databases. After searching for IAA genes, bioinformatics tools, such as DNASTAR and FGENESH (http://linux1.softberry.com/berry) were used to analyze and predict those unknown IAAs.
Isolation of the open reading frame (ORF) cDNA sequences
To identify the homologue unigenes or ESTs for the AUX/IAA family genes in tomato, the available SGN database was searched, and then 16 SlIAAs ORF sequences were found in the unigenes. Since only partial cDNAs sequences of SlIAA2, 11, 18, 20 and 26 were found existing in the unigene or EST database of SGN database and the full-length of those five genes and other two SlIAA genes (SlIAA15 and SlIAA17) without homologous EST or unigene in the SGN database were identified using BLASTN against ITAG Release 2 predicted CDS (SL2.31). Since SlIAA12, SlIAA13, and SlIAA16 showed no significant homology to any known sequences using the BLAST approaches, they were amplified through RT-PCR using the primers designed by FGENESH (Table S1). Total RNA was extracted from tomato variety “Micro-Tom” young ovaries using TRIZOL reagent (Invitrogen, Germany) according to the manufacturer’s instructions. After DNAse (Qiagen, Germany) treatment, RNA was reverse transcribed to cDNA using the Improm-TM Reverse Transcription system (Promega, Madison, USA) following the manufacture’s protocol. RT-PCR was performed as described in our previous study (Wu et al. 2011). In potato, homologue unigenes or ESTs were found from potato unigene database by BLASTN.
Mapping SlIAA genes on chromosomes
To determine the location of SlIAA genes on tomato chromosomes, each above SlIAA cDNA sequence was further used as query sequence for the BLASTN search against SGN tomato whole genome scaffolds data (2.30) (http://www.sgn.cornell.edu/tools/blast/).
Multiple-sequence alignments and phylogenetic analysis
All the identified SlIAA DNA sequences were analyzed by DNASTAR software and the net service ExPASy Proteomics Server (http://ca.expasy.org). All the conserved domains were investigated by multiple alignment analyses using ClustalX v1.81 (Hompson et al. 1997). Phylogenetic analysis was performed using MEGA 4.1 program by the neighbor-joining (NJ) method (Saitou and Nei 1987). MEME utility was used to display motifs of Aux/IAA proteins from tomato, maize, rice and Arabidopsis (http://meme.nbcr.net) (Bailey et al. 2009). Parameters were set as the following: (i) the occurrence of a single motif distributed among the sequences was zero or one per sequence; (ii) the motif width ranged from 10 to 300 amino acids; (iii) the maximum number of motifs to find was five. Other parameters were defaulted.
Promoter regions analysis of SlIAA genes
To investigate cis-elements in promoter sequences of tomato Aux/IAA genes, 2,000 bp of genomic sequences upstream of the initiation codon from the SGN database were analyzed for cis regulatory elements (Suppl Fig. 1). Only 1,098 bp and 896 bp for SlIAA18 and SlIAA19, respectively were used for analysis because of the unavailability of their 5′-upstream full DNA sequences in SGN database. The PLACE website (http://www.dna.affrc.go.jp/PLACE/) was applied to identify putative cis-regulatory elements along the promoter sequences of each SlIAA family gene (Higo et al. 1999).
Plant growth and treatments
Tomato (S. lycopersicum L. cv. Micro-Tom) seeds were obtained from Tomato Genetics Resource Center (University of California, Davis, USA). All the plants were grown in a temperature-controlled chamber until flowering at the experimental farm in Zhejiang University. To analyze tissue or organ-specific expression, leaves, stems, roots, and flower buds were collected from flowering plants, meanwhile the various floral organs (sepal, petal, stamen, and ovary) were collected from the flower buds (about 3 days before opening). To analyze the expression pattern during early flower developmental stages, flower buds were collected at three stages of early floral development (before flowering), which was roughly defined by the length of flower buds as follows: stage I: 3–4 mm, stage II: 5–6 mm, and stage III at 7–8 mm (Brukhin et al. 2003). In addition, the ovaries were sampled at 0, 3, 6, and 9 days after the flower fully opened.
For stress treatments, ‘Micro-Tom’ tomato plants were grown in a growth chamber at 28 ± 1°C with a photoperiod of 14 h light and 10 h dark. The 3-week-old seedlings with three fully opened leaves were selected for different stress treatments. For heat stress (HS) treatment, the seedlings were treated with 42 ± 1°C for 1 h. Then the leaves were sampled immediately. Drought stress was initiated by withholding water supply to 3-week-old seedlings after seedlings were fully watered. On the 6th day after stress, the leaves were harvested when some of them started to curl due to the drought stress. Salt stress was performed by the addition of 200 mM sodium chloride to the planter box and the seedlings were sampled after 6 h. For IAA treatment, 3-week-old seedlings were sprayed with 100 mM IAA, and then the leaves were sampled at 0, 6, and 12 h after spraying. The seedlings without treatment grown at 28 ± 1°C with normal irrigation were used as control (CK). Each above experiment was repeated two times. Fifteen seedlings were used in each treatment in each replication. All the samples were stored at −75°C.
QRT-PCR analysis
Total RNA and the first cDNA strand were prepared as described above. QRT-PCR was carried out using the primer pairs listed in Table S2. Specificity of each primer to its corresponding gene was checked using the BLASTN program of the NCBI. One milligram aliquots of cDNA was subjected to each qRT-PCR reaction in a final volume of 20 μl containing 12.5 μl SYBR Green Master Mix Reagent (Takara, Japan) and specific primers (3 pmol). QRT-PCR reactions were carried out in a StepOne real-time PCR machine (Applied Biosystems, USA) as described by Wu et al. (2011). Two biological replicas were performed with three technical replicates for each sample. To normalize the total amount of cDNA present in each reaction, the Ubi3 gene (accession number X58253) was co-amplified as an endogenous control for calibration of relative expression. The comparative Ct method (ΔΔCT method) of relative gene quantification recommended by Applied Biosystems (CA, USA) was used to calculate the expression levels of different treatments.
Results
Identification and isolation of IAA family genes in Solanaceae species
To identify the IAA family genes in Solanaceae species, BLAST searches of the SGN database were performed using the whole amino acid sequences of all four conserved IAA domains of the Arabidopsis, rice, Populus and maize protein as a query sequence. A total of 40 genomic DNA sequences and 63 unigenes in tomato were obtained from tomato genome database and unigene database using the TBLASTN program with an e value cutoff of 1e −1. Meanwhile, 45 candidates in potato were also found by TBLASTN against potato genome sequence with an e value cut-off 1e −1 and 35 homologous DNA sequences were indentified in tobacco.
Different sequences presenting on the same or overlapping contigs in different databases were identified and removed to obtain a set of nonredundant IAA sequences. All predicted sequences were confirmed by FGENESH (http://www.softberry.com/berry.phtml?topic=fgenesh). These predicted amino acid sequences were analyzed by ExPASy Proteomics server to find their conserved domains, followed by homologous alignment with known IAA genes in Solanaceae species. The overall revealed that the tomato genomes appeared to have 26 members which contained the IAA domains. In addition to the 11 previously known SlIAA genes (Nebenführ et al. 2000; Wang et al. 2005), 15 other putative novel SlIAA genes were found (Table 1). Meanwhile, a total of 27 StIAA family genes in potato and 18 NtIAA family genes in tobacco were identified (Table 2; Table S3).
The open reading frames (ORFs) of 16 SlIAAs were identified from the 23 unigenes. Meanwhile, the full-length cDNA sequences of SlIAA12, SlIAA13, and SlIAA16 were isolated by PCR-based methods. The primers for PCR reaction are listed in Table S1. The ORF sequences of the other seven SlIAA genes were obtained based on ITAG Release 2 predicted CDS (SL2.31) of SGN database. The peptide sequences of all 26 SlIAAs were listed in Suppl Fig. 2. All the results were verified by BLASTN against the ITAG Release 2 predicted CDS (SL2.31) of SGN database. The ORF length of 26 SlIAAs genes varied from 228 bp (SlIAA13) to 1,050 bp (SlIAA4), encoding polypeptides of 75–349 aa, with a predicted molecular mass range of 8.6–37.4 kDa. The theoretical pI ranged from 4.39 to 9.30 (Table 1), similar with the IAAs polypeptides previously determined in other plants (Nebenführ et al. 2000; Wang et al. 2010a, b).The whole-length ORFs of 11 StIAAs were identified from the 11 unigenes in potato unigene database, the CDS of other StIAAs were predicted from PGSC DM v3.4 CDS sequences (Table 2).
Chromosomal locations of SlIAAs
All SlIAA clones from tomato genome have been anchored to tomato chromosomes. The chromosomal locations and transcription directions of 26 SlIAA genes were demonstrated using BLASTN analysis on Tomato WGS Chromosomes (Fig. 1). Like in maize and rice, tomato SlIAA family genes were distributed over 9 of the 12 tomato genomes, except chromosomes 2, 10 and 11. The number of SlIAAs genes per chromosome ranged from one to six. Six SlIAA genes were anchored on chromosomes 3 and 6. Five SlIAA genes were anchored to chromosome 9, while three of them were clustered on the same region with the different transcriptional orientation. Two SlIAA genes were found on chromosomes 4, 7 and 12 each with the same transcriptional orientation. Chromosomes 1, 5, and 8 only carried one SlIAA gene each (Table 1; Fig. 1).
It is worth mentioning that the nomenclature system for SlIAAs is used in the present study. Since sequence analysis indicated that the similarity between the SlIAA in tomato with AtIAAs in Arabidopsis was quite low, it is difficult to assign all SlIAA names based on their homolog proteins in Arabidopsis. So the established names were used for 11 previously known genes or sequences in Genbank (SlIAA1-11). Other SlIAA genes (SlIAA12-26) were named according to their position from the top to the bottom on the tomato chromosomes 1–12. All the genes in other Solanaceae crops were named according to their homology with SlIAAs (Suppl Fig. 11).
Sequence analysis of IAAs proteins
All the tomato SlIAAs protein sequences were found to contain domain III and IV which act as C-terminal dimerization domains, mediating homodimerization and heterodimerization among Aux/IAA family members and between the Aux/IAA proteins and auxin response factors (ARFs). Among 26 SlIAAs, 23 proteins contain domain I and II, only three including SlIAA13, SlIAA16 and SlIAA20, lack these two domains (Table 1; Fig. 2). In potato, all the IAA proteins, except StIAA13, StIAA15, StIAA16, StIAA18, StIAA20 and StIAA27, contain all four conserved domains (Table 2; Suppl Fig. 3). Putative domain of NtIAA family genes in tobacco were also analyzed (Table S3; Suppl Fig. 4).
a Alignment of tomato Aux/IAA proteins obtained with the ClustalX program. The height of the bars indicates the number of identical residues per position. b Multiple alignments of the domains I–IV of the tomato Aux/IAA proteins obtained with ClustalX and manual correction. Black and light gray shading indicates identical and conversed amino acid residues, respectively. Conserved domains are also underlined and correspond to part (a). The LxLxLx and LxLxLxLxLx motif were also marked
Gene structure and phylogenetic analysis
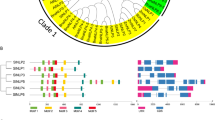
A comparison of the full-length cDNA sequences with the corresponding genomic DNA sequences revealed the numbers and positions of exons and introns for each individual SlIAA and StIAA gene. The coding sequences of all the SlIAAs and StIAAs except SlIAA13 and StIAA20 were disrupted by introns. The number of introns varied from 1 to 5 both in tomato and potato (Fig. 3). The IAA genes possessed a complex distribution of exons and introns both in tomato and potato. The IAA members displayed different structural pattern of exon–intron junctions even within the same phylogenetic subgroup.
a Left part illustrates the phylogenetic relationships among the tomato Aux/IAA proteins. b Left part illustrates the phylogenetic relationships among the patoto Aux/IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap supports from 1,000 replicates are indicated at each branch. Right part illustrates the exon–intron organization of corresponding IAA genes. The exons and introns are represented by black boxes and lines, respectively
There are two types of putative nuclear localization signals (NLS) detected in most of the identified Aux/IAA proteins. A bipartite NLS containing two stretches of K/R residues was found between a conserved basic doublet KR and basic amino acids in domain II, whereas a SV40-like NLS was located in domain III, consisting of one cluster of positively charged amino acid residues such as lysine (K) or/and arginine (R) (Fig. 2; Suppl Fig. 3; Suppl Fig. 4) (Raikhel 1992). These putative NLSs may direct Aux/IAA proteins to the nucleus.
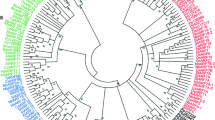
An unrooted phylogenetic tree was generated from the alignment of full-length protein sequences of all SlIAAs. The 26 SlIAA protein sequences could be divided into two major groups (group A and B) with well-supported bootstrap value, which was similar to rice (Jain et al. 2006) and Arabidopsis (Remington et al. 2004). Group A was further divided into five subgroups (Fig. 3a). Among them, subgroup A1 and A5 each contained seven members, but subgroup A2 and A6 only contained one IAA member each. Group B was further divided into three subgroups: B1, B2 and B3, which contained 3, 1 and 6 SlIAA proteins, respectively. Unlike in rice, where many sister pairs were found in 31 OsIAA proteins (Jain et al. 2006), the 26 SlIAA proteins only formed four sister pairs (Fig. 4a) with strong bootstrap support (>90%). All the 27 StIAAs formed two major groups (group A and B), containing 16 and 11 members, respectively. The phylogenetic relationships between the predicated StIAAs protein sequences were also analyzed. Similarly, group A and group B could be further divided into five and three subgroups, respectively (Fig. 3b).
Left part illustrates the phylogenetic relationships among tomato, rice, maize and Arabidopsis IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap supports from 1,000 replicates are indicated at each branch. Right part shows motif distribution in tomato (Sl), maize (Zm), rice (Os) and Arabidopsis (At) Aux/IAA proteins. Motifs of Aux/IAA proteins were investigated by MEME web server. Five motifs representing four domains I, II, III and IV were not observed in all Aux/IAA proteins. The heights of each box represent the conservation of each motif
Evolution relationships analysis that 117 IAA protein sequences including 29 AtIAAs, 31 OsIAAs, 31 ZmIAAs, and 26 SlIAAs (Fig. 4) fell into three broad groups, namely group A, B and C including 65, 46, and 6 IAA proteins, respectively. Group A was further divided into six subgroups A1, A2, A3, A4, A5, and A6, containing 12, 11, 6, 11, 14, and 11 members, respectively. Group B was further divided into three subgroups, namely B1, B2 and B3 (Fig. 4). This classification is consistent with previous analyses (Remington et al. 2004; Jain et al. 2006). All the IAAs proteins contained conserved motifs except SlIAA13, ZmIAA3, ZmIAA13, ZmIAA25 and ZmIAA30 (Fig. 4).
In this joint phylogenetic tree, a total of 21 sister pairs were found, including 2 SlIAA–SlIAA pairs, 4 ZmIAA–ZmIAA pairs, 7 AtIAA–AtIAA pairs, 2 OsIAA–OsIAA pairs, 4 OsIAA-ZmIAA pairs and 2 SlIAA-AtIAA pairs. Interestingly, subgroups A2, A5, A6, B1, B2 and B3 contained IAA genes from all the four species, but subgroup A3 only contained the IAAs from dicotyledon (Arabidopsis and tomato), meanwhile, all the IAA proteins except ZmIAA26 presented in subgroup A1 came from dicotyledon plants. By contraries, all IAA proteins in subgroup A4 and B2 except SlIAA14 were from monocot (rice or maize), while the group C, the most divergent part, only contained IAAs from maize (Fig. 4).
The motif distributions in maize, rice, tomato, and Arabidopsis Aux/IAA proteins were analyzed using Multiple Expectation Maximization for Motif Elicitation (MEME) tool. Four conserved domains of Aux/IAA proteins were divided into five motifs by MEME tool. Domain I, II and IV were represented by motif 4, 3 and 1, while domain III was constituted by motif 2 and 5 (Fig. 4; Suppl Fig. 5). The number of sites and e value for each motif were also presented in Suppl Fig. 5.
Cis-elements in promoter sequences of SlIAA genes
The investigation of 5′-upstream sequences of SlIAAs representing their promoter regions by PLACE and manual search revealed the presence of four types of cis-elements including seven auxin signaling transduction-related cis-element, 13 drought stress-related cis-element, one salt stress-related cis-element and one heat shock element (Table S4). All the locations of three AuxREs and other auxin signaling transduction-related cis-element were presented in Suppl Fig. 6. Three auxin-responsive elements (AuxREs)-S000026, S000234, and S000270, have been defined within upstream promoter regions of AUX/IAA genes. S000270 could be found in the promoter regions of 11 SlIAAs, while S000026 only in SlIAA5 and SlIAA25, S000234 in SlIAA14 and SlIAA21. However, no auxin signaling transduction-related cis-element was found in the 1,098 bp upstream region of SlIAA18.
Expression characterization of SlIAA genes
Most of the SlIAAs could be detected in root, stem, leaf, flower bud and ovary using qRT-PCR (Fig. 5). Some SlIAAs showed organ/tissue-specific expression pattern in tomato. SlIAA2, SlIAA12, SlIAA13, SlIAA15, SlIAA16, SlIAA20 and SlIAA25 were highly expressed in tomato roots, especially SlIAA16 and SlIAA20 exhibited root-specific expression in tomato. SlIAA4, SlIAA5, SlIAA6, SlIAA17, SlIAA21, and SlIAA26 exhibited a higher expression level in stem than in the other organs. SlIAA1, SlIAA7, SlIAA19 and SlIAA24 mRNA were highly expressed in leaves. Interestingly, SlIAA19 only expressed in leaf and root and SlIAA24 only in leaf and stem. SlIAA1 and SlIAA15 especially expressed in vegetative organs. In general, most of the SlIAAs exhibited relatively low expression level in reproductive organs, except that SlIAA8 and SlIAA18 highly expressed in ovary.
Expression profiles of all the 26 SlIAA genes in different tomato organs. QRT-PCR analyses of total RNA isolated from root (R), stem (S), leaf (L), buds (B), and ovary (O) were used to assess SlIAA transcript levels in flowering tomato plants. The data on represented mean ± SD normalized relative to the Ubi3 (accession number X58253) related protein transcript levels. All samples were run in triplicate and the entire assay was performed twice for each biological pool
The mRNA expression of most SlIAA genes could be detected in different tissues of the tomato flower (Suppl Fig. 7). In general, the transcript levels of most SlIAA genes were higher in petal than in other parts. However, higher mRNA levels of SlIAA3, SlIAA13, SlIAA14, SlIAA16, SlIAA18, and SlIAA19 were detected in stamen, while SlIAA20, SlIAA21 and SlIAA25 mRNA exhibited a higher transcript level in ovary than in other tissues. In contrast, only SlIAA24 and SlIAA26 exhibited relatively strong expression in sepal.
Most of the SlIAA family genes exhibited a similar expression pattern of mRNA accumulation during flower development (Suppl Fig. 8). However, SlIAA2, SlIAA22, SlIAA24 and SlIAA25 mRNA were markedly down-regulated at stage II and then up-regulated at stage III. Different expression patterns of SlIAA genes during the early development of tomato fruit were found using qRT-PCR analysis (Fig. 6). The relative mRNA levels of most SlIAA genes increased during flowering and reached the maximum value on the 3rd or 6th day after the flower opened (DAF), and finally drastically decreased again till the end of observation. However, the relative mRNA levels of SlIAA6, SlIAA11, SlIAA13, SlIAA15, SlIAA16, SlIAA18, SlIAA19, SlIAA21 and SlIAA25 continuously decreased during the early fruit development. Higher expression levels were observed in SlIAA5, SlIAA10 and SlIAA20 at the time of flower opening, then their mRNA levels were markedly decreased at 3 DAF, but increased at 6 DAF and drastically decreased again at 9 DAF. The expression level of SlIAA17 also decreased at 3 DAF, but it continuously increased during the early fruit development.
Expression profiles of all the 26 SlIAA genes during tomato early fruit developmental stages. QRT-PCR analyses were performed using RNA generated from tomato ovaries at different number of days (0, 3, 6, and 9 days) after the flower opening. For other details see Fig. 5
Expression of SlIAAs genes in response to IAA and stress treatments
QRT-PCR was performed with total RNA isolated from the tomato leaves treated with IAA. As expected, most of SlIAA genes were activated by IAA treatment (Fig. 7). The mRNA levels of these SlIAAs were increased from less than onefold (SlIAA14) to more than tenfolds (SlIAA16) at 6 h after the IAA treatment. Among them, most SlIAA genes were down-regulated at 12 h except SlIAA4, SlIAA8, SlIAA9, and SlIAA14. It is worth mentioning that SlIAA20, SlIAA21 and SlIAA22 were markedly down-regulated just after IAA treatment (Fig. 7; Table S5). SlIAA7 and SlIAA26 mRNA abundance was also slightly decreased in IAA-treated seedlings.
Expression profiles of all the 26 SlIAA genes in response to IAA treatment. QRT-PCR analyses were used to assess SlIAA transcript levels in the leaves sampled at 0, 6, and 12 h after spraying 100 mM IAA in 3-week tomato seedlings. For other details see Fig. 5
The SlIAA genes exhibited altered responses to salt, drought and heat stress. SlIAA20, SlIAA21 and SlIAA22 showed no significant changes after all stress treatment (Suppl Fig 9; Table S5). Most SIIAA mRNA transcripts were enhanced under the drought treatment, whereas SlIAA3, SlIAA11, and SlIAA14 were down-regulated after drought treatment. Salt stress treatment increased the transcript levels of 19 out of 26 SlIAA genes in tomato seedlings, especially SlIAA15 mRNA was increased even more than sixfolds. In contrast, SlIAA3, SlIAA11, and SlIAA14 were down-regulated, especially the transcript level of SlIAA11 was decreased over 46-fold subjected to salt treatment. Heat treatment significantly enhanced the accumulation of SlIAA6, SlIAA8, SlIAA15 and SlIAA16 mRNA over twofolds, but caused a decrease in the transcript level of SlIAA3, SlIAA11 and SlIAA14 by 12-, 4- and 2.4-fold, respectively. Other 14 SlIAA genes, including SlIAA2, SlIAA5, SlIAA9, SlIAA10, SlIAA12, and SlIAA18-26 seemed to be not significantly regulated by heat stress treatment (Suppl Fig. 9; Table S5).
Discussion
In this study, a comprehensive set of 26 and 27 non-redundant AUX/IAA genes were identified and characterized from the current version of the SGN database for tomato and potato genome, respectively. The number of SlIAAs and StIAAs members from tomato and potato is comparable to that of Arabidopsis (29), rice (31) and maize (31), although the genome size of tomato, potato, maize, Arabidopsis and rice is quite different. These partially accounts for the AUX/IAA conservation in these five species during the evolutionary process (Liscum and Reed 2002; Jain et al. 2006; Wang et al. 2010a, b). The phylogenetic analysis showed that subgroups A2, A5, A6, B1, B2 and B3 contained IAA genes from all four species, which implies that those genes originated prior to the divergence of monocots and dicots. IAAs in subgroup A1 and A3 were from dicot crops, while subgroup A4 only contained the IAAs from monocotyledon, which indicated that those proteins were either lost or evolved after the divergence of monocots and dicots, consequently, they might play an important role in the development of dicotyledonous or monocotyledonous plants. Interestingly, the SlIAA proteins in main group C all came from maize, indicating that they may play an important role in determining species-specific traits and functions. Furthermore, the phylogenetic analysis indicated that there were four sister pairs between OsIAAs and ZmIAAs and two sister pairs between SlIAAs and AtIAAs, but no sister pairs between dicotyledon and monocotyledon were found in the phylogenetic tree (Fig. 4), which is also consistent with the evolutionary relationships of IAA family genes among these four species.
Twenty of the 28 Aux/IAA loci formed 10 sister pairs in the neighbor-joining reconstructions, nine of which had strong bootstrap support (≥96% in all three trees) (Remington et al. 2004). In this study, six sister pairs of AtIAAs, five sister pairs of ZmIAAs, seven sister pairs and two triplets of OsIAAs were found (Suppl Fig. 12–14). However, only two sister pairs of SlIAAs and one sister pair of StIAAs were found (Fig. 3), indicating that AUX/IAA genes in tomato and potato may play non-redundant roles during plant development. As expectedly, SlIAA3, 23 and 24 distributed in chromosome 9 formed clusters, were comparative with their closely related genes SlIAA7, 10 and 17 with highly similar sequences, the clusters of in chromosome 6, which indicated that they might originate from local duplication events (Fig. 2). This pattern probably reflects the series of chromosomal and large segmental duplication events existing in tomato genome. Some ancient tandem duplications which preceded the divergence of some chromosomal segments were reported by Ku et al. (2000). However, although there are some other SlIAA paralogs displaying high levels of sequence similarity, they are distributed all across the genome. Conversely, some closely linked SlIAAs, such as SlIAA6, 8,11,14 on chromosome 3, SlIAA10 and SlIAA17 on chromosome 6, and SlIAA23 and SlIAA24 on chromosome 9 were not grouped in sister pairs. Consequently, we may conclude that the contribution of whole genome and chromosomal segment duplications in tomato was not as obvious as in other three species (Liscum and Reed 2002; Jain et al. 2006; Wang et al. 2010a, b).
Transcript abundance in particular organs at a given time is an important prerequisite to subsequent elucidation of the corresponding protein required for proper execution of developmental, metabolic and signaling processes. Virtually all 26 SlIAA genes were expressed in all organs/tissues analyzed, but their expression levels varied considerably. The mRNA levels of SlIAA12, SlIAA13, SlIAA15, SlIAA16, SlIAA20 and SlIAA25 in root were significantly higher than other organs (Fig. 5), implying that they might play an important role in the development of root. SlIAA4, SlIAA5, SlIAA6, SlIAA17, SlIAA21, and SlIAA26 might play a crucial role in stem due to the higher expression levels than the other organs (Fig. 5). Similarly, SlIAA1, SlIAA7, SlIAA19 and SlIAA24 mRNA might have important functions in leaf, and SlIAA11, SlIAA8 and SlIAA18 might affect the development of fruit (Fig. 5). Chaabouni et al. (2009) proved that the relative mRNA level of SlIAA13 in tomato stamen, leaf, and flower was much higher than root. Similar expression pattern was also found in the present study, although much lower in flower buds due to the different flower stage. When compared to the gene expression data of ARF family genes which are proved to interact with AUX/IAA (Kumar et al. 2011; Wang et al. 2007; Wu et al. 2011), Several SlIAA genes showed additional tissue-specific expression patterns suggesting the complexity of the Aux/IAA and ARF interactions (Jain et al. 2006; Wang et al. 2010a, b).
Aux/IAA genes encoding short-lived nuclear proteins are responsive primarily to auxin induction. The Aux/IAA genes in Arabidopsis, rice, maize and sorghum were induced by exogenous auxin, but displayed differential expression pattern (Yamamoto et al. 1992; Thakur et al. 2001). According to previous microarray data, AtIAA6 (At1g52830) and AtIAA3 (At1g04240) were up-regulated by IAA treatments (Nemhauser et al. 2004). IAA treatment up-regulated ten AUX/IAA genes in Arabidopsis (IAA1, 2, 3, 5, 6, 7, 11, 13, 19 and 26) (Goda et al. 2004). In sorghum, most SbIAA genes were slightly up-regulated under IAA treatment, especially in roots. But SbIAA3, SbIAA4, SbIAA5, SbIAA6, SbIAA14, SbIAA15, and SbIAA21 in leaves and SbIAA21 in roots were down-regulated under IAA treatment (Shibasaki et al. 2009). In tomato, SlIAA11, SlIAA15, SlIAA16, SlIAA17, SlIAA19 and SlIAA23 were up-regulated (over fourfolds), whereas SlIAA20, SlIAA21, and SlIAA22 were drastically down-regulated after IAA treatment in leaves (Fig. 6). Our promotor analysis identified seven auxin signaling transduction-related cis-elements presenting in the promoter regions of all the SlIAA genes except SlIAA18. The diversity of numbers and locations of their auxin signaling transduction-related cis-elements may partially account for the different expression patterns of SlIAAs under IAA treatment. However, although none of the auxin signaling transductions-related cis-elements were found in the promoter of SlIAA18 (Table S4), the relatively mRNA level of SlIAA18 increased after the IAA treatment (Fig. 7).
Interestingly, the investigation of conserved AUX/IAA domains indicated that three SlIAA genes (SlIAA13, SlIAA16 and SlIAA20) and six StIAA genes (StIAA13, StIAA15, StIAA16, StIAA18, StIAA20 and StIAA27) lacked domain I and domain II (Tables 1, 2; Fig. 2; Suppl Fig. 3). Domain I is a repression domain (Szemenyei et al. 2008), and domain II physically interacts with TIR1 which leading to the degradation of AUX/IAA proteins under a high level of auxin (Gray et al. 2001). It has been shown that the half-lives of these proteins which lack domain II are much longer than those of the canonical Aux/IAA proteins (Sato and Yamamoto 2008). It seems that these nine SlIAA genes should be insensitive to IAA treatment since they have longer half-lives than other SlIAAs proteins. However, SlIAA13 and SlIAA16 mRNA were both drastically up-regulated when exposed to exogenous IAA, especially SlIAA16 transcript level increased 9- to 15-fold at 6 and 12 h, respectively, after IAA treatment. On the other hand, SlIAA20 mRNA was found to be markedly down-regulated (23-fold at 12 h) after the IAA treatment. Similar results have been reported in previous researches where proved that OsIAA8, as a domain II-lacking IAA gene (Jain et al. 2006), was highly sensitive to IAA treatment (Song et al. 2009). So a possible unknown auxin-related pathway may exist in the degradation of the proteins encoded by these genes and the negative (SlIAA20) or positive (SlIAA13 and SlIAA16) regulations in the gene expression. These noncanonical Aux/IAA genes (IAA20, IAA30, and IAA31) could cause auxin-related aberrant phenotypes in Arabidopsis, which suggests that these noncanonial genes have potential functions in auxin signaling (Sato and Yamamoto 2008). Recently, Li et al. (2011) found that when the first Leu was replaced by Ala in the LxLxL motif of AtIAA3, AtIAA6 and AtIAA19, the “low-auxin” phenotypes were repressed and “high-auxin” phenotypes were activated. However, in addition to a single Leu-to-Ala substitution of AtIAA12 in LxLxLxLxL motif, a second and third Leu residues in the LxLxLxLxL motif should be replaced by Ala to active the “high-auxin” phenotypes. In tomato, most of the IAA genes contain the LxLxL motif (Fig. 2). However, AtIAA12, SlIAA20, 22 and 25 were found to contain the LxLxLxLxL motif (Fig. 2), indicating that these two motifs might play similar function in auxin signaling.
Extensive researches have showed that various environmental signals are integrated into changes in auxin homeostasis, redistribution, and signaling (Park et al. 2007; Shibasaki et al. 2009). There are increasing evidences that the auxin-response genes, auxin/indole-3-acetic acid (Aux/IAA), auxin-response factor (ARF), Gretchen Hagen 3 (GH3) are involved in stress/defense responses in Arabidopsis, rice, maize and sorghum (Ghanashyam and Jain 2009; Jain and Khurana 2009; Wang et al. 2010a, b). In our study, many different SlIAAs showed similar expression patterns under diverse stress treatments, the expression levels of most of SlIAAs increased after drought, salt and heat treatments when compared with control treatment. However, SlIAA3, SlIAA11, and SlIAA14 expression levels were down-regulated after abiotic stress treatments (Suppl Fig. 9). Similar results have been reported in other species. The transcript levels of most OsIAA genes in rice were up-regulated by both drought and salt stresses, but OsIAA31 was down-regulated (Song et al. 2009). In the present study, 13 drought stress-related cis-element, 1 salt stress-related cis-element and 1 heat shock element were found and distributed differently in the promoter regions of most SlIAAs, those abiotic stress-related cis-elements combining with the cis-elements involved in the auxin signaling regulation pathway may lead to the specific expression and function of SlIAA genes. However, the results of qRT-PCR were not always consistent with those of the promoter region analysis on SlIAA genes. Although there were some stress-related cis-elements present in the SlIAA20, SlIAA21 and SlIAA22, no significant changes of their expression levels were detected in response to abiotic stress treatment. On the other hand, there were no salt stress-related cis-elements detected in the promoter regions of SlIAA11 (Table S4), but the real-time PCR data showed that it was significantly stress responsive (Suppl Fig. 9). Although the putative salt stress-related cis-element (S000453) and heat shock element (S000030) were found in promoters of SlIAA14 and SlIAA3 (Table S4), previously proved to enhance the expression levels of relevant genes (Park et al. 2004; Rieping and Sehfffl 1992), their expression levels were significantly down-regulated under the heat or salt stresses (Suppl Fig. 9). This might indicate that some unidentified cis-regulated elements may play an important role in regulating the expression of those SlIAAs in stress response in tomato.
In general, no significant expression similarity in terms of tissue distribution and in response to stresses were exhibited between SlIAAs and AtIAAs in the same group of the phylogenetic tree based on the comparison of the expression pattern of IAA genes in tomato and Arabidopsis (Winter et al. 2007) (Figs. 5, 6; Suppl Figs. 7–9; Tables S6, S7). However, the SlIAAs and AtIAAs from the same group, showed similar sensibility to auxin treatment. Consistent with analysis by Goda et al. 2004, the IAA genes in group A1 and A2 showed high sensibility to auxin, but the expression level of the genes from group A3, A5, B2 changed slightly after the auxin treatment (Figs. 5, 6; Suppl Figs. 7–9; Tables S6, S7). These findings suggest that although the diverse levels of IAA genes expressed in different species, a similarity in response to auxin was inherited from their common ancestors through the long evolution.
Conclusion
Overall, 26 and 27 non-redundant AUX/IAA genes were identified and characterized in Solanaceae species, tomato and potato, respectively. A comprehensive genome-wide analysis of SlIAA gene family is presented, including the gene structures, chromosome locations, phylogeny, and conserved motifs. Our work demonstrates that different spatio-temporal transcript accumulation patterns exist for most members of the SlIAA gene family in tomato. The tomato SlIAAs could be transcriptionally induced by exogenous auxin, and most of them could also be induced by drought, salt and heat treatments in tomato leaves. However, it is needed to note that our qRT-PCR analysis only provides an estimate of the whole organ/tissue expression of AUX/IAA family genes, more detailed or exact expression pattern analysis of these IAA genes will be required using in situ hybridization or promoter–reporter fusion system. The final challenge is to define the specific functions of each individual AUX/IAAs gene during plant development and in response to environment stress.
Abbreviations
- ARF:
-
Auxin response factor
- Aux/IAA:
-
Auxin/indoleacetic acid
- AuxRE:
-
Auxin responsive elements
- NLS:
-
Nuclear localization signal
- qRT-PCR:
-
Quantitative real-time PCR
References
Abel S, Theologis A (1995) A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J 8:87–96
Bailey TL, Boden M, Buske FA, Frith M, Grant E, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:202–208
Berleth T, Krogan NT, Scarpella E (2004) Auxin signals—turning genes on and turning cells around. Curr Opin Plant Biol 7:553–563
Brukhin V, Hernould M, Gonzalez N, Chevalier C, Mouras A (2003) Flower development schedule in tomato Lycopersicon esculentum cv. Sweet Cherry. Sex Plant Reprod 15:311–320
Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, Frasse P, Latche A, Pech JC, Bouzayen M (2009) Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot 60(4):1349–1362
Cle’ment B, Pollmann S, Wielder E, Urbanczyk-Wochniak E, Otter L (2006) The Agrobacterium vitis T-6b oncoprotein induces auxin-independent cell expansion in tobacco. Plant J 45:1017–1027
Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445
Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29:153–168
Ghanashyam C, Jain M (2009) Role of auxin-responsive genes in biotic stress responses. Plant Signal Behav 4:846–848
Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134:1555–1573
Gray WM, Kepinski S, Rouse D, Leyser D, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 15:414
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385
Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T (2004) Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131:1089–1100
Hayashi K, Tan X, Zheng N, Hatate T, Kimura Y, Kepinski S, Nozaki H (2008) Small-molecule agonists and antagonists of F-box protein–substrate interactions in auxin perception and signaling. Proc Natl Acad Sci USA 105:5632–5637
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Hompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J 276:3148–3162
Jain M, Kaur N, Garg R, Jitendra K, Thakur, Akhilesh K, Tyagi, Jitendra P, Khurana (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6:47–59
Kalluri UC, DiFazio SP, Brunner AM, Tuskan GA (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7:59
Kim J, Harter K, Theologis A (1997) Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94:11786–11791
Kloosterman B, Visser RGF, Bachem CWB (2006) Isolation and characterization of a novel potato Auxin/indole-3-acetic Acid family member (StIAA2) that is involved in petiole hyponasty and shoot morphogenesis. Plant Physiol Biochem 44:766–775
Ku HM, Vision T, Liu JP, Tanksley SD (2000) Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci USA 97:9121–9126
Kumar R, Tyagi AK, Sharma AK (2011) Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol Genet Genomics 285:245–260
Leyser O (2002) Molecular genetics of auxin signalling. Ann Rev Plant Biol 53:377–398
Li H, Tiwari SB, Hagen G, Guilfoyle TJ (2011) Identical amino acid substitutions in the repression domain of auxin/indole-3-acetic acid proteins have contrasting effects on auxin signalling. Plant Physiol 155:1252–1263
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49:387–400
Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M (2006) Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J 46:297–306
Nebenführ A, White TJ, Lomax TL (2000) The diageotropica mutation alters auxin induction of a subset of the Aux/IAA gene family in tomato. Plant Mol Biol 44:73–84
Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2(9):e258
Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13:829–841
Park JY, Kim HJ, Kim J (2002) Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J 32:669–683
Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, Yoon HW, Lee S, Chung WS, Lim CO, Lee SY, Hong JC, Cho MJ (2004) Pathogen- and NaCl-induced expression of the scam-4 promoter is mediated in part by a gt-1 box that interacts with a gt-1-like transcription factor. Plant Physiol 135:2150–2161
Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282:10036–10046
Quint M, Gray WM (2006) Auxin signaling. Curr Opin Plant Biol 9:448–453
Raikhel N (1992) Nuclear targeting in plants. Plant Physiol 100:1627–1632
Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6(9):420–425
Remington DL, Vision TJ, Guilfoyle TJ, Reed WJ (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135:1738–1752
Rieping M, Sehfffl F (1992) Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol Gen Genet 231(2):226–232
Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13:465–480
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sato A, Yamamoto KT (2008) Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant 2(133):397–405
Shibasaki K, Uemura M, Tsurumi S, Rahman A (2009) Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell 21:3823–3838
Song Y, Wang L, Xiong LZ (2009) Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229:577–591
Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384
Tan X, Luz IA, Villalobos C, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–645
Terrile MC, Fiol DF, Casalongue CA (2010) Solanum tuberosum Aux/IAA family: new members and characterization of StIAA1 interacting proteins. Plant Growth Regul 62:93–99
Thakur JK, Tyagi AK, Khurana JP (2001) OsIAA1: an Aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Res 8:193–203
Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13:2809–2822
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543
Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16:533–543
Ulmasov T, Hagen G, Guilfoyle TJ (1997a) ARF1, a transcription factor that binds to auxin response elements. Science 276:1865–1868
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997b) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971
Walker JC, Key JL (1982) Isolation of cloned cDNAs to auxinresponsive poly (A)+RNAs of elongating soybean hypocotyls. Proc Natl Acad Sci USA 79(23):7185–7189
Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech JC, Bouzayena M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17:2676–2692
Wang DK, Pei KM, Fu YP, Sun ZX, Li SJ, Liu H, Tang K, Han B, Tao YZ (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394:13–14
Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latche A, Pech JC, Fernie AR, Bouzayen M (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21:1428–1452
Wang SK, Bai YH, Shen CJ, Wu YR, Zhang SN, Jiang D, Guilfoyle TJ, Chen M, Qi YH (2010a) Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics 10:533–546
Wang Y, Deng D, Bian Y, Lv Y, Xie Q (2010b) Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol Biol Rep 37:3991–4001
Winter D, Vinegar B, Nahal H, Ammar R, Wilson G, Provart N (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718
Wu J, Wang F, Cheng L, Kong F, Peng Z, Liu S, Yu X, Lu G (2011) Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep 30:2059–2073
Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J et al (2011) Genome sequence and analysis of the tuber crop potato. Nature 475(7355):189–195
Yamamoto KT (1994) Further characterization of auxin-regulated mRNAs in hypocotyl sections of mung bean [Vigna radiata (L.) Wilczek]: sequence homology to genes for fatty-acid desaturases and atypical late-embryogenesis-abundant protein, and the mode of expression of the mRNAs. Planta 192:359–364
Yamamoto KT, Mori H, Imaseki H (1992) cDNA cloning of indole-3-acetic acid regulated genes: Aux22 and SAUR from mung bean (Vigna radiata) hypocotyls tissue. Plant Cell Physiol 33:93–97
Yang XP, Lee SS, So J, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle1 M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J 40:772–782
Zanetti ME, Terrile MC, Godoy AV, San Segundo B, Casalongue CA (2003) Molecular cloning and characterization of a potato cDNA encoding a stress regulated Aux/IAA protein. Plant Physiol Biochem 41:755–760
Acknowledgments
This work was supported by the State Key Basic Research and Development Plan of China (2009CB119000), the National Natural Science Foundation of China (30771470) and Zhejiang Provincial Natural Science Foundation of China (R3110209 and 2009C32025). Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: JN043331(SlIAA12), JN043332(SlIAA13), JN043333(SlIAA16).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
438_2012_675_MOESM6_ESM.xls
Table S6. The expression analysis of AtIAA family genes in various Arabidopsis tissues. The expression data were searched against Arabidopsis eFP Browser using the AtIAA gene accession number (XLS 31 kb)
438_2012_675_MOESM7_ESM.xls
Table S7. The expression analysis of AtIAA family genes in response to auxin and abiotic treatments. The expression data were serached against Arabidopsis eFP Browser using each AtIAA gene accession number. (XLS 23 kb)
438_2012_675_MOESM10_ESM.jpg
Supplemental Figure 10. Alignment of potato Aux/IAA proteins obtained with the ClustalX program. The height of the bars indicates the number of identical residues per position. b: Multiple alignments of the domains I to IV of the tomato Aux/IAA proteins obtained with ClustalX and manual correction. Black and light gray shading indicate identical and conversed amino acid residues, respectively. Conserved domains are also underlined and correspond to part a. (JPEG 1417 kb)
438_2012_675_MOESM11_ESM.jpg
Supplemental Figure 11 a. Alignment of tobacco Aux/IAA proteins obtained with the ClustalX program. The height of the bars indicates the number of identical residues per position. b: Multiple alignments of the domains I to IV of the tobacco Aux/IAA proteins obtained with ClustalX and manual correction. Black and light gray shading indicate identical and conversed amino acid residues, respectively. Conserved domains are also underlined and correspond to part a. (JPEG 936 kb)
438_2012_675_MOESM12_ESM.jpg
Supplemental Figure 12. The five conserved consensus motifs in AUX/IAA of tomato, rice, maize and Arabidopsis IAA proteins were found by MEME. The symbol heights represent the relative frequency of each residue. The numbers of sites and e-value for each motif are also shown. (JPEG 432 kb)
438_2012_675_MOESM14_ESM.jpg
Supplemental Figure 14. Expression profiles of all the 26 SlIAA genes in different tomato flower tissues. qRT-PCR analyses using RNA generated from sepal (Se), petal (P), stamen (St), and ovary (O) were performed. For other details see Figure 5. (JPEG 557 kb)
438_2012_675_MOESM15_ESM.jpg
Supplemental Figure 15. Expression profiles of all the 26 SlIAA genes at different tomato flower developmental stages. QRT-PCR analyses were performed using RNA generated from tomato flower buds at three stages of early floral development, roughly defined by the length of flower buds as follows: stage I: 3-4 mm, stage II: 5-6 mm, and stage III at 7-8 mm. (JPEG 485 kb)
438_2012_675_MOESM16_ESM.jpg
Supplemental Figure 16. Expression profiles of all the 26 SlIAA genes in response to environmental stress. QRT-PCR analyses were used to assess SlIAA transcript levels in tomato seedlings exposed to drought (D), salt (S), heat (H) stresses when compared to control treatment. (JPEG 604 kb)
438_2012_675_MOESM17_ESM.jpg
Supplemental Figure 17. Phylogenetic relationships among the potato, tomato and tobacco IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap support from 1000 replicates are indicated at each branch. (JPEG 90 kb)
438_2012_675_MOESM18_ESM.jpg
Supplemental Figure 18. Phylogenetic relationships among the Arabidopsis IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap support from 1000 replicates are indicated at each branch. (JPEG 37 kb)
438_2012_675_MOESM19_ESM.jpg
Supplemental Figure 19. Phylogenetic relationships among the maize IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap support from 1000 replicates are indicated at each branch. (JPEG 34 kb)
438_2012_675_MOESM20_ESM.jpg
Supplemental Figure 20. Phylogenetic relationships among the rice IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap supports from 1000 replicates are indicated at each branch. (JPEG 38 kb)
Rights and permissions
About this article
Cite this article
Wu, J., Peng, Z., Liu, S. et al. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol Genet Genomics 287, 295–311 (2012). https://doi.org/10.1007/s00438-012-0675-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-012-0675-y