Abstract
Dirigent is proposed to be involved in either lignan or lignin biosynthesis. In the present study, sequences for dirigent and dirigent-like proteins from the wheat genome were analyzed. We obtained wheat dirigent (TaDIR13), and dirigent-like (TaDIR4 and TaJA1) recombinant proteins used for biochemical assays. It was shown that TaDIR13 could effectively direct coniferyl alcohol coupling into (+)-pinoresinol. In contrast, TaDIR4 and TaJA1 did not exhibit this activity. In accordance with this role of TaDIR13, overexpression of TaDIR13 in transgenic tobacco increased total lignan accumulation. Lignan extracts from TaDIR13 transgenic plants had enhanced antibacterial effects to Pseudomonas syringae. Furthermore, these plants did show strong resistance to P. syringae and Phytophthora parasitica. On the contrary, TaDIR4 transgenics did not show noticeable changes in either lignan accumulation or pathogen resistance. Overexpression of TaJA1 did not increase lignan accumulation either. Unlike TaDIR4 plants though, TaJA1 plants exhibited high pathogen resistance; yet, this was achieved through its jacalin-related lectins domain and not its dirigent domain. Lignin content and gene expression were not affected in all transgenic plants. Interestingly, expression of the pinoresinol-lariciresinol reductase gene (NtPLR) was increased in TaDIR13 transgenic plant; this is apparently owing to a favored metabolic flux from coniferyl alcohol into the lignan pathway. These data collectively suggest that TaDIR13 is mainly involved in regulating lignan biosynthesis which, in turn, responsible for the observed roles in pathogen resistance. TaDIR4 and TaJA1, both being dirigent-like proteins, have little functional overlap with dirigent proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignans are phenylpropanoid dimers synthesized via the phenylpropanoid pathway They occur naturally in a number of plant families and are thought to play the important physiological and ecological roles in the interaction with pathogens and insects, due to their antibacterial and antifeedant activities (Harmatha and Dinan 2003; Schroeder et al. 2006). Coniferyl and sinapyl alcohols are the direct precursors to form lignan dimers. Since coniferyl and sinapyl alcohols, together with other monolignol, p-coumaryl alcohol, are also polymerized into macromolecule lignin, lignans are overlapped with lignin in certain biosynthetic pathways (Davin and Lewis 2003). Much is known about lignin biosynthesis, including the action of oxidative enzymes to convert monolignols into corresponding free radicals, but one of the unknowns is how coupling radicals to produce a functional lignin molecule (Boerjan et al. 2003). There are two hypotheses to address this process. The random coupling hypothesis suggests that lignin formation proceeds through coupling of individual monolignols to the growing lignin polymer in a nearly random fashion. Therefore, the amount and species of individual monolignols available will be the main regulators for lignin formation and its final component (Sederoff et al. 1999). In an alternative hypothesis, there is a strict regulation for the formation of individual bonds and hence, the final component of lignin monolignol composition of the resultant lignin molecules (Lewis 1999). The discovery of dirigent proteins provides molecular underpinning for this latter hypothesis (Davin et al. 1997).
A 78-kDa dirigent was first isolated from Forsythia suspensa (Davin et al. 1997). An in vitro biochemical analysis demonstrated that this protein, in the presence of an oxidase or one electron oxidant, could stereoselectively couple two coniferyl alcohol molecules to give rise to a (+)-pinoresinol (Davin et al. 1997). This dimer, known as lignan, was proposed to couple more monolignols and then form the polymer of lignin (Davin et al. 1997). A dirigent cDNA from Forsythia intermedia was found to contain an open-reading frame for 186 amino acids (Gang et al. 1999). Therefore, the native dirigent protein is a trimer molecule, with each subunit being 26 kDa (Gang et al. 1999). Homologs of dirigent genes were recently found in a few other plants, including cotton (Zhu et al. 2007), and Boea hygrometrica (Wu et al. 2009). So far, at least 104 more dirigent and dirigent-like protein gene sequences have been discovered from other species (25 from Arabidopsis, 54 from rice, and 35 from spruce, Ralph et al. 2006; Ralph et al. 2007). A phylogenetic analysis indicated that these proteins form a super-family consisting of distinct subfamilies (Ralph et al. 2007).
According to gene expression analyses, many dirigent and dirigent-like genes are stress-inducible. Upregulation has been documented for a number of stresses, such as wounding (Ralph et al. 2007), stressed by ABA, dehydration, and low temperature (Wu et al. 2009). Upregulation upon attacks by insects (Ralph et al. 2007) and fungi (Zhu et al. 2007) are of particular interest. The involvement of dirigents in pathogen resistance can be better understood if lignans are brought into the picture. Lignans often exist in high quantities in the heartwood region of trees, potentially preventing degradation by heart-rot fungi (Suzuki and Umezawa 2007). There is a strong implication for dirigents’ roles in directing the monolignol-to-pinoresinol (lignan) conversion.
Direct experimental evidence to substantiate dirigent proteins’ contributions to lignin biosynthesis is still rare (Davin et al. 2008; Önnerud et al. 2002). Neither is known whether or not dirigent and dirigent-like proteins of different subfamilies play similar roles. In this report, (1) gene sequences for wheat dirigent and dirigent-like proteins were analyzed, (2) corresponding cDNAs were isolated and recombinant proteins were examined for their coupling properties on monolignols, and (3) functions of dirigents in lignan/lignin biosynthesis and stress responses were studied in transgenic tobacco plants.
Materials and Methods
Plant Materials
Wheat plants (Triticum aestivum L. cv. H4564) were grown in a naturally lit glasshouse. Total RNA was isolated using TRI reagent (Molecular Research Center, Inc., Cincinnati, OH). Genomic DNA was isolated as described by Edwards et al. (1991).
Phylogenetic Analysis of Dirigent and Dirigent-Like Proteins
Dirigent and dirigent-like protein sequences from Arabidopsis, rice, and spruce plants were obtained from the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/). A similarity search was conducted in a wheat EST database (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=wheat). For each plant species, partial sequences and incorrect or repetitive sequences were discarded. Sequences with more than 97 % identity within any species were treated as one entity. Sequence were compared using the SIM alignment tool (Altschul et al. 1997) with an open gap penalty of 10 and an extend gap penalty of 0.05. Phylogenetic analysis was performed by the maximum parsimony method using the PAUP 4.0 software (Sinauer Associates, Sunderland, MA).
Heterologous Expression and Purification of Dirigent and Dirigent-Like Proteins
TaDIR13 cDNA was amplified using primers 5′-CGgaattcATGCAAGGCCTCGCGGCATC-3′ and 5′-CCCaagcttTCAGATGTAGCACTCGTAGAG-3′ (with an EcoRI and a HindIII site, respectively). Similarly, TaDIR4 cDNA was amplified using primers 5′-CGgaattcATGGCTTGCTTCAAGCTCAAC-3′ and 5′-CCCaagcttTCATGGGTTGACGTAGAGTC-3′. Each PCR mixture contained 5 ng template, 1 μmol/L each primer, 0.4 mmol/L each dNTP, and 2.5 U Taq DNA polymerase (Gibco, Beijing). Each PCR product was then cloned into vector pET-32a (Novagen, Beijing), and the recombinant plasmids were designated as pET-TaDIR13 and pET-TaDIR4, respectively. Each recombinant plasmid was introduced into competent E. coli strain BL21 cells. Bacterial culture and protein extraction-purification were conducted as reported previously (Ma and Tian 2005). TaJA1, another wheat dirigent-like protein of the subclade C, was expressed as previously reported (Ma et al. 2013).
Analysis of Coupling Properties of Dirigent and Dirigent-Like Proteins
Coupling assays were performed by following the method of Davin et al. (1997). The reaction mixture consisted of ammonium peroxydisulfate (2 μmol/mL), coniferyl alcohol (10 μmol/mL), and recombinant protein (1.5 nmol/mL). After 1-h incubation at 30 °C, the mixture was extracted with ethyl acetate for three times. The extract was evaporated to dryness under vacuum, and the residue was re-dissolved in 50 % methanol. Reversed-phase column chromatography (HPLC) was performed, following the procedure of Zheng et al. (2011). A Dionex HPLC system (Sunnyvale, California) was used, which was equipped with a P680 HPLC pump, an UltiMate 3000 autosampler, a TCC-100 thermostated column compartment, a Dionex PDA100 photodiode array detector, and a C18 column of ODS 80Ts QA (150 mm × 4.6 mm, 5 μm i.d., Tosoh, Tokyo).
Generation of Transgenic Tobacco
The full-length TaDIR13 cDNA was PCR-amplified using primers 5′-GCCaagcttCTGGTAATTAATAGTTGCG-3′ and 5′-ATActcgagGGCACATCACTCCTTATT-3′ (with HindIII and XhoI sites, respectively). The full-length TaDIR4 cDNA was amplified using primers 5′-TCTaagcttGTCTTCCACTCATCCCG-3′ and 5′-TTTctcgagGTAATGCTGCCACAGGT-3′. After digestion with HindIII and XhoI, the product was ligated into vector pKYLX71 (An et al. 1985), resulting in binary plasmids designated as pKYLX-TaDIR13 and pKYLX-TaDIR4, respectively. The insert was subcloned into E. coli DH5α cells and confirmed by enzymatic digestion and sequencing. The vector was transferred into Agrobacterium tumefaciens strain LBA4404 by the freeze-thaw method, and tobacco (Nicotiana tabacum cv. Wisconsin 38) was transformed (Ma et al. 2008). Transgenic tobacco with over-expressed TaJA1 gene, for subclade C of dirigent from wheat, has been reported previously (Ma et al. 2013). Tobacco plants were also transformed with an empty pKYLX71 vector to generate controls.
The two groups of transgenic plants were identified by PCR with two sets of specific primers (for TaDIR13, 5′-CTGGTAATTAATAGTTGCG-3′ and 5′-GGCACATCACTCCTTATT-3′; for TaDIR4, 5′-GTCTTCCACTCATCCCG-3′ and 5′-GTAATGCTGCCACAGGT-3′). The abundance of the TaDIR13 and TaDIR4 transcripts in transgenic plants were examined by RT-PCR, using the same primer sets.
Semi-quantitative RT-PCR analysis for lignin and lignan biosynthetic genes was conducted following the procedure of Ma et al. (2011). For lignin genes, primer sequences for cinnamyl alcohol dehydrogenase (NtCAD, Genbank accession no. X62343), cinnamoyl-CoA reductase (NtCCR, A47101), and caffeic acid O-methyltransferase (NtCOMT, X74453) can be found in Ma et al. (2011). For lignan biosynthetic genes, pinoresinol-lariciresinol reductase (NtPLR) and piperitol-sesamin synthase (NtPSS) primer sequences were as follows: 5′-GCTGTCAAACAGGTGGAT-3′ plus 5′-CTTGTTCAGTGTCCTTGG-3′ and 5′-TCTGCAAGAATCAGAACCAG-3′ plus 5′-TCTCCTTCCCATTCCAAAT-3′, respectively. The tobacco actin gene was used as a reference, with primers 5′-CCAATCGAGAGAAGATGACCCA-3′ and 5′-CCATCTGGCAGCTCATAGCTCT-3′. The expression intensity of each target gene relative to the actin gene was estimated on the images from electrophoresis.
Quantitative RT-PCR reactions were conducted using a SYBR Green RT-PCR Master Mix (Toyobo, Tokyo) on a Stratagene Mx3000P thermocycler (Stratagene, La Jolla, CA) under conditions of an initial denaturation at 95 °C for 2 min followed by 40 cycles of denaturing at 95 °C for 15 s, annealing at 55 °C for 20 s, and extending at 72 °C for 15 s. Dissociation curves were verified for each reaction. Quantification was performed using the 2−ΔΔCt method (Livak and Schmittgen 2001) using tobacco actin as a reference gene. The relative value of the gene expression was normalized through the reference gene and calculated as the mean of three biological replicates.
Lignin and Lignan Content Analyses for Transgenic Plants
Samples from air-dried stems were subjected to quantitative analysis for lignin by the Klason method (Kirk and Obst 1988). Lignin content was expressed relative to the cell wall residue (%CWR). Total lignan was extracted from leaf samples using the method of Chen et al. (2010). The extract was evaporated to dryness under vacuum, and the residue was re-dissolved in water. Total lignan was determined using a chromotropic acid spectrophotometric method (Lu et al. 2011) with schisantherin A as a standard.
Determination of Antibacterial Effects of Lignan Extracts
A Pseudomonas syringae pv. tabaci liquid culture at the exponential phase (OD600 = 0.3) was diluted by 1:1,000, and a 5-μL diluted bacteria sample was spread on a LB plate supplemented with the lignan extract (9:1, v/v). After incubation for 24 h at 28 °C, bacterial colonies were counted.
Pathogen Resistance Assays
T2 generation plants homologous for the TaDIR13 and TaDIR4 transgenes, respectively, were used for disease resistance analyses. Two pathogens, Phytophthora parasitica var. nicotianae and P. syringae pv. tabaci, were used. These pathogens, on susceptible plants, cause tobacco black shank and wildfire disease, respectively. The fourth to sixth leaves from the top on 2-month-old plants were inoculated with either pathogen. Methods for pathogen growth, inoculation, and disease scoring were performed according to Guo et al. (2004). For tobacco black shank disease, the lesion diameter was recorded. For tobacco wildfire disease, bacterial population inside leaf disks was determined from cultures on King’s B plates. The Student’s t test for independent samples (Geng and Hills 1989) was applied to compare transgenic with control lines, with probability at P 0.05 and P 0.01.
Results
Dirigent and Dirigent-Like Proteins in Wheat
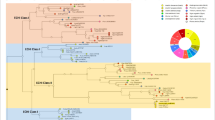
In the wheat EST database, 12 dirigent and dirigent-like proteins were identified. Along with sequences from Arabidopsis, rice, and spruce, a phylogenetic tree was constructed (Fig. 1). These proteins from wheat are members of either the dirigent subfamily (also referred to as the DirA subclade) or the dirigent-like subfamily. In the dirigent-like subfamily, DirB/D subclade proteins are commonly found in all seed plants listed. Other subclades of the dirigent-like subfamily exhibit narrower distributions (DirE in Arabidopsis and rice, DirF in spruce, and DirG in rice). DirC is only found in monocot plants. The functions of DirC proteins have been investigated in maize, rice, and wheat (see a review by Ma 2013).
Molecular phylogenetic tree for dirigent and dirigent-like proteins from Arabidopsis, rice, spruce, and wheat. The tree was constructed using Clustal W with PAM 250 residue weight table. The bootstrap values were shown in each branch. AtDIR25 was selected as out-group and FiDIR from Forsythia was used for landmark of DirA subclade. FiDIR, TaDIR13, and TaDIR4 were highlighted in the figure. GenBank and EST accession numbers for these sequences are as follows: Forsythia x intermedia FiDIR (AAF25357); Arabidopsis thaliana AtDIR1 (ABR46205), AtDIR2 (AAM13094), AtDIR3 (NP_199715), AtDIR4 (NP_179707), AtDIR5 (BAD43018), AtDIR6 (AAO00799), AtDIR7 (BAC42662), AtDIR8 (NP_187976), AtDIR11 (BAD44205), AtDIR12 (AAO42052), AtDIR13 (AAP88352), AtDIR14 (NP_192860), AtDIR15 (NP_195582), AtDIR19 (BAC42538), AtDIR20 (AAM91539), AtDIR21 (NP_176762), AtDIR23 (AAO64191), and AtDIR25 (AAP49521); Picea spp. PDIR1 (ABD52112), PDIR6 (ABD52117), PDIR7 (ABD52118), PDIR9 (ABD52120), PDIR10 (ABD52121), PDIR11 (ABD52122), PDIR12 (ABD52123), PDIR13 (ABD52124), PDIR14 (ABD52125), PDIR15 (ABD52126), PDIR16 (ABD52127), PDIR17 (ABD52128), PDIR18 (ABD52129), PDIR32 (ABR27728), and PDIR33 (ABR27729); Oryza sativa OsDIR11 (NP_001060395), OsDIR12 (NP_001060396), OsDIR13 (NP_001060404), OsDIR14 (BAC19943), OsDIR15 (NP_001060409), OsDIR19 (NP_001051654), OsDIR31 (NP_001066288), OsDIR32 (NP_001065892), OsDIR33 (NP_001065888), OsDIR34 (AAX96293), OsDIR35 (NP_001065890), OsDIR36 (NP_001065889), OsDIR52 (AAX96290), OsDIR53 (ABA93522), and OsDIR54 (AAX96314); and Triticum aestivum TaDIR1 (TC307806), TaDIR2 (TC309798), TaDIR3 (TC362616), TaDIR4 (TC297337), TaDIR5 (TC326720), TaDIR6 (TC429903), TaDIR7 (TC424593), TaDIR8 (CV777987), TaDIR12 (TC317574), TaDIR13 (TC299725), TaDIR14 (BJ284006), and TaDIR15 (CK154646)
Phylogenetic analysis showed that members of the DirA subclade had higher degrees of sequence conservation than those of DirB/D. Members of DirA found in eudicots (Arabidopsis), monocots (rice), and gymnosperms (spruce) were clustered to the different branches (Fig. 1). Members of the DirB/D subclade exhibited higher sequence similarities in inter-taxon than intra-taxon; for example, AtDIR1 and AtDIR2 were more close to OsDIR32 and OsDIR34, while AtDIR4 was more close to TaDIR1. This implies that DirB/D may be a very ancient group of genes, which diverged into different classes with eudicots, monocots, and gymnosperms.
In the current study, we chose TaDIR13 and TaDIR4 as representatives of the DirA and DirB/D subclades, respectively, for further analysis. The full length TaDIR13 cDNA has 1,002 nucleotides, with a 609-nucleotide open-reading frame (ORF), flanked by 5′ and 3′ untranslated regions of 58 and 335 nucleotides, respectively. The predicted protein consists of 202 amino acids with a relative molecular mass of 22 kDa and a theoretical isoelectric point of 4.85. The TaDIR4 cDNA has 782 nucleotides, with a 498-nucleotide ORF, flanked by 5′ and 3′ untranslated regions of 52 and 232 nucleotides, respectively. The predicted protein consists of 165 amino acids with a relative molecular mass of 17 kDa and a theoretical isoelectric point of 7.34. One DirC isoform from wheat, TaJA1 (Wang and Ma 2005), was also included in this project.
Recombinant TaDIR13 Protein Directs Pinoresinol Formation
Recombinant TaDIR13 protein expressed and purified from E. coli (Fig. 2) was tested for activity to couple monolignols. In the presence of ammonium peroxydisulfate as an oxidant, TaDIR13 could direct conversion of coniferyl alcohol into (+)-pinoresinol (Fig. 3). The calculated specific activity for this conversion was about 3.4 nmol (+)-pinoresinol/nmol TaDir13 protein. This result is similar as that reported for F. suspensa (Davin et al. 1997). By contract, recombinant TaDIR4 and TaJA1 proteins did not show this activity in the same reaction condition (data not shown).
Gel electrophoresis images showing recombinant TaDIR13 expressed in E. coli. Proteins were separated from the total protein fractions of uninduced and IPTG-induced E. coli harboring pET-TaDIR13 expression plasmids, and after His-Tag resin purification of the soluble fraction from induced cells. Molecular markers are indicated on the right of the figure
TaDIR13 and TaDIR4 Transgenic Plants Exhibit No Morphological and Developmental Abnormalities
Transgenic plants carrying TaDIR13 or TaDIR4 were confirmed by PCR and RT-PCR, and four TaDIR13 transgenic lines (DIR13-1, DIR13-2, DIR13-3, and DIR13-4) (Fig. 4a) and four TaDIR4 transgenic lines (DIR4-1, DIR4-3, DIR4-4, and DIR4-9) (Fig. 4b) were examined in detail.
Seeds of T1 progeny of transgenic plants were screened on a kanamycin-complimented medium. The segregation ratio of drug resistance versus sensitivity was approximately 3:1 for three DIR13 lines (DIR13-2, DIR13-3, and DIR13-4) and three DIR4 lines (DIR4-3, DIR4-4, and DIR4-9). These transgenic lines were essentially identical to wide-type plants in morphology (height and leaf number) and flowering time when grown in greenhouse.
Overexpression of TaDIR13 Gene Increase Disease Resistance
Lignan extracts from leaves of transgenic plants were tested on P. syringae pv. tabaci. After incubation for 24 h, extracts from TaDIR13 transgenic plants significantly inhibited bacterial growth, compared with extracts from control tobacco (Fig. 5). Extracts from either TaDIR4 or TaJA1 transgenic plants did not show significant changes in bacterial growth (Fig. 5).
Antibacterial effects of the lignan extracts from different transgenic tobacco lines. A sample of 5-μL Pseudomonas syringae culture (104 CFU/mL) was spread on LB plate supplemented with the lignan extracts. The colonies were counted after 24 h. DIR13-2, DIR13-3, and DIR13-4 lines for TaDIR13 transgene; DIR4-3, DIR4-4, and DIR4-9 lines for TaDIR4 transgene; JA-2, JA-5 and JA-6 lines for TaJA1 transgene. Values are means of three different experiments, with SE bars. Probability values between the control and transgenic tobacco plants were estimated by the Student’s t test, with significant difference at **P 0.01
P. syringae and P. parasitica var. nicotianae were further used in disease-resistant bioassays. Growth of P. syringae was significantly inhibited in TaDIR13 transgenic lines (DIR13-2, DIR13-3, and DIR13-4) compared to the control (Fig. 6a). Pathogen growth in TaDIR4 transgenic lines (DIR4-3, DIR4-4, and DIR4-9) was not affected (Fig. 6a).
Analysis of pathogen resistances to Pseudomonas syringae pv. tabaci and Phytophthora parasitica var. nicotianae in transgenic plants. a Resistance to Pseudomonas syringae. b Resistance to Phytophthora parasitica. Other symbols are the same as to Fig. 5
A more careful examination revealed that TaDIR13 transgenic lines exhibited less severe symptoms of the disease than control after the inoculation of P. parasitica (Fig. 6b). Again, TaDIR4 transgenic lines showed similar disease lesion as the control tobacco (Fig. 6b). Taken together, the data indicate that overexpression of TaDIR13 in tobacco plants enhances resistance to both bacterial and fungal pathogens, while TaDIR4 is not involved in these processes.
As shown previously, TaJA1 gene enhances the disease resistance to both bacterial and fungal pathogens (Ma et al. 2010; Ma et al. 2013), although extracts from TaJA1 transgenic plants did not show antibacterial effects (Fig. 5).
Overexpression of TaDIR13 Gene Increases Lignan Accumulation
Compared to control plants, TaDIR13 transgenic lines exhibited 13.1 to 15.4 % increases in lignan content. TaDIR4 and TaJA1 lines showed little change in lignan content (Fig. 7a). By contrast, acid-insoluble lignin remained nearly unchanged in all transgenic plants relative to controls (Fig. 7b).
Lignan and lignin contents in TaDIR13, TaDIR4, and TaJA1 lines. a Lignan content. b Lignin content. Other symbols are the same as to Fig. 5
Overexpression of TaDIR13 Affects Lignan Biosynthetic Genes
Initiating from coupling two coniferyl alcohol molecules into pinoresinol, lignan biosynthesis branches into two pathways. The branch-point enzymes for which are piperitol-sesamin synthase (PSS) and pinoresinol-lariciresinol reductase (PLR), respectively (Kim et al. 2009). As detected by RT-PCR with RNA from tobacco leaf tissues, TaDIR13 tobacco lines exhibited strong expression of NtPLR but not NtPSS (Fig. 8a). The analysis from quantitative real-time PCR confirmed the elevated transcripts of NtPLR gene (Fig. 8b). These results were consistent with the observed lignan accumulation pattern (Figs. 3 and 7a). Overexpression of either TaDIR4 or TaJA1 showed no influence on NtPLR and NtPSS expressions (Fig. 8), consistent with the lignan measurement data for these transgenic plants (Fig. 7a).
Abundances of NtCAD, NtCCR, and NtCOMT transcripts were not affected in all transgenic plants, as detected either by RT-PCR (Fig. 9a) or quantitative real-time PCR (Fig. 9b) with RNA from tobacco stem tissues. This result was parallel to the unchanged lignin content in these plants (Fig. 7b). It is clear that although lignin and lignan pathways share coniferyl alcohol as the common substrate, our analyses suggest that they are not closely linked to each other.
Discussion
Dirigent Proteins Promote Lignan Accumluation and Enhance Pathogen Resistance
Although dirigent proteins were found for more than a decade (Davin et al. 1997), its concert biological roles are still ambiguous. The data mining showed that homologous genes of DirA and DirB/D existed in wheat. From the phylogenetic tree of dirigent proteins (Fig. 1), we chose wheat TaDIR13 and TaDIR4, as representatives of DirA and DirB/D, respectively, to investigate their functions.
We demonstrated that recombinant TaDIR13 was able to direct the coniferyl alcohol to pinoresinol conversion (Fig. 3b), as reported for the Forsythia dirigent (Davin et al. 1997), confirming that TaDIR13 is indeed a dirigent protein. In accordance with this result, overexpression of the TaDIR13 gene was found to increase lignan accumulation (Fig. 7a). Further analyses revealed that TaDIR13 was able to upregulate PLR expression, leading to a favored metabolite flux into the lignan pathway (Fig. 8). In other systems, there have been similar observations. During seed development in flax, accumulation of lignan secoisolariciresinol diglucoside was found to be parallel to PLR expression, suggesting PLR’s regulatory roles in lignan synthesis (Hano et al. 2006). Overexpression of Forsythia PLR gene in wheat elevated lignan levels for more than two times (Ayella et al. 2007). Mutation analysis in Arabidopsis showed the involvement of both dirigent protein and PLR in the enantiomeric control in lignan biosynthesis (Nakatsubo et al. 2008). Taken together, all these studies support the mechanism that dirigent regulates lignan synthesis together with activation of the PLR genes.
On the contrary, lignin biosynthesis was not affected by TaDIR13 overexpression, neither in lignin content (Fig. 7b) nor lignin biosynthetic genes (CAD, CCR, and COMT) (Fig. 9). This result provides further evidence that dirigent-directed pinoresinol formation is mainly, if not completely, involved in lignan biosynthesis. This is conceivable when spatial separation of lignin and lignan synthesis systems is considered; the former occurs in the cell wall compartment, while the latter in the cytoplasm (Davin et al. 2008; Vanholme et al. 2010; Bonawitz and Chapple 2010).
Lignans are thought to have the important physiological roles in defense process, particularly for their interaction with insects (Harmatha and Dinan 2003; Schroeder et al. 2006). However, their actions on pathogens remain to be elucidated. Constitutive expression of DRR206 (originally categorized as a disease resistance gene in authors’ paper, and we recognized it as a dirigent gene by sequence similarity analysis) increased resistance to blackleg disease (Leptosphaeria maculans) in transgenic canola (Wang et al. 1999). Our results demonstrated that lignan accumulation was increased when TaDIR13 was overexpressed (Fig. 7a). Intensive investigations confirmed that the increased lignan level in transgenic tobacco enhanced its resistance to pathogens, both in in vitro (Fig. 5) and in vivo assays (Fig. 6). The current study on TaDIR13 strengthens the functional connection of dirigent-regulated lignan synthesis to pathogen resistance.
Dirigent-Like Proteins Have Different Mechanisms for Stress Responses
TaDIR4 and TaJA1, both being dirigent-like proteins, did not exhibit to couple coniferyl alcohol into (+)-pinoresinol and promote lignan accumulation (Fig. 7a). Clearly, these proteins are functionally distinct to dirigent protein TaDIR13.
Many members of the DirB/D subclade were speculated to play roles in plants’ responses to biotic and abiotic stresses. Examples include biotic and abiotic, such as Gbd1 and Gbd2 from cotton (Gossypium hirsutum) (Zhu et al. 2007), BhDir1 from B. hygrometrica (Wu et al. 2009), and some pDIRs from spruce (see Fig. 1; Ralph et al. 2007). However, all pieces of evidence were from association analysis by gene expression with relation to stress responses; hence, the roles that DirB/D proteins may play in stress responses are not clear. We demonstrated that TaDIR4, a dirigent-like protein of the DirB/D subclade, did not enhance pathogen resistance (Figs. 5; and 6). This is in sharp contrast to the functions of the dirigent protein TaDIR13 but simply supports the proposed correlation between lignan and pathogen resistance. On the other hand, we observed that the transgenic TaDIR4 plants exhibited longer roots than controls did on media supplemented with NaCl or mannitol (unpublished data), indicating better resistance to osmotic stress. Detailed studies for biotic and abiotic challenges are needed to understand the underlying mechanisms of DirB/D functions.
TaJA1, another dirigent-like protein, yet from the DirC subclade, was found to be responsible for the increased pathogen resistance in transgenic plants (Ma et al. 2010); but this function is achieved through TaJA1’s agglutinating activity by its jacalin domain (Ma et al. 2013), rather than through lignan metabolism (Fig. 7a). Most DirC proteins contain a dirigent-related domain in the N-terminus and a jacalin-related lectin (JRL) domain in the C-terminus (see review by Ma (2013) and references therein). Besides these chimeric DirC, some DirC sequences with only dirigent-related domain were also found in monocot plants, such as SoDIR1 (Casu et al. 2004) and ShDIR16 (Damaj et al. 2010) from sugarcane (Saccharum hybrid), and OsDIRs from rice (Ralph et al. 2007). What the dirigent-related domain does remains unknown.
In conclusion, our results suggest that dirigent and dirigent-like proteins are distinct in their biochemical action, in which dirigent proteins direct the coupling reaction of two molecules of coniferyl alcohol into pinoresinol, and this leads to increase lignan accumulation and pathogen resistance. Dirigent-like proteins have distinct subclades, in which DirB/D may be involved in stress response with an unknown mechanism; DirC increase disease resistance to pathogen by its jacalin-related lectin domain.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman JD (1997) Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
An G, Watson BG, Stachel S, Gordon MP, Nester EW (1985) New cloning vehicles for transformation of higher plants. EMBO J 4:277–284
Ayella AK, Trick HN, Wang W (2007) Enhancing lignan biosynthesis by over-expressing pinoresinol lariciresinol reductase in transgenic wheat. Mol Nutr Food Res 51:1518–1526
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44:337–63
Casu RE, Dimmock CM, Chapman SC, Grof CPL, McIntyre CL, Bonnett GD, Manners JM (2004) Identification of differentially expressed transcripts from maturing stem of sugarcane by in silico analysis of stem expressed sequence tags and gene expression profiling. Plant Mol Biol 54:503–517
Chen Y, Shi Z-W, Huang X-M (2010) Ultrasonic assisted extraction of total lignans and its content analysis in soybeans. Soybean Sci 29:168–170
Damaj MB, Kumpatla SP, Emani C, Emani C, Beremand PD, Reddy AS, Rathore KS, Buenrostro-Nava MT, Curtis IS, Thomas TL, Mirkov TK (2010) Sugarcane dirigent and O-methyltransferase promoters confer stem-regulated gene expression in diverse monocots. Planta 231:1439–1458
Davin LB, Lewis NG (2003) An historical perspective on lignan biosynthesis: monolignol, allylphenol and hydroxycinnamic acid coupling and downstream metabolism. Phytochem Rev 2:257–288
Davin LB, Wang H-B, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, Lewis NG (1997) Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275:362–366
Davin LB, Jourdes M, Patten AM, Kim K-W, Vassaǒ DG, Lewis NG (2008) Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat Prod Rep 25:1015–1090
Edwards K, Johnstone C, Thompson C (1991) A simple rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19:1349
Gang DR, Costa MA, Fujita M, Dinkova-Kostova AT, Wang H-B, Burlat V, Martin W, Sarkanen S, Davin LB, Lewis NG (1999) Regiochemical control of monolignol radical coupling: a new paradigm for lignin and lignan biosynthesis. Chem Biol 6:143–151
Geng S, Hills FG (1989) Biometrics in agriculture science. Kendell Hunt Publisher, Dubuque
Guo Z-J, Chen X-J, Wu X-L, Ling J-Q, Xu P (2004) Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol Biol 55:607–618
Hano C, Martin I, Fliniaux O, Legrand B, Gutierrez L, Arroo RRJ, Mesnard F, Lamblin F, Lainé E (2006) Pinoresinol–lariciresinol reductase gene expression and secoisolariciresinol diglucoside accumulation in developing flax (Linum usitatissimum) seeds. Planta 224:1291–1301
Harmatha J, Dinan L (2003) Biological activities of lignans and stilbenoids associated with plant-insect chemical interaction. Phytochem Rev 2:321–330
Kim HJ, Ono E, Morimoto K, Yamagaki T, Okazawa A, Kobayashi A, Satake H (2009) Metabolic engineering of lignan biosynthesis in Forsythia cell culture. Plant Cell Physiol 50:2200–2209
Kirk TK, Obst JR (1988) Lignin determination. Methods Enzymol 161:87–101
Lewis NG (1999) A 20th century roller coaster ride: a short account of lignification. Curr Opin Plant Biol 2:153–162
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25:402–408
Lu X, Li J, Jiang X-J, Zhang K-F, Duan X-Q (2011) Content determination of lignans in Kadsura coccinea (Lem.) A. C. Smith. J Anhui Agri Sci 39:12152–12153
Ma Q-H (2013) Monocot chimeric jacalins: a novel subfamily of plant lectins. Crit Rev Biotechnol. doi:10.3109/07388551.2013.793650
Ma Q-H, Tian B (2005) Biochemical characterization of a cinnamoyl-CoA reductase from wheat. Biol Chem 386:553–560
Ma Q-H, Wang X-M, Wang Z-M (2008) Expression of isopentenyl transferase gene controlled by seed-specific lectin promoter in transgenic tobacco influences seed development. J Plant Growth Regul 27:68–76
Ma Q-H, Tian B, Li Y-L (2010) Overexpression of a wheat jasmonate-regulated lectin increases pathogen resistance. Biochimie 92:187–193
Ma Q-H, Wang C, Zhu H-H (2011) TaMYB4 cloned from wheat regulates lignin biosynthesis through negatively controlling the transcripts of both cinnamyl alcohol dehydrogenase and cinnamoyl-CoA reductase genes. Biochimie 93:1179–1186
Ma Q-H, Zhen W-B, Liu Y-C (2013) Jacalin domain in wheat jasmonate-regulated protein Ta-JA1 confers agglutinating activity and pathogen resistance. Biochimie 95:359–365
Nakatsubo T, Mizutani M, Suzuki S, Hattori T, Umezawa T (2008) Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J Biol Chem 283:15550–15557
Önnerud H, Zhang L, Gellerstedt G, Henriksson G (2002) Polymerization of monolignols by redox shuttle-mediated enzymatic oxidation: a new model in lignin biosynthesis I. Plant Cell 14:1953–1962
Ralph SG, Park JY, Bohlmann J, Mansfield SD (2006) Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.). Plant Mol Biol 60:21–40
Ralph SG, Jancsik S, Bohlmann J (2007) Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.). Phytochemistry 68:1975–1991
Schroeder FC, Del Campo ML, Grant JB, Weibel DB, Smedley SR, Bolton KL, Meinwald J, Eisner T (2006) Pinoresinol: a lignol of plant origin serving for defense in a caterpillar. Proc Natl Acad Sci U S A 103:15497–15501
Sederoff RR, MacKay JJ, Ralph J, Hatfield RD (1999) Unexpected variation in lignin. Curr Opin Plant Biol 2:145–152
Suzuki S, Umezawa T (2007) Biosynthesis of lignans and norlignans. J Wood Sci 53:273–284
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153:895–905
Wang X-M, Ma Q-H (2005) Characterization of a jasmonate-regulated wheat protein related to a beta-glucosidase-aggregating factor. Plant Physiol Biochem 43:185–192
Wang Y, Nowak G, Culley D, Hadwiger LA, Fristensky B (1999) Constitutive expression of pea defense gene DRR206 confers resistance to blackleg (Leptosphaeria maculans) disease in transgenic canola (Brassica napus). Mol Plant-Microbe Interact 12:410–418
Wu R, Wang L, Wang Z, Shang H, Liu X, Zhu Y, Qi D, Deng X (2009) Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog Nat Sci 19:347–352
Zheng J, Li H, Ding CX, Suo YR, Wang LS, Wan HL (2011) Anthocyanins composition and antioxidant activity of two major wild Nitraria tangutorun Bobr. variations from Qinghai–Tibet Plateau. Food Res Int 44:2041–2046
Zhu L, Zhang X, Tu L, Zeng F, Nie Y, Guo X (2007) Isolation and characterization of two novel dirigent-like genes highly induced in cotton (Gossypium barbadense and G. hirsutum) after infection by Verticillium dahliae. J Plant Pathol 89:41–45
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 31170277, No. 31370336, and No. 31070235), the Natural Science Foundation of Beijing (No. 5122024), and the Innovation Project of Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ma, QH., Liu, YC. TaDIR13, a Dirigent Protein from Wheat, Promotes Lignan Biosynthesis and Enhances Pathogen Resistance. Plant Mol Biol Rep 33, 143–152 (2015). https://doi.org/10.1007/s11105-014-0737-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0737-x