Abstract
Water-deficit is the major abiotic stress factor that limits the yield and quality of cotton produced around the world. We observed earlier that a CCL (CCR-like; cold-circadian rhythm-RNA binding like) gene has been differentially expressed during boll development in cotton under drought stress in the field. Isolation and functional characterization of GhCCL from upland cotton (Gossypium hirsutum L. cv. Bikaneri Narma) was carried out in the present study. We studied the GhCCL gene structure and organization and demonstrated for the first time that GhCCL may be involved in abiotic stress tolerance response in plants. RT-PCR analysis indicated that GhCCL is differentially regulated in cotton seedlings by abiotic stresses such as salt, mannitol, cold, heat, dehydration, wounding and jasmonic acid, salicylic acid, and hydrogen peroxide. In silico and subcellular localization analysis suggested that GhCCL is localized in the chloroplasts. Constitutive expression of GhCCL in tobacco (Nicotiana tabacum var. Petit Havana) conferred tolerance to water-deficit stress and salt stresses during seed germination on amended MS media. The transgenic plants showed better growth performance and increased fresh weight under long-term stress. The transgenic plants grown in the glass house tolerated water-deficit stress (by withholding water) and recovered upon rewatering (45 days) whereas WT plants did not survive. Characterization of the GhCCL promoter sequence by in silico analysis showed various light, stress, tissue specific, and hormone responsive cis-elements. The present study suggested that GhCCL positively regulates the response to abiotic stresses, especially water-deficit stress in transgenic plants and that the overexpression of GhCCL may enhance the stability of mRNA thereby conferring tolerance to abiotic stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cotton is the major source of natural fibers used in the textile industry and is cultivated across the globe (Smith and Cothren 1999). Among the four cultivated species, Gossypium arboreum and G. herbaceum are diploids with A genome and G. hirsutum and G. barbadense are allotetraploid species with AD genome. G. hirsutum represents more than 95 % of the cultivated cotton worldwide (National Cotton Council, 2012, http:/www.cotton.org/econ/cropinfo/index.cfm). Cotton fiber development includes four distinct, but overlapping stages: fiber cell initiation, elongation/primary cell wall (PCW) synthesis, secondary cell wall (SCW) biosynthesis, and maturation (Basra and Malik 1984; Ruan and Chourey 1998). The fiber cell initiation usually occurs from 2–3 days before anthesis to 2–3 days post anthesis (dpa) and fiber cell elongation occurs up to 20 dpa. However, fast elongation of fiber cell occurs between 5 and 15 dpa. Secondary cell wall synthesis starts at about 20 dpa and continues up to 45 dpa. During this period, large amount of cellulose (>90 %) deposition takes place, and the fiber cell wall becomes thick. In the final maturation stage (45–50 dpa), fibers undergo dehydration and produce mature cotton lint (Wilkins and Arpat 2005; Arpat et al. 2004; Ji et al. 2003). The quality of fiber, which is based on its final length and strength, is determined mainly during the elongation and secondary cell wall deposition stage (Li et al. 2007; Deng et al. 2012).
Abiotic stresses remain the greatest constraint to crop production. Worldwide, it has been estimated that approximately 70 % of yield reduction is the direct result of abiotic stresses (Acquaah 2007). It reduces plant productivity by inhibiting growth and photosynthesis (Taiz and Zeiger 1998). Adaptation to drought stress is undoubtedly one of the most complex biological processes. It involves numerous changes including reduced growth, transcriptional activation, inactivation of specific genes, transient increases in abscisic acid (ABA) levels, accumulation of compatible solutes and protective enzymes, increased levels of antioxidants, and suppression of energy-consuming pathways.
Various abiotic stresses severely impair growth and yield of cotton plants. Severe drought stress will slow cotton plant development and cause small bolls and squares to shed. Establishment and pre-bloom irrigations affect total yield, but water deprivation following bloom and into boll development also affects lint quality. Cotton is known to contain intrinsic mechanisms that allow it to partially withstand water-deficit and heat stresses by its deep and extensive root system and adaptive osmoregulation mechanisms (Ackerson and Hebert 1981; Grimes and Yamada 1982). However, as water-deficit stress progressively continues, the development and reproduction of cotton are severely affected, along with significant reduction in quality and yield of cotton fiber (Cothren 1999; Bradow and Davidonis 2010). Therefore, to achieve maximal cotton yield, it is essential to understand proper management strategies between water demand, irrigation, and plant responses that are seasonally variable (Martin et al. 2007).
High-throughput screening techniques such as microarray analysis have been used for efficient profiling of abiotic stress-responsive genes including drought stress in model and agronomically important crop species (Ji et al. 2003; Kawaguchi et al. 2004; Zhou et al. 2007; Aprile et al. 2009; Cohen et al. 2010; Gong et al. 2010; Padmalatha et al. 2012). Comparative transcriptomic analysis of gene expression profiles in cotton leaf and root tissues under well-watered and water-deficit conditions (Payton et al. 2011), boll developmental stages under field drought stress condition (Padmalatha et al. 2012), and root and leaf samples from drought-resistant cultivar grown under water-deficit stressed and well-watered field conditions (Park et al. 2012) were studied extensively, to discover tissue specific and stress-responsive changes in gene expression under water-deficit stress condition in cotton. Recently, Zhu et al. (2013) identified genes involved in diverse stresses including ABA, cold, drought, salinity, and alkalinity of cotton seedling by comparative microarray analysis, and they suggested that there was crosstalk of responsive genes and pathways to multiple abiotic or even biotic stresses in cotton.
So far, a limited number of genes/transcripts which are involved in drought tolerance were isolated from cotton and functionally characterized either in heterologous systems like tobacco or in cotton to validate their role in imparting drought tolerance. G. hirsutum 64-amino acid type 3 metallothionein protein that was isolated through complementary DNA (cDNA) library screening showed accumulated higher levels of mRNA under salinity, drought, low temperature, heavy metal ions, ABA, ethylene, and reactive oxygen species (ROS) in cotton seedlings and overexpression of cDNA in transgenic tobacco plants conferred increased abiotic stress tolerance compared to wild-type (WT) plant (Xue et al. 2009). Transgenic cotton plants expressed with tobacco osmotin showed a slower rate of wilting during drought and fast recovery upon rewatering in glass house conditions; relative water content and proline levels were higher and showed reduced H202 levels, lipid peroxidation, and electrolyte leakage as a result of which, fiber yield of transgenic plants did not suffer as much as that of their non-transgenic counterparts under drought conditions (Parkhi et al. 2009). GHSP26 cDNA was isolated from G. arboreum and overexpressed in G. hirsutum, and the transgenic plants showed an enhanced drought tolerance (Maqbool et al. 2010). Transgenic cotton was developed by overexpression of Annexin gene from Brassica juncea which showed abiotic stress tolerance and improved fiber development and quality (Divya et al. 2010). Overexpression of Arabidopsis vacuolar H+-pyrophosphatase (AVP1) gene in cotton plants showed more vigorous growth, displayed significantly improved tolerance to both drought and salt stress and 20 % higher fiber yield than that of WT plants in the field under dry land conditions. They concluded that AVP1 may improve crop performance under drought and saline conditions in areas where water and salinity are limiting factors for agricultural production (Pasapula et al. 2011; Zhang et al. 2011a). Constitutive overexpression of G. hirsutum mitogen-activated protein kinase (GhMPK2) in Nicotiana tabacum conferred reduced sensitivity to ABA during both seed germination and vegetative growth and decreased rate of water loss and exhibited enhanced drought and salt tolerance which strongly suggests that GhMPK2 positively regulates salt and drought tolerance in transgenic plants (Zhang et al. 2011b). GhMKK1 is one of the mitogen-activated protein kinases, and it is a crucial regulator of the response to environmental stresses, overexpression in N. benthamiana enhances the tolerance to salt and drought stresses (Lu et al. 2013). Overexpression of Arabidopsis LOS5 cDNA in cotton plants showed 20, 25, and 13 % of more proline content, endogenous ABA level after 3 days water withholding and increase fresh weight under 8 weeks water-deficit condition, respectively, in transgenic plants compared with WT plants (Yue et al. 2012). Overexpression of isopentenyl transferase gene (IPT) from Agrobacterium tumefaciens driven by senescence-associated receptor-like kinase gene (SARK) promoter in G. hirsutum cv. Coker 312 showed increased drought tolerance under water deficit condition (Kuppu et al. 2013). Calcium neurin B-like interacting protein kinase (CIPK6) gene was isolated from cotton somatic embryogenesis and overexpressed in transgenic Arabidopsis were significantly enhances the salt, drought, and ABA stress tolerance and indicating that GhCIPK6 acts as a positive regulator in response to salt and drought stress tolerance (He et al. 2013). Transgenic Arabidopsis with constitute overexpression of G. barbadense receptor-like kinase gene exhibited a reduced rate of water loss in leaves in vitro, along with improved salinity and drought tolerance and increased sensitivity to ABA compared with wild-type Arabidopsis (Zhao et al. 2013). Transgenic tobacco with constitute expression of G. barbadense mitogen-activated protein kinase 3 (GbMPK3) was conferred with enhanced drought tolerance, reduced water loss during drought treatment, and improved plant height and survival rates after rewatering, and they suggested that GbMPK3 may positively regulate drought tolerance through enhanced ROS scavenging ability (Long et al. 2013).
In the previous study, we reported the gene expression profiles under drought stress during the fiber developmental stages (0, 5, 10, and 20 dpa) in the field-grown cotton plants using Affymetrix cotton GeneChip Genome arrays (http://www.affymetrix.com/estore/browse/products.jsp?productId=131430#1_1) and identified a number of differentially expressed transcripts with P value ≤0.01 and log2 fold change ≥2 at various stages analyzed. Based on the transcriptome study, we observed that one of the transcripts (Unigene id: Ghi.10692) encoding CCL (CCR-like) gene was upregulated by 2.5, 239.0, 4.9, and 2.3 folds at 0, 5, 10, and 20 dpa, respectively, as compared to the respective control samples (Padmalatha et al. 2012). The high level upregulation of GhCCL transcript was observed during fiber elongation stage (5 dpa).
In this study, we have chosen the gene GhCCL for molecular characterization and functional analysis in transgenic tobacco. In silico analysis of the GhCCL upstream sequence was carried out to find the various cis-regulatory elements. Ectopic expression of GhCCL in tobacco resulted in water-deficit stress tolerance upon withholding water for 45 days and the transgenic plants made fast recovery upon rewatering. GhCCL transgenic tobacco plants showed similar phenotype as that of WT plants. Further, we observed early germination and faster growth rate in the transgenic plants compared to those in WT plants. This is the first report that demonstrates the involvement of cotton CCL gene in abiotic stress tolerance. We suggest that the ectopic expression of GhCCL gene in transgenic crops will confer tolerance to an array of abiotic stresses.
Materials and Methods
Plant Materials, Growth Conditions, and Stress Treatments
Cotton (G. hirsutum L. cv. Bikaneri Narma) seedlings were raised in a culture room at 25 °C with 16 hrs light: 8 h dark cycles. Seven-day-old cotton seedlings were used for different stress treatments. Seedlings were dipped in 0.5× MS (Murashige and Skoog 1962) medium containing 15 % polyethylene glycol (PEG)-6000, 200 mM each sodium chloride (NaCl), and mannitol. For methyl jasmonate (MeJA), salicylic acid (SA), hydrogen peroxide (H2O2), and ABA treatments, leaves were sprayed with 10 mM each of MeJA, SA, H2O2, and ABA. To investigate the effect of temperature stress, the seedlings were placed in a growth chamber at either high temperature (42 °C) or low temperature (4 °C). For wounding experiments, the seedlings were punched by forceps, and dehydration-stress seedlings were removed from the bottles, and the roots were washed with water then placed on tissue paper under culture room conditions. The samples were collected from all the treatments after 1 and 12 h, frozen in liquid nitrogen, and stored at −70 °C for later use.
For expression profiling of the genes, cotton samples at different boll developmental stages under drought stress conditions were collected with their respective controls. Drought induction under field conditions and related parameters were described previously (Padmalatha et al. 2012). Arabidopsis thaliana ecotype Columbia seeds were sown in pots filled with a coconut pulp/cocofeed, and pots were kept in culture room at 22 °C and 16: 8 h light sources for 2 weeks, and then healthy leaf samples were collected and frozen in liquid nitrogen and stored at −70 °C for later use.
Tobacco (Nicotiana tabacum L. var. Petit Havana) plants have been maintained by subculturing the healthy greenish stem portion in MS medium without any phytohormones in tissue culture room at 25–27 °C temperature with a light intensity of 100 μmol m−2 s−1 with 16 h light: 8 h dark cycles. One-month-old healthy tobacco leaves were used for plant transformation studies. In transgenic glass house, tobacco plants were grown at 25–27 °C temperature with 16 h light: 8 h dark cycle at 300 μmol m−2 s−1 light and 70–80 % relative humidity (RH).
Total RNA and RT-PCR analysis
Total RNA was isolated from cotton bolls collected from control and drought-induced plots at different fiber development stages and also from cotton seedlings subjected to various stresses using Spectrum plant total RNA extraction kit (Sigma, USA). Semiquantitative reverse transcription-polymerase chain reaction (RT-PCR) was employed to investigate the expression profiles of GhCCL. Of total RNA, 1.0 μg was converted to single stranded cDNA using by AffinityScript QPCR cDNA Synthesis Kit (Stratagene, USA). The resulting cDNA was used as a template to perform the RT-PCR analysis using gene specific primer. The PCR conditions were as follows: 94 °C for 1 min, initial denaturation; 30 cycles of 94 °C for 40 s, 60 °C for 40 s, and 72 °C for 60 s, and a final extension at 72 °C for 10 min. The RT-PCR for the house-keeping Ubiquitin1 (accession number EU604080) (Li et al. 2009) gene from cotton was performed under the same conditions for normalization of the samples.
The Full-Length cDNA Cloning of GhCCL and Construction of Phylogenetic Tree
Total RNA was extracted from 5 dpa cotton bolls subjected to drought using the method discussed above. The full-length GhCCL cDNA was amplified by rapid amplification of cDNA ends-PCR (5′ RACE kit, Invitrogen, USA) and reverse transcription-PCR (RT-PCR). The amplified cDNA was cloned in pGEM T-Easy vector and sequenced. The full-length cDNA sequence and coding sequence (CDS) were identified by the FGENESH analysis of softberry software (www.softberry.com/) and the sequence has been submitted to the GenBank. The primer sequences used in this study are listed in Table 1. Phylogenetic tree was constructed using protein sequence of GhCCL and those of the closely related plant species by using MEGA software version 5.20 (Tamura et al. 2011).
Amplification of the Genomic and Upstream Sequence of GhCCL
Genomic DNA was extracted from the cotton leaves using a modified cetyltrimethylammonium bromide (CTAB) method (Porebski et al. 1997). The full-length DNA sequence was amplified by PCR with the primers 5′-CGGGATCCATGCAGTCAGCAGCAGCAAC-3′ (BamHI site underlined) and 5′-CGAGCTCCTAATACACTCCCCTTTGATAC-3′ (SacI site underlined) designed for full-length GhCCL cDNA sequence. The promoter region was amplified with the genome walking method as per the manufacturer’s instructions (Genome walking kit, Clonetech, USA) using primary PCR with gene specific primer (GhCCLGSP2) and genome walker adapter primer1 (GWAP1) and for nested PCR with gene specific primer (GhCCLGSP3) and genome walker nested adaptor primer2 (GWAP2). Genome walking product was cloned in pGEMT-Easy vector and sequenced. The sequences obtained from the different libraries were aligned by Lasergene software to find out the upstream promoter sequence. The sequence has been submitted to the GenBank. The program PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) was used to analyze the cis-elements in the promoter sequences.
Identification of GhCCL Copy Number in Cotton (G. hirsutum L. cv. Bikaneri Narma)
Twenty micrograms of genomic DNA was digested with BamHI, HindIII, and EcoRV (New England Biolabs, USA). The electrophoresis of nucleic acids was performed as described (Sambrook and Russell 2001; Southern 1975), blotted overnight onto a Hybond-N+ membrane (Amersham Biosciences, UK), and fixed at 22 J/cm for 2 min using Stratalinker UV crosslinker (Stratagene, USA). Immobilized nucleic acids were hybridized with α [32P]-dCTP labeled 623-bp GhCCL genomic fragment using mega prime DNA labeling system (Amersham Biosciences, UK). Subsequent to the hybridization and stringent washes the membrane was exposed to the X-ray film with an intensifying cassette in dark and was placed at −70 °C for a week. The exposed X-ray film was developed to visualize the results.
Subcellular Localization of GhCCL-GFP Fusion Proteins and Agroinfiltration Study
The full-length GhCCL coding region was amplified without stop codon using forward primer 5′-GGAAGATCTGATGCAGTCAGCAGCAGCAAC-3′ (BglIII site underlined) and reverse primer 5′-CGGACTAGTATACACTCCCCTTTGATACTCTCG-3′ (SpeI I site underlined). The amplified fragment was restricted and ligated to the binary vector pCAMBIA1302-mGFP, which generated a C-terminal fusion with the modified green fluorescence protein (mGFP) gene driven by the Cauliflower Mosaic Virus (CaMV) 35S promoter. The binary vector was mobilized to A. tumefaciens strain LBA4404 by freeze-thaw method (Hofgen and Willmitzer 1988). Tobacco leaves were infiltrated with induced Agrobacterium culture prepared with 50 mM MES buffer (pH 5.2), 100 mM MgCl2, and 100 μM acetosyringone (Gelvin 2006) by a syringe. The infiltrated plants were kept in the culture room for 4 days. The leaf portions were observed under Leica TCS SP6 confocal microscope (Germany) using bright light, GFP channels and merged to localize the expressed 35S-GhCCL::mGFP fusion and 35S-mGFP proteins.
Construction of Plant Transformation Vector and Tobacco Transformation
Full-length coding sequence of GhCCL cDNA was isolated from drought-induced 5 dpa cotton bolls of G. hirsutum L. (cv. Bikaneri Narma) with the primers of 5′-CGGGATCCATGCAGTCAGCAGCAGCAAC-3′ (BamHI site underlined) and 5′-CGAGCTCCTAATACACTCCCCTTTGATAC-3′ (SacI site underlined), and the amplified gene fragment was inserted in the corresponding sites of pBI121 binary vector (Clontech Laboratories, USA) by replacing the GUS reporter gene under the control of constitutive CaMV35S promoter. The derived vector was named as pBI121-35S::GhCCL. Another binary vector was constructed with Arabidopsis thaliana rd29A gene specific promoter (Shinozaki and Shinozaki 1994) which was amplified by PCR with primer pair 5′-CCCAAGCTTGCCATAGATGCAATTC-3′ (HindIII site underlined) and 5′-GCTCTAGATTTCCAAAGATTTTTTTC-3′ (XbaI site underlined) from the genomic DNA isolated from of A. thaliana ecotype Columbia using modified CTAB method (Porebski et al. 1997) and ligated to replace CaMV35S promoter in pBI121-35S::GhCCL binary vector. The derived vector was named pBI121-rd29A::GhCCL. These binary vectors were individually mobilized into A. tumefaciens strain LBA4404 by freeze-thaw method (Hofgen and Willmitzer 1988). Agrobacterium-mediated transformation was carried out with co-cultivation of leaf discs obtained from in vitro grown tobacco (Nicotiana tabacum L. var. Petit Havana) plants, for 15 min with Agrobacterium strain LBA4404 harboring the recombinant binary vectors. Transformed leaf discs were selected on MS agar supplemented with 6-benzylaminopurine (2.5 mg l−1), naphthalene acetic acid (0.1 mg l−1), cefotaxime (250 mg l−1), and kanamycin (200 mg l−1). Regenerated shoots were rooted on phytohormone-free MS basal medium containing cefotaxime (250 mg l−1) and kanamycin (200 mg l−1) (Sunilkumar et al. 1999). The profusely rooted plantlets were further subcultured and maintained in tissue culture condition for further molecular and other stress-related studies. The clones were transferred to soil and grown in a glasshouse. The WT and the transgenic plants were studied under similar conditions for comparative molecular and physiological analyses.
Molecular Analysis of Transgenic Plants
PCR, Southern, and Northern Blot Analysis
Five lines of 35S::GhCCL (named as A1, A2, A3, A5, and A6) and 9 lines of rd29A::GhCCL (named as B1, B3, B6, B11, B15, B17, B18, B19, and B20) of transgenic and WT-rooted plantlets which were maintained in the glass house were selected for the isolation of the genomic DNA from leaf samples by following the method of Rogers and Bendish (1994). PCR analysis of the transgenic plants (T 0) was performed using NPTII gene specific primers (forward primer 5′-CCGGAATTCATGATTGAACAA-3′ and reverse primer 5′-CCCAAGCTTCAGAAGAACTC-3′) and GhCCL gene specific primers (forward primer 5′-CGGGATCCATGCAGTCAGCAGCAGCAAC-3′ (BamHI site underlined) and 5′-CGAGCTCCTAATACACTCCCCTTTGATAC-3′) (SacI site underlined). PCR analysis was also carried out with selected T 1 transgenic plants (A1, A2, A3, A5, A6, B1, B3, B6, B18, and B20) to amplify the specific 700-bp and 432-bp amplification fragments of NPTII and GhCCL genes, respectively. The binary vectors pBI121-35S::GhCCL and pBI121-rd29A::GhCCL were used as positive controls, and non-transformed plants (WT) and water control (W) were used as negative and environmental controls, respectively. PCR amplification was carried out in a thermal cycler (Eppendorf, Germany) programmed with an initial denaturation of DNA at 94 °C for 5 min, followed by 30 cycles of 94 °C for 1 min, an annealing step at 60 °C for 1 min and 72 °C for 1 min, and a final extension step at 72 °C for 10 min. The amplified products were analyzed by electrophoresis on 1.0 % agarose gel.
Southern blot analysis was performed to confirm the stable integration and copy number of the NPTII gene. A 10-μg of the genomic DNA from WT tobacco and T 1 transgenic plants along with pBI121-35S::GhCCL binary plasmid was digested with BamHI, which restricts T-DNA at single site. The digested genomic DNA was resolved on 0.8 % agarose gel and blotted on a nitrocellulose membrane (Hybond-N+, Amersham Pharmacia, UK). The blot was hybridized at 65 °C with α [32P]-dCTP labeled 700-bp NPTII fragment using mega prime DNA labeling system (Amersham Biosciences, UK). The processing of the membrane was carried out as described before. Total RNA was isolated from T 1 transgenic and WT tobacco plants using TRIzol reagent (Invitrogen Life technologies) as per manufacturer’s instructions. Fifteen micrograms of total RNA was resolved on a 1.2 % formaldehyde denaturing gel and transferred to the nitrocellulose membrane (Hybond-N+, Amersham Biosciences, UK) using diethylpyrocarbonate (DEPC) treated 20× SSC. Pre-hybridization and hybridization were performed with ULTRAhyb hybridization buffer (Ambion, USA) according to the manufacturer’s instructions. The blot was hybridized at 42 °C with α [32P]dCTP labeled 432-bp GhCCL fragment labeled with mega prime DNA labeling system (Amersham Biosciences, UK). The hybridized membrane was washed at 42 °C, and the membrane was exposed to X-ray film with an intensifying cassette under dark conditions. The cassette was placed at −70 °C for a day, and the exposed X-ray film was developed to visualize the results.
Analysis of Transgenic Tobacco Plants for Abiotic Stress Tolerance
Leaf Disc Assay
Leaf discs (1.0 cm2) were excised from healthy and fully expanded tobacco leaves of 4–6-week-old WT and T 0 transgenic plants (A3, A6, B6, and B20) using a cork borer. Leaf discs were floated on 20 ml solutions of 10 % PEG, 200 mM each of mannitol for dehydration stress and NaCl for salinity stress. The experiments were carried out in three replicates. The plates were placed in the culture room for 72 h, and subsequently, the number of greenish and yellowish leaf disc were scored and plotted.
Seed Germination Assay Under Stress
For germination studies, WT and T 1 transgenic tobacco seeds were surface sterilized and germinated on 90-mm-Petri dishes containing half strength MS agar medium supplemented with 10 % infused PEG-6000, 200 mM each mannitol or NaCl. Germination assays were carried out in three replicates, and the germination rate was scored after 20 days. Tolerance to PEG-6000, mannitol and NaCl stresses was judged based on the ability of the seeds to germinate and grow on the amended media. The number of germinated seeds was expressed as a percentage of the total number of seeds sown, and the performance of germination was compared to that of WT seeds.
Long-Term Growth Performance of Transgenic Plants Under Stress
Long-term stress effect was evaluated by subculturing the stem portion of WT and transgenic plants in glass bottles containing MS agar media supplemented with 10 % PEG, 200 mM each of mannitol and NaCl. The growth performance was observed after 45 days. The fresh weights were measured and plotted.
Water-Deficit Stress Tolerance of Transgenic Tobacco
WT and T 1 transgenic tobacco plants were transferred from the culture conditions to soil in the glass house for water-deficit stress experiment. Water was withheld for 45 days at the time of flowering. The relative water content (Weatherley 1950, 1951) and total chlorophyll (Arnon 1949) were analyzed at 0 and 15 days after withholding water. Plants were allowed to recover for 20 days post 45 days of water withholding to observe the recovery in WT and transgenic plants.
Statistical Analyses
Each experiment was replicated three times, and the values were expressed as means ± SE. All mean comparisons were done using paired t test for independent samples. The measurements for determining different treatments or times were analyzed by one-way analysis of variance (ANOVA).
Results
Isolation, Molecular Cloning, and Structural Organization of GhCCL
In the present study, we analyzed the possible role of CCL gene in water-deficit stress tolerance. In Affymetrix GeneChip, the probe for the transcript representing CCL gene was obtained from the cotton EST (GenBank Acc. No ES816870.1) which is 605 nucleotide in length with poly-A tail at 3′ end. In order to isolate the complete gene sequence, the 5′ RACE PCR technique was followed using degenerate primers to sequence 5′ region. The RACE PCR product was cloned in pGEMT-Easy vector and sequenced. The sequence obtained was 671-bp in length with a 48-bp of 5′ untranslated region (UTR) and 173-bp of 3′ UTR. Further, coding region of the gene was identified by FGENESH of softberry online software using aligned sequence. The cDNA contained a 432-bp open reading frame (ORF) encoding a protein of 144 amino acids with a calculated molecular weight of about 15.43-kDa and an isoelectric point of 4.72. The full-length cDNA sequence of GhCCL was submitted to NCBI GenBank (accession number KF553636). The coding region (ORF of 432 bp) was amplified using specific primers having restriction sites and cloned in pGEMT-Easy vector and sequenced. Further, the ORF of GhCCL was cloned in a binary vector pBI121 with two different promoters viz., the constitutive Cauliflower Mosaic Virus 35S promoter and stress specific Arabidopsis thaliana rd29A promoter. The recombinant binary vectors were named pBI121-35S::GhCCL and pBI121-rd29A::GhCCL, respectively (Fig. S1) and mobilized to Agrobacterium strain LBA4404 by freeze-thaw method for plant transformation.
Identification of Introns and Copy Number Determination of GhCCL Gene
The primers used to amplify the CDS were employed to isolate the genomic fragment of GhCCL from cotton (G. hirsutum L. cv. Bikaneri Narma) DNA and the genomic fragment was cloned in pGEMT-Easy vector and sequenced. The comparison of the full-length cDNA sequence with the corresponding genomic DNA sequence revealed that the coding region of the GhCCL is interrupted by two introns (Fig. 1a). The 844-bp long sequence comprises of 5′UTR (1–48 bp), Exon 1 (49–239 bp), Intron 1 (240–334 bp), Exon 2 (335–494 bp), Intron 2 (495–595 bp), Exon 3 (596–671 bp), and 3′UTR region (672–844 bp).
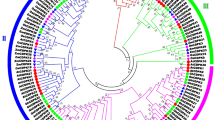
Organization of GhCCL gene. a Schematic representation of the GhCCL and exon-intron organization. b Detection of copy number of GhCCL by southern blot analysis. Genomic DNA (20 μg) from cotton (cv. Bikaneri Narma) leaves was digested with BamHI, HindIII, and EcoRV respectively, separated on a 0.8 % agarose gel and hybridized with α [32P] dCTP–labeled 623-bp of GhCCL as probe. Molecular marker in kb is indicated on the left. Note that BamHI and EcoRV have no restriction sites and HindIII have single restriction site within the probe sequence. c The phylogenetic tree showing the relationship between Gossypium hirsutum CCL and other plant proteins. Phylogram was generated based on protein sequences using the neighbor joining algorithm of MEGA software, version 5.05. Numbers above or below branches indicate bootstrap values (>50 %) from 1,000 replicates. The plant name is followed by the protein ID
To characterize the GhCCL in the genome, we performed Southern blotting, which could be used to obtain preliminary information on organization of the corresponding genomic sequence and the number of gene copies in cotton genome. Genomic DNA of G. hirsutum L. cv. Bikaneri Narma was digested with three different restriction enzymes and blotted to Hybond-N+ membrane (Amersham Biosciences, UK). The blot was hybridized with α [32P] dCTP-labeled 623-bp GhCCL fragment under high stringency conditions. The restriction enzyme HindIII has single site in the probe region, yielded two bands, whereas BamHI and EcoRV having no restriction sites resulted in a single band, indicating that GhCCL is probably present as a single copy within the G. hirsutum genome (Fig. 1b).
Sequence Analysis of GhCCL
The sequence of GhCCL was aligned with Arabidopsis CCL sequence using BLAST algorithm to identify the homology between these sequences. We observed 69 and 48 % identity in nucleotide and protein sequences, respectively. Nucleotide composition was analyzed using BioEdit program. The percentages of A, C, G, and T nucleotides are 27.08, 28.24, 23.61, and 21.06, respectively with 51.85 % of GC content in GhCCL nucleotide sequence whereas the protein sequence has a high content of cysteine (28.24 %) followed by alanine (27.08), glycine (23.61), and threonine (21.06). Further, putative conserved domain called lir1 (light, induced rice 1) superfamily (Fig. S2) was identified by NCBI-Conserved Domain Database using protein sequence of GhCCL.
BLASTP program was employed to identify the homology of the GhCCL protein sequence with that of other plant species. The alignment results showed maximum identity of 78 % with Ricinus communis putative light regulated protein precursor followed by 76 % with Bruguiera gymnorrhiza CCR protein, 75 % with Lotus japonicus unknown protein, 72 % with light regulated protein of Vitis vinifera and Eutrema halophilum. Phylogenetic tree showed that most of the sequences are aligned with light-regulated protein sequences of different plant species within two major clusters (Fig. 1c)
Expression Profiles of GhCCL in Cotton Leaves and Boll Developmental Stages
Expression profiles of GhCCL gene were also studied in leaves and bolls of cotton at different stages (−2 days pre-anthesis to 0, 2, 3, 4, 5, 6, 7, 8, 10, and 20 days post anthesis) under control and drought induced in field conditions. Expression of the gene was observed in leaves and up to 8 dpa boll samples in control and drought-induced samples. Surprisingly, the expression of the gene was observed in 10 and 20 dpa of drought-induced bolls whereas there was no expression in respective control samples (Fig. 2a). The samples used for RT-PCR and earlier microarray experiments were collected from two different seasons. In earlier microarray experiments (Padmalatha et al. 2012), maximum fold changes were observed in 5 dpa cotton bolls, but the RT-PCR study showed differential expression of GhCCL only in 10 and 20 dpa cotton bolls. This observation suggests that the circadian clock and environmental conditions may affect the expression levels in a stage-specific manner in plants. It also indicates that GhCCL gene may play an important role during drought stress at fiber elongation stage of cotton boll development.
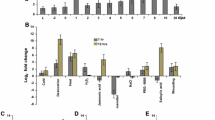
Expression profiles of GhCCL in stage-specific control and drought-induced cotton tissues and seedlings subjected to various stresses. a Expression profile of GhCCL in field-grown cotton during different boll developmental stages under drought stress and control conditions. L leaf,−2 days pre anthesis, 0–20 days post anthesis, and lower panel indicates the expression of cotton GhUBI1 internal gene, respectively. b Expression profile of GhCCL in response to different stresses and phytohormone treatments in cotton seedlings. One-week-old cotton seedlings were subjected to 200 mM NaCl, 200 mM mannitol, 15 % PEG-6000, cold (4 °C), heat (42 °C), dehydration, wounding, 10 mM methyl jasmonic acid, 10 mM salicylic acid, and 10 mM hydrogen peroxide for 1 and 12 h. Above panel and lower panel indicate the expression of GhCCL gene and cotton GhUBI1 internal gene, respectively
Expression Profiles of GhCCL in Response to Diverse Abiotic Stresses in Cotton Seedlings
To determine the expression pattern of the GhCCL gene under various abiotic stress conditions, the seven-day-old cotton seedlings were exposed to PEG, mannitol, NaCl, cold, heat, dehydration, and wounding treatments for 1- and 12-hour periods to evaluate the effect of abiotic stresses. Total RNA isolated from treated cotton seedlings was used for semiquantitative RT-PCR. In mannitol, NaCl- and cold-treated seedlings, transcripts of GhCCL were observed within 1 h, and the expression level declined at 12 h. In the case of seedlings exposed to heat and wounding, transcripts of GhCCL accumulated within 1 h, and their expression is even seen at 12 h. In dehydration- and H2O2-treated seedlings, low level of expression of transcripts was observed within 1 h, and high level of its expression was observed at 12 h. There was very low or no expression of the gene upon treatment with PEG for minimum and maximum periods tested in this study. We also observed the expression of the GhCCL when the cotton seedlings were treated with MeJA and SA, which are plant signal molecules that may accumulate upon pathogen infection and are involved in plant defense-signaling pathways (Agrawal et al. 2001). The expression of the gene was detected in both MeJA- and SA-treated seedlings (1 and 12 h periods) (Fig. 2b).
In Silico Prediction and Subcellular Localization of GhCCL
GhCCL is localized in chloroplasts as predicated by in silico analysis of GhCCL protein sequence using ProtComp v. 9.0 (www.softberry.com/) and ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP/) programs (Table S1). To validate this prediction, mGFP was fused in frame to the C-terminal region of GhCCL under the control of CaMV 35S promoter. The binary vectors 35S-mGFP (Fig. 3a) and 35S-GhCCL::mGFP (Fig. 3b) were used for transient expression studies in tobacco leaves by agroinfiltration method. The injected leaf portion was visualized under TCS SP6 confocal microscope (Leica, Germany) after 4 days. As shown in Fig. 3c, the 35S-mGFP construct showed homogenous green fluorescence throughout the cells, whereas in 35S-GhCCL::mGFP, green fluorescence of GFP was localized in the chloroplasts, confirming the chloroplast localization.
Subcellular localization of the GhCCL protein in tobacco leaf tissue. a, b T-DNA regions of pCAMBIA1302 binary vector carrying 35S-mGFP construct and 35S-GhCCL::mGFP binary fusion construct, respectively. c Transient expression of 35S-mGFP and 35S-GhCCL::mGFP in tobacco leaf tissue by Agroinfiltration method. The tissues were analyzed by Leica TCS SP6 confocal microscope after 4 days of agroinfiltration (scale bars in the lower right corners represent 20 μm)
GhCCL Promoter Analysis
To explore the mechanism underlying GhCCL expression patterns, a 1,028-bp 5′ flanking region of GhCCL was isolated by using Genome Walker kit (Clontech, USA) following the manufacturer’s instructions. The genomic DNA sequence was submitted to the NCBI GenBank (accession number KF547867). Cis-acting regulatory elements were predicted using PlantCARE online tool (Lescot et al. 2002). A number of potential regulatory motifs corresponding to several known cis-acting elements related to light, stress and hormone responsive, and tissue specific expression elements were predicted (Table 2). Light-responsive elements such as G-box, I-box, as-2-box, rbcS-CMA7a among other related motifs which are involved in light response were identified. Interestingly, MRE-motif with a MYB binding site was also identified within the promoter region. Cis-regulatory elements responsive to various stresses such as elicitor (EIRE), heat stress (HSE), low temperature (LTR), and MYB binding site involved in drought inducibility were also identified. The motifs CGTCA, TGACG, and P-box elements were predicted to involve in MeJA and Gibberellic acid (GA) response, respectively. Cis-elements involved in tissue specific expression such as vacuole, meristem, endosperm, and shoot were predicted by in silico analysis of the GhCCL promoter sequence. Interestingly, the circadian element which is involved in the endogenous mechanism that allows the plant species to time their physiological changes to predict day/night cycles was also predicted. Taken together, the upstream sequence analysis showed that GhCCL gene may be under the control of a light, biotic, and abiotic stress-responsive promoter which is also expressed in a tissue specific manner in plants.
Abiotic Stress Tolerance of Transgenic Tobacco
Overexpression of GhCCL in Transgenic Plants Shows no Negative Effect on Plant Growth
To evaluate the effect of over expression of GhCCL in tobacco, the GhCCL cDNA was fused to Cauliflower Mosaic Virus 35S promoter and Arabidopsis thaliana rd29A promoter and transferred to tobacco by Agrobacterium-mediated transformation. After selection on kanamycin-containing medium, the putative transgenic tobacco plants were further analyzed by PCR in T 0 and T 1 plants (Fig. S3a, b, c). Southern blot analysis of T 1 transgenic plants demonstrated that the transgene was stably integrated into the genome of transgenic tobacco lines (Fig. 4a) with each carrying one or multiple copies of the transgene. RNA gel-blot analysis of the transgenic plants grown under normal and dehydration stress conditions confirmed that the transgene is expressed constitutively and in a stress-inducible manner in CaMV35S and rd29A promoter transgenic plants, respectively, whereas no expression was observed in the WT plants, as expected (Fig. 4b). No growth inhibition or phenotypic alterations were observed in transgenic plants compared to the WT. The transgenic tobacco plants developed normally and did not exhibit any growth retardation. Seed setting was also normal.
Southern and Northern blot analysis of transgenic tobacco plants. a Southern blot analysis of GhCCL expressing transgenic T 1 tobacco plants showing the copy number of NPTII. Genomic DNA of transgenic events was digested with BamHI that has no restriction site in the probe and single cutter in the T-DNA region. The blot was hybridized with α [32P] dCTP–labeled with NPTII probe. Molecular weight marker in kb indicated on the left. b RNA gel-blot analysis of wild-type and GhCCL expressing T 1 tobacco plants. The ethidium bromide-stained gel demonstrates equivalent RNA quantities loaded in each lane (A3 and A6 are 35S::GhCCL, B6 and B20 are rd29A::GhCCL independent transgenic lines)
Leaf Disc and Germination Assay for Dehydration, Osmotic, and Salt Stresses
The WT and transgenic plants were exposed to dehydration, osmotic, and salt stresses, and their response was examined by leaf-disc senescence assay under PEG-6000, mannitol and salt treatments, respectively. The leaf discs excised from mature leaves of WT and T 0 transgenics of 35S::GhCCL (A3, A6) and rd29A::GhCCL (B6, B20) were used for the stress analysis. The maximum percentage of green leaf discs was observed in line B6 (88 %) followed by A3 (85 %) and A6 (84 %) after 72 h of treatment with 10 % PEG solution. The lines A3 and A6 showed 86.6 % of green leaf discs followed by B20 (81.1 %) and B6 (76.6 %) after 72 h of treatments with 200 mM mannitol solution. During the 72 h period of incubation in PEG-6000 and mannitol solution, the leaf discs from WT plants bleached drastically compared to the transgenic leaf discs (Fig. 5a–c). Interestingly, in NaCl-induced salinity stress samples, the WT tobacco plant performed better than the different transgenic lines tested (Fig. 5e). Leaf discs treated with distilled water devoid of PEG-6000, mannitol, and salt appeared green in both WT and the transgenics, and the percentage of green and bleached yellow leaf discs was calculated (Table S2) by ANOVA test and plotted (Fig. 5b–f).
Leaf disc senescence assay of WT and T 0 Transgenic plants. Photograph and bars showing the phenotypic differences and percentage response of the effect of a, b 10 % PEG-6000 c, d 200 mM mannitol and e, f 200 mM NaCl on the leaf-discs floated on distilled water from WT and T 0 transgenic tobacco plants and leaf discs were observed after 72 h, above and below panel as wild-type, GhCCL transgenic tobacco lines, respectively. Each bar value represents the mean ± of triplicates (ANOVA test), and means are significantly different from control at P < 0.05 according to Dunnett’s test at each treatment
To test the ability of seeds to germinate under dehydration, osmotic, and salt stresses, seeds from WT and transgenics of 35S::GhCCL (A3, A6) and rd29A::GhCCL (B6, B20) were surface sterilized and placed on 0.5× MS agar medium supplemented with 10 % PEG-6000, 200 mM mannitol and 200 mM NaCl. The germination rate was observed (Fig. 6), and the percentage of germination was calculated (Table S3) after 20 days of germination. Higher percentage of germination was observed in the case of transgenic seeds (A3, A6, B6, and B20) as compared to the WT grown in MS media supplemented with PEG, mannitol whereas no significant differences were observed between transgenic seedlings and WT plants grown in MS agar plates without any stress. Further, seedlings of transgenic lines showed faster growth rate and produced thick green leaves compared to seedlings of WT which showed stunted growth and developed pale green leaves in the later stages. However, under NaCl-induced salinity stress, the germination rate of transgenic plants showed varied results.
Performance and percentage (%) of WT and T 1 transgenic seed germination on amended media. Sterilized seeds of WT and transgenics were germinated in petriplates having a, b 10 % PEG-6000 c, d 200 mM mannitol and e, f 200 mM NaCl on MS agar medium and observed growth performance and calculated percentage of germination after 20 days. Each bar value represents the mean ± of triplicates (ANOVA test), and means are significantly different from control at P < 0.05 according to Dunnett’s test at each treatment
Higher percentage of germination in MS media (10 % PEG-6000) was observed in A3 (97.3 %) and A6 (95 %) lines of 35S::GhCCL followed by B6 (82.6 %) and B20 (49.6 %) lines of rd29A::GhCCL whereas only 41.3 % germination was observed in WT tobacco seeds (Fig. 6a, b). Similarly, higher percentage of germination in MS media supplemented with 200 mM mannitol was observed in A3 (97.3 %) and A6 (86.3 %) lines of 35S::GhCCL followed by B6 (67.6 %) and B20 (66.6 %) lines of rd29A::GhCCL whereas only 33.6 % germination was observed in WT plants (Fig. 6c, d).
For salt tolerance analysis, surface-sterilized seeds of WT and transgenics were germinated on MS agar supplemented with 200 mM NaCl. The percentage of germination was observed and calculated after 20 days of sowing (Fig. 6e, f). Higher germination percentage was observed in A6 of 35S::GhCCL (76.6 %) followed by B6 of rd29A::GhCCL (53.3 %), A3 of 35S::GhCCL (41.3 %) and B20 of rd29A::GhCCL (27.6 %), whereas 25.6 % of germination was observed in WT plants. However, in the case of transgenic lines, we observed poor growth rate of seedlings grown under salt stress (200 mM NaCl) as compared to that of the seedlings grown under dehydration and osmotic stresses. Transgenic lines showed better tolerance to salt stress (200 mM NaCl) in seed germination assay as compared to leaf disc senescence assay.
In another set of experiment, stem portions of the WT and transgenic plants (A3, A6, B6, and B20) were subcultured in MS media supplemented with 10 % infused PEG-6000, 200 mM each of mannitol and NaCl for 45 days. Transgenic plants grew normally and initiated early root formation whereas the WT plants showed stunted growth, delayed root formation, and pale green leaves (Fig. 7a–e). Percentage of fresh weight was calculated in the WT and transgenic plants after 45 days of exposure to the stress media (Table S4). In PEG-6000, higher fresh weight was observed in B6 (67.8 %) of rd29A::GhCCL followed by A6 (65.9 %) of 35S::GhCCL, B20 (62.8 %) of rd29A::GhCCL and A3 (57.4 %) of 35S::GhCCL, whereas only 48.5 % of fresh weight was observed in WT plants (Fig. 7b). In 200 mM mannitol, higher fresh weight was observed in A6 (64.4 %) of 35S::GhCCL followed by B6 (63 %) of rd29A::GhCCL, B20 (59.1 %) of rd29A::GhCCL and A6 (51.1 %) of 35S::GhCCL, but in WT plants, only 31.7 % accumulation of fresh weight was observed (Fig. 7d). Similarly, in 200 mM NaCl, higher fresh weight was observed in B20 (71.1 %) of rd29A::GhCCL followed by A3 (68.5 %) of 35S::GhCCL, B6 (57.9 %) of rd29A::GhCCL and A6 (49.5) of 35S::GhCCL whereas only 40.4 % fresh weight accumulated in WT plants (Fig. 7f). Further prolonged growth under stress resulted in stunting and bleaching of the WT plants.
Long-term growth performance and fresh weight of WT and transgenic tobacco plants under stress medium. Stem portion of WT and transgenic tobacco were subcultured to bottles having a, b 10 % PEG-6000 c, d 200 mM mannitol and e, f 200 mM NaCl on MS agar medium and growth phenotypes were observed after 40 days. Each bar value represents the mean ± of triplicates (ANOVA test), and means are significantly different from control at P < 0.05 according to Dunnett’s test at each treatment
Stress Tolerance Study Under Glasshouse Conditions
The WT and T 1 transgenic lines (A3, A6, B3, and B20) were maintained in a glass house and water-deficit stress was imposed by withholding the water at the flowering stage (Fig. 8a) as described in “Material and methods”. Severe leaf wilting was observed in the case of WT plants but not in the case of transgenic plants even after 30 days of withholding the water (Fig. 8b). The stress was prolonged further to another 15–20 days, and we observed complete wilting of WT plants after 45 days whereas transgenic plants retained few green leaves, and even the stems remained green (Fig. 8c). We also tested the recovery capacity of the plants by providing water after water-deficit stress treatment for 45 days. Transgenic plants recovered within 20 days of providing the water and produced new branches and leaves. Flowering was also observed in the later stage whereas WT plants did not show any recovery as they wilted completely after the stress treatment (Fig. 8d).
Evaluation of water-deficit stress tolerance, relative water content, and total chlorophyll content analysis in WT and transgenic plants under glass house conditions. a WT and transgenic plants at flowering stage and 0 days of withholding water. b After 30 days of withholding water. c After 45 days of withholding water. d Plants recovered after 20 days of rewatering. e Percentage of relative water content at 0 days and after 15 days of withholding water. f Total chlorophyll content at 0 days and after 15 days of withholding water. Each bar value represents the mean ± of triplicates (ANOVA test), and means are significantly different from control at P < 0.05 according to Dunnett’s test at each treatment
Relative water content (RWC) and chlorophyll content were estimated in WT and transgenic plants under water-deficit stress conditions at 0 and 15 days after withholding water. The percentage of RWC (Table S5) was in the range of 50.1 to 55.2 % in transgenic plants and 49.7 % in WT at 0 days of water withholding and the percentage of RWC in transgenics almost remained the same as at 0 dpa, but in WT, it declined from 49.7 to 43.1 % after 15 days of withholding water (Fig. 8e). Similarly, total chlorophyll content (Table S6) was in the range of 3.38 to 3.79 mg/g fw in transgenics and 3.41 mg/g fw in WT at 0 days of water withholding, and the total chlorophyll content was estimated in the range of 3.34 to 3.7 mg/g fw in the transgenics and 3.04 mg/g fw in WT after 15 days of withholding water (Fig. 8f). We observed the total chlorophyll content declined drastically in WT whereas in the transgenics, slight reduction was observed after 15 days of water-deficit stress. These results clearly suggested that the overexpression of GhCCL can impart tolerance to water-deficit stress in transgenic tobacco.
Discussion
Cotton is the world’s primary fiber crop and is a major agricultural commodity in over 30 countries. Like many other global commodities, sustainable cotton production is challenged by restricted natural resources. In response to the anticipated increase of agricultural water demand and constrains in water availability, research efforts in developing drought tolerant crops that can efficiently use water are being extensively carried out. Water-deficit stress is one of the most important factors that affect plant growth and development (Boyer 1982). Worldwide crop losses due to drought have multi-billion dollar impact on the economies (Mittler 2006; Mittler and Blumwald 2010). Severe drought-induced yield reduction has been reported in crops like maize, barley, wheat, rice, and cotton (Wahid et al. 2007; Frederick et al. 2001; Pettigrew 2004). Although cotton is considered relatively drought-tolerant crop, water deficit reduces the lint quality and yield (Antony and Kutty 1975; Guinn and Mauney 1984a, b). Reduction in lint yield in cotton is due to reduced boll production because of fewer flowers and greater boll abortion when drought intensity is greater.
Drought stress impacts a network of plant gene expression mechanisms that lead to the reprogramming of a variety of physiological and metabolic processes in accordance with the stress response. Early studies primarily used model plant species to identify a wide spectrum of genes that are involved in different levels of metabolism, signal transduction, osmotic regulation, stress response, and gene regulation (Seki et al. 2002; Rabbani et al. 2003; Umezawa et al. 2006). Compared to the conventional breeding methods, the genetic transformation technology has become an efficient way to accelerate the process of improving agronomic traits and economic characteristics of crops by incorporating exogenous genes encoding the desired traits (Zhang et al. 2011c).
Based on the transcriptome analysis of cotton during fiber development stages under drought-stress condition, we selected a transcript representing CCL gene which is highly expressed at early fiber elongation stage (5 dpa) for molecular characterization and functional analysis in transgenic tobacco. In the present study, we developed transgenic tobacco plants by overexpressing the coding region of GhCCL gene under constitutive (CaMV35S) and stress-specific (rd29A) promoters. The full-length gene and its 5′ upstream gene sequence were isolated using 5′ RACE and genome walking PCR, respectively. The full-length gene is 844 bp in length with the coding region of 432 bp. Amplification of GhCCL gene from genomic DNA shows the presence of two introns. Further, subcellular localization studies showed that GhCCL is localized in chloroplasts. The gene was annotated earlier in TAIR database (http://arabidopsis.org/servlets/TairObject?accession=locus:2090837). GhCCL gene is present as single copy in the cotton (G. hirsutum L.) genome as identified by Southern hybridization analysis. We also observed that the GhCCL gene expression is modulated when cotton seedlings were exposed to various biotic and abiotic stresses. Analysis of 5′ upstream sequence showed the presence of a number of potential regulatory motifs involved in regulation of light (G-box, I-box, as-2-box, rbcS-CMA7a), stress (EIRE, HSE, LTR, and MYB), hormone responses (Methyl Jasmonic acid and Gibberellic acid) and tissue specific expression elements. Analysis of the promoter region shows that the biotic and abiotic stresses can alter the expression of GhCCL gene. Analyzed sequence showed conserved domain for light regulation protein lir1 family members. Reimmann and Dudler (1993) demonstrated that the expression of lir1 mRNA is controlled by light and circadian clock. The mRNA accumulates in the light, reaching maximum and minimum steady-state levels at the end of the light and dark period, which regulates growth and development and resets the circadian clock (Harmer et al. 2000). It is also reported that differential expression of many genes such as CCL is upregulated under low temperature stress in Arabidopsis (An et al. 2012). In support of this, our studies also proved that the GhCCL gene and its promoter region contain light regulatory, low temperature, and circadian clock responsive elements.
Cotton is most sensitive to cold at the beginning of the day and to high temperatures in the evening (Rikin et al. 1993). It has been suggested that this circadian-controlled gating of the timing of sensitivity to extreme temperatures might be a way for the plant to distinguish between changes in temperatures during the course of the day and seasonal changes in temperature. Microarray experiments have shown that around 70 % of the known clock-controlled genes may also be regulated by cold, salt, or drought stresses and regulate the mRNA levels of some pathogen-related genes in Arabidopsis as indication of the importance of the circadian system in regulating stress response (Kreps et al. 2002; Yakir et al. 2007b). An important adaptive feature of the circadian clock is its ability to be entrained by environmental signals such as changes in light or temperature. In general, RNA stability has an important role in the regulation of gene expression in eukaryotic cells (Ross 1995; Meyer et al. 2004). Several putative entrainment points have been described for the Arabidopsis oscillator. One such entrainment point is the activation of CCA1 and LHY transcription by light (Wang and Tobin 1998; Kim et al. 2003). Another well-characterized effect that light has on the Arabidopsis oscillator is the regulation of proteasomal degradation of TOC1 and APRR5 proteins that is mediated by ZEITLUPE (ZTL), an F-box motif and Kelch domain protein (Mas et al. 2003; Kiba et al. 2007). ZTL itself is stabilized by a blue light-mediated interaction with GI (Kim et al. 2007). Finally, the translation rate of LHY may be light regulated; light causes an increase in the translation of LHY in transgenic plants expressing LHY from a constitutive promoter (Kim et al. 2003). Concluded that, CCA1 transcript stability and its degradation rate is regulated by light and is critical to ensure the circadian oscillator is accurately entrained by environmental changes (Yakir et al. 2007a).
The coding region of GhCCL was cloned in a binary vector under constitutive CaMV35S (35S::GhCCL) and stress-responsive rd29A promoter (rd29A::GhCCL) and overexpressed in transgenic tobacco. The leaf discs of the transgenic plants showed better response under 10 % PEG and 200 mM mannitol stress. However, the results showed that the transgenic lines were more prone to salinity stress.
Transgenic tobacco plants (lines # A3, A6, B6, and B20) grown in MS media supplemented with 10 % PEG, 200 mM each of mannitol and NaCl showed better growth compared to that of WT plants which suggested that high level expression of the gene might confer stress tolerance. Similarly, pot-grown transgenic and WT tobacco plants were subjected to water-deficit stress at the flowering stage. Transgenic plants could produce a few green leaves, and their stems remained green even after 45 days of withholding water whereas the WT plants completely dried. The transgenic plants recovered quickly upon rewatering and flowered normally, but the WT plants did not exhibit any signs of recovery demonstrating that the overexpression of GhCCL gene improved the plant growth, development, and tolerance to water-deficit stress.
Molecular mechanisms and other biological processes by which CCL gene is involved in stress tolerance and/or adaptation mechanism need further investigations. An earlier study reported that the CCL protein was probably involved in various physiological functions related to the adaptation of the plants to water deficit and/or saline conditions and has a vital role in the salt adaptive physiological processes in the halophyte Suaeda maritima exposed to NaCl treatment (Sahu and Shaw 2009). CCL gene encodes a highly unstable mRNA, the stability being regulated by the circadian clock. The transcript of this gene is significantly more stable in the morning than in the afternoon (Lidder et al. 2005). A high accumulation of the transcripts of the gene was observed when the plants were harvested at the evening time. It appeared that the salt treatment either had increased the stability of the CCL transcripts or had enhanced the expression of the gene in the plant (Sahu and Shaw 2009). Interestingly, several genes of the circadian clock were demonstrated to be involved in abiotic stress responses, and an intimate relationship between the circadian clock and ABA regulation was established (Sanchez et al. 2011). Since ABA-induced responses play a major role in conferring water-induced stress whether CCL gene also acts through ABA dependent pathway has to be looked into. However, the proteins of circadian clock like TOC1 and PRR5 are localized to the nucleus (Wang et al. 2010) in contrast to CCL protein which is found to be localized in the chloroplast. Also, CCL gene in Arabidopsis is under the control of circadian clock through selective mRNA degradation (Lidder et al. 2005). Whether CCL gene functions as a signal from or to the chloroplast in regulating its functions is not clear at this stage. Nevertheless, the steady-state levels of chlorophyll that were evident from our studies indicate that water-deficit stress-induced degradation of chlorophyll was absent in CCL overexpressed lines indicating that CCL transcripts can confer stability to the chloroplasts during abiotic stress conditions.
In conclusion, our results suggested that GhCCL is differentially expressed during boll developmental stages under drought stress in the field-grown cotton plants. GhCCL gene is also upregulated in cotton seedlings treated with different abiotic stresses. Complete molecular analysis such as gene organization, copy number, phylogenetic relationships, subcellular localization, and in silico analysis of the promoter sequence were carried out. Overexpression of the GhCCL gene positively regulated water-deficit stress and conferred abiotic stress tolerance to the transgenic plants which is the first demonstration of its kind. The gene is expressed better under the control of constitutive CaMV35S promoter as compared to the stress-responsive rd29A promoter. The ectopic expression of GhCCL in cotton would provide useful insights that would enhance our understanding of its function, regulation, and involvement in fiber quality under drought stress. Molecular mechanisms involved in the abiotic stress tolerance and the genome wide changes that are affected by the overexpression of GhCCL in cotton need further investigation.
References
Ackerson RC, Hebert RR (1981) Osmoregulation in cotton in response to water stress: I. Alterations in photosynthesis, leaf conductance, translocation and ultrastructure. Plant Physiol 67:484–488
Acquaah G (2007) Principles of plant genetics and breeding. Blackwell, Oxford, UK
Agrawal GK, Jwa NS, Rakwal R, Agrawal VP (2001) Signalling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: a model illustrating components participating during defense/stress response. Plant Physiol Biochem 39:1095–1103
An D, Yang J, Zhang P (2012) Transcriptome profiling of low temperature treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics 13:64
Antony AK, Kutty KE (1975) Effects of soil and climatic conditions on yield and fibre properties of improved strains of upland cotton. Ind J Agri Sci 45:199–203
Aoki H, Tanaka K, Ida S (1995) The genomic organization of the gene encoding a nitrate-inducible ferredoxin-NADP + oxidoreductase from rice roots. Biochim Biophys Acta 1229:389–392
Aprile A, Mastrangelo AM, De Leonardis AM, Galiba G, Roncaglia E, Ferrari F, De Bellis L, Turchi L, Giuliano G, Cattivelli L (2009) Transcriptional profiling in response to terminal drought stress reveals differential responses along the wheat genome. BMC Genomics 10:279
Arnon DI (1949) Copper enzymes in intact chloroplast. Polyphenoxidase in Beta vulgaris. Plant Physiol 24:1–15
Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54:911–929
Astorga GA, Estrella LH (1998) Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol 49:525–555
Basra AS, Malik CP (1984) Development of the cotton fibre. Int Rev of Cytol 87:65–113
Boyer JS (1982) Plant productivity and environment. Science 218:443–448
Bradow JM, Davidonis GH (2010) Effects of environment on fiber quality. In: Stewart JM, Oosterhuis DM, Heitholt JJ, Mauney JR (eds) Physiology of cotton. Springer, Heidelberg, pp 229–245
Brown APC, Dunn MA, Goddard NJ, Hughes MA (2001) Identification of a novel low-temperature-response element in the promoter of the barley (Hordeum vulgare L.) gene blt101.1. Planta 213:770–780
Chattopadhyay S, Hong Ang L, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. The Plant Cell 10:673–683
Cohen D, Bogeat-Triboulot MB, Tisserant E, Balzergue S, Martin-Magniette ML, Lelandais G, Ningre N, Renou JP, Tamby JP, Le Thiec D, Hummel I (2010) Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11:630
Cothren J (1999) Physiology of the cotton plant. In: Smith CW, Cothren J (eds) Cotton: origin, history, technology and production. Wiley, New York, pp 207–268
Dabrowska G, Adamska AM, Goc A (2012) Plant metallothioneins: putative functions identified by promoter analysis in silico. Acta Biologica Cracoviensia series Botanica 54:109–120
Deng F, Tu L, Tan J, Li Y, Nie Y, Zhang X (2012) GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol 158:890–904
Divya K, Jami SK, Kirti PB (2010) Constitutive expression of mustard annexin, AnnBj1 enhances abiotic stress tolerance and fiber quality in cotton under stress. Plant Mol Biol 73:293–308
Dunn MA, White AJ, Vural S, Hughes MA (1998) Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol Biol 38:551–564
Fang W, Zhang Y, Zhou L, Wang W, Li X (2013) Isolation and characterization of Histone1 gene and its promoter from tea plant (Camellia sinensis). Mol Biol Rep 40:3641–3648
Frederick JR, Camp CR, Bauer PJ (2001) Drought- stress effects on branch and main stem seed yield and yield components of determinate soybean. Crop Sci 41:759–763
Fukuda Y (1997) Interaction of tobacco nuclear protein with an elicitor-responsive element in the promoter of a basic class I chitinase gene. Plant Mol Biol 34:81–87
Gelvin SB (2006) Agrobacterium virulence gene induction. Methods Mol Biol 343:77–84
Gomez-Maldonado J, Canovas FM, Avila C (2004) Molecular analysis of the 5′-upstream region of a gibberellin-inducible cytosolic glutamine synthetase gene (GS1b) expressed in pine vascular tissue. Planta 218:1036–1045
Gong P, Zhang J, Li H, Yang C, Zhang C, Zhang X, Khurram Z, Zhang Y, Wang T, Fei Z, Ye Z (2010) Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J Exp Bot 61:3563–3575
Grimes DW, Yamada H (1982) Relation of cotton growth and yield to minimum leaf water potential. Crop Sci 22:134–139
Gu L, Han Z, Zhang L, Downie B, Zhao T (2013) Functional analysis of the 5′ regulatory region of the maize GALACTINOL SYNTHASE2 gene. Plant Sci 213:38–45
Guinn G, Mauney JR (1984a) Fruiting of cotton. I. Effects of moisture status on flowering. Agro J 76:90–94
Guinn G, Mauney JR (1984b) Fruiting of cotton. II. Effects of plant moisture status and active boll load on boll retention. Agro J 76:94–98
Harmer SL, Hogenesch JB, Straume M, Chang H, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110–2113
He L, Yang X, Wang L, Zhu L, Zhou T, Deng J, Zhang X (2013) Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem Biophys Res Commun 30:1–9
Hofgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucl Acids Res 16:9877
Ji SJ, Lu YC, Feng JX, Wei G, Li J, Shi YH, Fu Q, Liu D, Luo JC, Zhu YX (2003) Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucl Acids Res 31:2534–2543
Karimzadeh M, Nematzadeh G, Hashemi SH (2013) Study of 5′UTR’s cis-elements region of some plants P5CS gene. Intl J Agri Crop Sci 6:623–626
Kawaguchi R, Girke T, Bray EA, Serres BJ (2004) Differential mRNA translation contributes to gene regulation under non stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38:823–839
Kiba T, Henriques R, Sakakibara H, Chua NH (2007) Targeted degradation of PSEUDO-RESPONSEREGULATOR5 by a SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19:2516–2530
Kim JY, Song HR, Taylor BL, Carre IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22:935–944
Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449:356–360
Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic and cold stress. Plant Physiol 130:2129–2141
Kumar GM, Mamidala P, Podile AR (2009) Regulation of polygalacturonase-inhibitory proteins in plants is highly dependent on stress and light responsive elements. Plant Omics J 2:238–249
Kuppu S, Mishra N, Hu R, Sun L, Zhu X, Shen G, Blumwald E, Payton P, Zhang H (2013) Water-deficit inducible expression of a cytokinin biosynthetic gene IPT improves drought tolerance in cotton. PLoS One 8(5):e64190
Lam E, Chua NH (1989) ASF-2: A factor that binds to the cauliflower mosaic virus 35 s promoter and a conserved GATA motif in cab promoters. The Plant Cell 1:1147–1156
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucle Acids Res 30:325–327
Li HB, Qin YM, Pang Y, Song WQ, Mei WQ, Zhu YX (2007) A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol 175:462–471
Li DD, Wu YJ, Ruan XM, Li B, Zhu L, Wang H, Li XB (2009) Expressions of three cotton genes encoding the PIP proteins are regulated in root development and in response to stresses. Plant Cell Rep 28:291–300
Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ (2005) Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol 138:2374–2385
Long L, Gao W, Xu L, Liu M, Luo X, He X, Yang X, Zhang X, Zhu L (2013) GbMPK3, a mitogen-activated protein kinase from cotton, enhances drought and oxidative stress tolerance in tobacco. Plant Cell, Tissue Organ Cult. doi:10.1007/s11240-013-0392-1
Lu W, Chu X, Li Y, Wang C, Guo X (2013) Cotton GhMKK1 induces the tolerance of salt and drought stress and mediates defence responses to pathogen infection in transgenic Nicotiana benthamiana. PLoS One 8(7):e68503
Maqbool A, Abbas W, Rao AQ, Irfan M, Zahur M, Bakhsh A, Riazuddin S, Husnain T (2010) Gossypium arboreum GHSP26 enhances drought tolerance in Gossypium hirsutum. Am Inst Chem Eng Biotech Prog 26:21–25
Martin EC, Stephens W, Wiedenfeld R, Bittenbender HC, Beasley JPJ, Moore JM, Neibling H and CGallian JJ (2007) Sugars, oil & fiber crops. In Irrigation of agricultural crops. 2nd edition. Edited by Lascano RJ and Sojka RE: Madison, Wisconsin: ASA-CSSA-SSSA30:279-335
Mas P, KimWY SDE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426:567–570
Meyer S, Temme C, Wahle E (2004) Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol 39:197–216
Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19
Mittler R, Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol 61:443–462
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nejad ES, Askari H, Hamzelou S, Gholami M (2013) Regulation of core cell cycle genes by cis-regulatory elements in Arabidopsis thaliana. Plant Knowl J 2:69–75
Olive MR, Walker JC, Singh K, Dennis ES, Peacock WJ (1990) Functional properties of the anaerobic responsive element of the maize Adh1 gene. Plant Mol Biol 15:593–604
Padmalatha KV, Dhandapani G, Kanakachari M, Kumar S, Dass A, Patil DP, Rajamani V, Kumar K, Pathak R, Rawat B, Leelavathi S, Reddy PS, Jain N, Powar KN, Hiremath V, Katageri IS, Reddy MK, Solanke AU, Reddy VS, Kumar PA (2012) Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Mol Biol 78:223–246
Park W, Scheffler BE, Bauer PJ, Campbell BT (2012) Genome-wide identification of differentially expressed genes under water deficit stress in upland cotton (Gossypium hirsutum L.). BMC Plant Biol 12:90
Parkhi V, Kumar V, Sunilkumar G, Campbell LM, Singh NK, Rathore KS (2009) Expression of apoplastically secreted tobacco osmotin in cotton confers drought tolerance. Mol Breeding 23:625–639
Pasapula V, Shen G, Kuppu S, Valencia JP, Mendoza M, Hou P, Chen J, Qiu X, Zhu L, Zhang X, Auld D, Blumwald E, Zhang H, Gaxiola R, Payton P (2011) Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol J 9:88–99
Payton P, Kottapalli KR, Kebede H, Mahan JR, Wright RJ, Allen RD (2011) Examining the drought stress transcriptome in cotton leaf and root tissue. Biotech Lett 33:821–828
Pettigrew WT (2004) Physiological consequences of moisture deficit stress in cotton. Crop Sci 44:1265–1272
Piechulla B, Merforth N, Rudolph B (1998) Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol Biol 38:655–662
Porebski S, Bailey G, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15:8–15
Rabbani AM, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Shinozaki KY (2003) Monitoring expression profiles of rice genes under cold, drought and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Reimmann C, Dudler R (1993) Circadian rhythmicity in the expression of a novel light-regulated rice gene. Plant Mol Biol 22:165–170
Rikin A, Dillwith JW, Bergman DK (1993) Correlation between the circadian rhythm of resistance to extreme temperatures and changes in fatty acid composition in cotton seedlings. Plant Physiol 101:31–36
Rogers SO, Bendish AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA (eds) Plant Mol Biol Manu. Kluwer Academic Publishers, London
Rose A, Meier I, Wienand U (1999) The tomato I-box binding factor LeMYB1 is a member of a novel class of Myb-like proteins. The Plant J 20:641–652
Ross J (1995) mRNA stability in mammalian cells. Microbiol Rev 59:423–450
Roy S, Choudhury SR, Singh SK, Das KP (2012) Functional analysis of light-regulated promoter region of AtPolλ gene. Planta 235:411–432
Ruan YL, Chourey PS (1998) A fibreless seed mutation in cotton is associated with lack of fibre cell initiation in ovule epidermis and alterations in sucrose synthase expression and carbon partitioning in developing seeds. Plant Physiol 118:399–406
Sahu BB, Shaw BP (2009) Isolation, identification and expression analysis of salt-induced genes in Suaeda maritima, a natural halophyte, using PCR-based suppression subtractive hybridization. BMC Plant Biol 9:69
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Sanchez A, Shin J, Davis SJ (2011) Abiotic stress and the plant circadian clock. Plant Signal Behav 6:223–231
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enjul A, Sakurai T, Satou M, Akiyama K, Taji T, Shinozaki KY, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Shinozaki KY, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251–264
Smith CW, Cothren JT (1999) Cotton: origin, history, technology and production. Wiley, New York
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Staiger D, Kaulen H, Schell J (1989) A CACGTG motif of the Antirrhinum majus chalcone synthase promoter is recognized by an evolutionarily conserved nuclear protein. Proc Natl Acad Sci U S A 86:6930–6934
Sunilkumar G, Vijayachandra K, Veluthambi K (1999) Preincubation of cut tobacco leaf explants promotes Agrobacterium-mediated transformation by increasing vir gene induction. Plant Sci 141:51–58
Taiz L and Zeiger E (1998) Stress physiology. In: Plant physiology, 2nd edn. Sunderland, MA: Sinauer Associates Inc, 725-757
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thanh T, Chi VTQ, Omar H, Abdullah MP, Napis S (2012) Sequence Analysis and Potentials of the Native RbcS Promoter in the Development of an Alternative Eukaryotic Expression System Using Green Microalga Ankistrodesmus convolutus. Int J Mol Sci 13:2676–2691
Umezawa T, Fujita M, Fujita Y, Shinozaki KY, Shinozaki K (2006) Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol 17:113–122
Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199–223
Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93:1207–1217
Wang L, Fujiwara S, Somers DE (2010) PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J 29:1903–1915
Wang K, Zhang X, Zhao Y, Chen F, Xia G (2013) Structure, variation and expression analysis of glutenin gene promoters from Triticum aestivum cultivar Chinese Spring shows the distal region of promoter 1Bx7 is key regulatory sequence. Gene 527:484–490
Weatherley PE (1950) Studies in the water relations of the cotton plant I. The field measurements of water deficits in leaves. New Phytol 49:81–97
Weatherley PE (1951) Studies in the water relations of the cotton plant II. Diurnal and seasonal fluctuations and environmental factors. New Phytol 50:36–51
Whitelaw CA, Le Huquet JA, Thurman DA, Tomsett AB (1997) The isolation and characterization of the type II metallothionein-like genes from tomato (Lycopersicon esculentum L.). Plant Mol Biol 33:503–511
Wilkins TA, Arpat AB (2005) The cotton fiber transcriptome. Physiol Plant 124:295–300
Xue T, Li X, Zhu W, Wu C, Yang G, Zheng C (2009) Cotton metallothionein GhMT3a, a reactive oxygen species scavenger, increased tolerance against abiotic stress in transgenic tobacco and yeast. J Exp Bot 60:339–349
Yakir E, Hilman D, Harir Y, Green RM (2007a) Regulation of output from the plant circadian clock. FEBS J 274:335–345
Yakir E, Hilman D, Hassidim M, Green RM (2007b) CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol 145:925–932
Yue Y, Zhang M, Zhang J, Tian X, Duan L, Li Z (2012) Overexpression of the AtLOS5 gene increased abscisic acid level and drought tolerance in transgenic cotton. J Exp Bot 63:3741–3748
Zhang H, Shen G, Kuppu S, Gaxiola R, Payton P (2011a) Creating drought- and salt-tolerant cotton by overexpressing a vacuolar pyrophosphatase gene. Plant Signal Behav 6:861–863
Zhang L, Xi D, Li S, Gao Z, Zhao S, Shi J, Wu C, Guo X (2011b) A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol Biol 77:17–31
Zhang X, Zhen JB, Li ZH, Kang DM, Yang YM, Kong J, Hua JP (2011c) Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.). Plant Mol Biol Rep 29:626–637
Zhang LF, Li WF, Han SY, Yang WH, Qi LW (2013) cDNA cloning, genomic organization and expression analysis during somatic embryogenesis of the translationally controlled tumor protein (TCTP) gene from Japanese larch (Larix leptolepis). Gene 529:150–158
Zhao W, Wang S, Li X, Huang H, Sui X, Zhang Z (2012) Molecular cloning and characterization of the light-regulation and circadian-rhythm of the VDE gene promoter from Zingiber officinale. Plant Cell Rep 31:1381–1392
Zhao J, Gao Y, Zhang Z, Chen T, Guo W, Zhang T (2013) A receptor-like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought-stress tolerance in Arabidopsis. BMC Plant Biol 13:110
Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, Chen C, Ma L, Wang J, Xiong L, Zhang Q, Fan L, Deng XW (2007) Global genome expression analysis of rice in response to drought and high salinity stresses in shoot, flag leaf and panicle. Plant Mol Biol 63:591–608
Zhu YN, Shi DQ, Ruan MB, Zhang LL, Meng ZH, Liu J, Yang WC (2013) Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLoS One 8(11):e80218
Acknowledgments
This work was supported by funds from the Indian Council of Agricultural Research (ICAR) under the National Agricultural Innovation Project (NAIP/C4/C10103). Funds from the Department of Biotechnology (DBT), Government of India are gratefully acknowledged. We thank Dr. I.S Katageri, University of Agricultural Sciences, Dharwad for providing the drought-induced cotton boll samples used in the study and Dr. Ajay Jain, National Research Centre on Plant Biotechnology, New Delhi for providing the binary vector pCAMBIA1302. We also thank Dr. Shelly Praveen and Mr. Vipin Kumar, Division of Plant Pathology, Indian Agricultural Research Institute (IARI), New Delhi for providing confocal microscope facility and technical help for subcellular localization study.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 21.9 kb)
Fig. S1
T-DNA regions of binary vectors used for tobacco transformation. a Cotton GhCCL gene was cloned in pBI121 binary vector in the place of GUS reporter gene driven by CaMV35S promoter. b pBI121 binary vector carrying cotton GhCCL gene driven by Arabidopsis thaliana rd29A promoter. The right border (RB) junction fragment of the T-DNA is >3.3, 3.4-kb in 35S::GhCCL and Atrd29A::GhCCL, respectively (the distance between BamHI and RB). (PPTX 54 kb)
Fig. S2
Predicted conserved domain in GhCCL protein by NCBI-Conserver domain database (CDD). (PPTX 38 kb)
Fig. S3
PCR analysis of transgenic tobacco plants. T 0 transgenic tobacco analysed with NPTII and GhCCL gene specific primer (a, b), respectively. c PCR analysis of selected T 1 transgenic tobacco plants with GhCCL gene specific primer. W-water control C-WT control, A-35S::GhCCL, B-rd29A::GhCCL, AP-35S::GhCCL binary plasmid, BP-rd29A::GhCCL and M1, M2:1-kb and 100-bp ladder, respectively. (PPTX 212 kb)
Rights and permissions
About this article
Cite this article
Dhandapani, G., Kanakachari, M., Padmalatha, K.V. et al. A Gene Encoding Cold-Circadian Rhythm-RNA Binding-Like Protein (CCR-Like) from Upland Cotton (Gossypium hirsutum L.) Confers Tolerance to Abiotic Stresses in Transgenic Tobacco. Plant Mol Biol Rep 33, 22–42 (2015). https://doi.org/10.1007/s11105-014-0729-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0729-x