Abstract
A novel stress tolerance cDNA fragment encoding GhDRIN1 protein was identified and its regulation was studied in cotton boll tissues and seedlings subjected to various biotic and abiotic stresses. Phylogenetic and conserved domain prediction indicated that GhDRIN1 was annotated with a hypothetical protein of unknown function. Subcellular localization showed that GhDRIN1 is localized in the chloroplasts. The promoter sequence was isolated and subjected to in silico study. Various cis-acting elements responsive to biotic and abiotic stresses and hormones were found. Transgenic tobacco seedlings exhibited better growth on amended MS medium and showed minimal leaf damage in insect bioassays carried out with Helicoverpa armigera larvae. Transgenic tobacco showed better tolerance to water-deficit and fast recovered upon rewatering. Present work demonstrated that GhDRIN1, a novel stress tolerance gene of cotton, positively regulates the response to biotic and abiotic stresses in transgenic tobacco.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stresses such as drought and salinity are the major constraints in modern agriculture. These stresses lead to a series of morphological, physiological, biochemical and molecular changes in plants that negatively affect plant growth and productivity (Wang et al. 2001). Water-deficit and salinity affects more than 10 % of arable land on our planet which results in a yield reduction of more than 50 % on average for most of the major crop plants (Bartels and Sunkar 2005). Cotton (Gossypium spp.) is the leading fiber crop worldwide and is an important source of oil and protein meal. Gossypium hirsutum L. is the most widely cultivated cotton species, dominating global cotton commerce with a share of more than 95 % of the world production. As with the other major fiber crops, cotton also suffers yield losses due to various biotic and abiotic stress factors. Enhancing tolerance to biotic and abiotic stresses is one of the key issues in augmenting cotton yield.
Many genes are expressed in response to various environmental stresses in plants (Horan et al. 2008) and different unknown and poorly characterized genes might be required for defence mechanisms involving a variety of complex signaling pathways (Gollery et al. 2007; Luhua et al. 2008). A large number of unknown genes is involved in responses of plants to environmental conditions such as less water availability, changes in atmospheric temperature, toxicity of salt and other minerals and limiting O2 conditions (Hirayama and Shinozaki 2010). With increase in the ease of use and decrease in the cost of various genomic tools such as expressed sequence tags (ESTs) generation, microarrays, next generation sequencing etc., used for studying the gene expression landscape during various conditions, abundant data generation on gene expression profiles during stress conditions is being actively pursued. Also, identification of key drought responsive genes that enhance broad spectrum drought tolerance in cotton is critical for its improvement (Zhang et al. 2009).
In a previous study, we used microarray gene expression analyses to study global transcriptomic changes taking place in cotton plants, subjected to drought under field conditions. A number of hypothetical uncharacterized/unpredicted proteins which are highly expressed in cotton boll tissues subjected to drought stress were identified (Padmalatha et al. 2012). Although the transcriptomic data can give us insights into various process and key genes involved in a particular response, it is important to validate the gene function to determine its ability to impart stress tolerance/resistance to the plants. In the present study, we investigated an uncharacterized hypothetical protein (Unigene ID: Ghi. 6656) from G. hirsutum L. (cv. Bikaneri Narma) which was upregulated during all boll developmental stages under drought in the field conditions. The gene was named as G. hirsutum drought-induced 1 (GhDRIN1). It was upregulated in drought-induced samples at 135-, 243- and 2.5-fold at 5, 10 and 20 dpa, respectively, compared to the controls (Padmalatha et al. 2012). The upregulation of the same transcript was also observed in bollworm (Helicoverpa armigera) infested cotton boll samples for 8-, 19.5-, 128-, 575-fold more at 0, 2, 5, 10 dpa (unpublished data), respectively, compared to the control samples as revealed in the transcriptome based microarray study. In order to determine the role of this transcript in multiple stresses, the full-length complementary DNA (cDNA) for a gene encoding GhDRIN1 protein was isolated and characterized.
A full-length cDNA of the gene encoding GhDRIN1 protein was isolated, cloned in a binary vector and overexpressed in transgenic tobacco to verify its efficacy in enhancing the tolerance to stresses such as dehydration, salinity and insect damage. The transgenic tobacco seedlings survived on MS media supplemented with 200 mM each of NaCl, mannitol and 10 % (v/v) PEG 6000 under in vitro conditions. The transgenic plants tolerated and retained the ability to survive under water-deficit stress in the glass house conditions. The promoter region was isolated and various cis-acting elements involved in biotic and abiotic stress response were identified by in silico analysis. Subcellular localization studies revealed that GhDRIN1::GFP fused protein was localized in chloroplasts of tobacco. The results showed that the gene GhDRIN1 may play an explicit role in biotic and abiotic stress tolerance in transgenic plants it could be a good candidate gene for the genetic manipulation of cotton and other crops to confer tolerance to drought and salinity as well as to the biotic stresses.
Materials and methods
Plant materials, growth conditions and stress treatments
Cotton (G. hirsutum L. cv. Bikaneri Narma) was used for all the experiments. For plant transformation, tobacco (Nicotiana tabacum var. Petit Havana) was used. The plant culture conditions and related parameters under in vitro and transgenic glass house conditions were as reported earlier (Dhandapani et al. 2014).
Seven-day-old cotton seedlings were used for different treatments. Seedlings were dipped in 0.5× MS medium containing 15 % (v/v) PEG 6000, 200 mM each of NaCl and mannitol. For methyl jasmonate (MeJA), salicylic acid (SA), H2O2 and abscisic acid (ABA) treatments, leaves were sprayed with each at 10 mM. To investigate the effect of temperature, the pots with germinated seedlings were placed in a growth chamber at high (42 °C) and low temperature (4 °C). For wounding, the seedlings were punched by forceps and the samples were collected at the appropriate time points. For dehydration-stress, the seedlings were removed from the bottles and placed on tissue paper under culture room conditions. The samples were collected from all the treatments after 1 and 12 h. Pathogen treatment was carried with Fusarium oxysporum f. sp. vasinfectum (F. oxysporum f. sp. vasinfectum) and Colletotrichum gloeosporioides (C. gloeosporioides) conidial suspensions were used to inoculate the cotton seedlings by the root dip method (Dowd et al. 2004). The inoculated plants were kept in a moist chamber and the seedlings were collected after 2, 4, 6, 8 and 12 days post-infection. The seedlings were harvested at indicated time period and the samples were immediately frozen in liquid N2 and stored at −70 °C for later use.
Cotton samples at different boll developmental stages under drought-stress condition were collected along with the respective controls for expression analysis of the gene by qPCR. Drought stress induction under field condition and related parameters were followed as described previously (Padmalatha et al. 2012). The bollworm infected cotton bolls were collected from the field-grown plants. Second–Third instar larvae of H. armigera were released on the day of pollination. The buds were covered with paper bag with proper aeration to prevent larvae movement from the buds ensuring the damage to the bolls. The samples were collected after 8 h, 2 and 5 days after infestation and labeled as 0, 2 and 5 dpa, respectively. After 5 days of infestation, insects were removed from the bolls to prevent complete damage. The bolls were allowed to grow up to 10 days. The infested boll samples were collected along with uninfected bolls from the control plot. The samples were labeled and frozen immediately in liquid N2 and stored at −70 °C for later use.
Cloning the full-length GhDRIN1 and sequence analysis
Total RNA was extracted from field-grown drought-induced 10 dpa cotton boll samples using Spectrum Plant total RNA extraction kit (Sigma) according to the manufacturer’s protocol. Full-length GhDRIN1 was obtained by 5′-rapid amplification of cDNA ends-PCR (5′-RACE kit, Invitrogen). The ORF was identified by the FGENESH analysis using softberry software (www.softberry.com). First stranded cDNA was synthesized from 1 µg of total RNA isolated from cotton boll with an AffinityScript QPCR cDNA Synthesis Kit (Stratagene, Agilent Technologies) and used as a template to perform the RT-PCR using gene specific primers of GhDRIN1-F and GhDRIN1-R. PCR conditions were: 94 °C for 4 min, initial denaturation; 30 cycles of 94 °C for 40 s, 60 °C for 40 s, and 72 °C for 60 s, and a final extension at 72 °C for 10 min. The amplified cDNA was cloned in pGEM-T Easy vector and sequenced.
Expression analysis of GhDRIN1
qRT-PCR analysis was carried out for the quantification of GhDRIN1 transcripts in cotton seedlings treated with various biotic and abiotic stresses. Cotton bolls from drought-induced and bollworm infested samples as well as the respective control samples were used. The gene specific qRT primers (GhDRIN1qRT-F and GhDRIN1qRT-R) were used to quantify the expression of GhDRIN1 transcript in cotton boll tissues collected from plants grown under drought-induced field conditions and bolls infested with H. armigera larvae, seedlings exposed to different stresses and fungal infection. GhUbiquitin1 (GhUBI1-F and GhUBI1-R) (Padmalatha et al. 2012) and GhPP2A1 (GhPP2A1-F and GhPP2A1-R) (Artico et al. 2010) gene specific primers from G. hirsutum, respectively were used as housekeeping genes to normalize the amount of template cDNA added in each reaction. The qPCR reactions consisted of a total volume of 25 µl with 0.4 µl each of 10 µM primer, 12.5 µl Brilliant-III SYBR Green qPCR master mix (Stratagene, Agilent Technologies), 0.375 µl ROX, 1 µl cDNA (1/10 dilution) and 10.325 µl RNase-Free water. The experiment was carried out with a MX3005P Real-time PCR system (Stratagene, Agilent Technologies). The qRT-PCR was performed as follows: 5 min at 95 °C, followed by 40 cycles of amplification with 30 s of denaturation at 95 °C, 30 s of annealing at 60 °C and 30 s of extension at 72 °C. Three biological replicates were used. Amplicons were subjected to the meltcurve analysis to check the specificity of the amplified products. The relative expression level of each gene was calculated by the 2−(ΔΔCt). All reactions were performed in triplicate.
Subcellular localization studies
To observe the cellular localization, GhDRIN1 coding region was amplified without the stop codon by PCR using primer pair GhDRIN1:GFP-F and GhDRIN1:GFP-R having a BglII and SpeI sites, respectively. The resulting fragment was inserted into the BglII/SpeI site of the binary vector pCAMBIA1302-mGFP, which generated a C-terminal fusion with the modified green fluorescence protein gene (mGFP) controlled by the CaMV 35S promoter. The recombinant vectors were transferred to Agrobacterium tumefaciens strain LBA4404. To monitor the transient expression, agroinfiltration study was performed by using induced Agrobacterium culture in tobacco leaf tissue. After 4 days, the leaf portion was visualized under a confocal microscope.
Isolation and analysis of the GhDRIN1 promoter
Genome walking was performed to isolate the upstream region of GhDRIN1 using a GenomeWalker Kit (Clontech). The upstream sequences were cloned in pGEM-T Easy vector, sequenced and the aligned sequences were analyzed using the PlantCARE database of cis-acting regulatory DNA elements (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for promoter prediction. The primer sequences used in this study are listed in Supplementary Table 1. The full-length genomic sequence has been submitted to NCBI.
Construction of binary vector and ectopic expression of GhDRIN1 in tobacco
The ORF of GhDRIN1 was amplified with GhDRIN1-F and GhDRIN1-R specific primers having BamHI and SacI sites, respectively; amplified coding sequence was ligated to corresponding sites of binary vector pBI121 under the control of the CaMV 35S promoter and NOS terminator. The recombinant binary vector was mobilized into A. tumefaciens strain LBA4404. The plant transformation was carried out using wild-type (WT) plants of tobacco (N. tabacum var. Petit Havana) by leaf disc method. The transformants were screened on 200 mg kanamycin/l and the putative transgenics were confirmed by PCR in T 0 generation with NPTII and gene specific primers. Molecular and physiological characterization was carried out in T 1 transgenic lines.
Southern and northern blot analysis
Genomic DNA was isolated from WT and nine T 1 transgenic tobacco plants using the CTAB method. Southern blot analysis was performed to confirm the stable integration and copy number of the NPTII gene. Genomic DNA, 10 µg, was digested with HindIII and digested genomic DNA samples was resolved on 0.8 % agarose gel and blotted on a Hybond-N+ Nylon membrane (Amersham Biosciences). The blot was hybridized at 65 °C with α [32P]-dCTP labeled 700-bp NPTII fragment using mega-prime DNA labeling system (Amersham Biosciences). Total RNA, 15 µg, was isolated from the wild type and eight T 1 transgenic tobacco plants using TRIzol and was resolved on a 1.2 % formaldehyde-denaturing agarose gel and transferred to the nylon membrane (Hybond-N+, Amersham Biosciences) using diethylpyrocarbonate (DEPC)-treated 20 × SSC. Pre-hybridization and hybridization were performed with ULTRAhyb hybridization buffer (Ambion) according to the manufacturer’s instructions. The blot was hybridized at 42 °C with α [32P]-dCTP labeled 390-bp GhDRIN1 fragment using with mega-prime DNA labeling system (Amersham Biosciences). The complete procedure for Southern and northern blot analysis was followed as described earlier (Dhandapani et al. 2014).
Stress tolerance analysis of transgenic tobacco
Growth performance of transgenic plants under stress
Long-term stress effect was evaluated by culturing the 15-days-old seedlings of WT and T 1 transgenic plants in culture bottles containing MS agar media supplemented with 200 mM each of NaCl, mannitol and 10 % (v/v) PEG 6000. The growth performance was observed after 30 days. The fresh weight was calculated and plotted.
Insect bioassay of transgenic tobacco
The larval population of cotton bollworm (H. armigera) was collected from the cotton and chickpea fields were maintained on artificial diet (Gupta et al. 2004) in environmentally-controlled growth chamber (ECG); (conditions: temperature 26 ± 1 °C, relative humidity 65 ± 5 % and photoperiod 16:8 h scoto/photo regime) in the insect laboratory. Insect bioassay was performed using detached leaves from 25-days-old in vitro-grown WT and transgenic plants (T5, T7 and T14) were placed in Petri dishes with a wet filter paper at the bottom. Five larvae of the second instar stage reared on artificial diet were used in each Petri dish. Two replicates were used in each experiment. Feeding was allowed up to 72 h, the leaf damage was observed and the images were captured.
Water-deficit stress tolerance of transgenic tobacco
WT and T 1 transgenic tobacco plants were transferred to soil in the glass house to carry out water-deficit stress experiment. Water was withheld for 35 days at the time of flowering. The relative water content (RWC) and total chlorophyll were analyzed at 0 and 15 days after withholding water. Plants were allowed to recover after 35 days of withholding water to observe the nature of recovery in WT and transgenic plants.
Statistical analyses
Each experiment was repeated two–three times and the values are expressed as mean ± SE. All mean comparisons were done using paired t test for independent samples. The measurements for determining different treatments or times were analyzed by one-way analysis of variance (ANOVA).
Results
Isolation and characterization of GhDRIN1
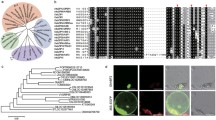
In Affymetrix cotton GeneChip, the probe for the transcript representing unknown hypothetical gene was obtained from the cotton EST (GenBank Acc. No CA993777.1) which is 644 nucleotide in length with poly-A tail at the 3′-end. In order to isolate the complete upstream gene sequence, the 5′-RACE PCR technique was followed using degenerate primers to sequence the 5′-region. The RACE PCR product was cloned in pGEM-T Easy vector and sequenced. The ORF of the gene was identified by FGENESH of softberry online software using aligned sequence. The 477-bp GhDRIN1 cDNA sequence has a 88-bp 5′-UTR and 390-bp coding sequence. The cDNA contained a 390-bp ORF encoding a protein of 129 deduced amino acid residues (AARs), with a calculated molecular mass of 14.477 kDa and a predicted average isoelectric point (pI) of 6.35. The genomic sequence was also amplified to analyze the structural properties of GhDRIN1. Its analysis suggested that GhDRIN1 did not have any introns and the amplified sequence showed similarity with the corresponding cDNA sequence. The full-length genomic DNA sequence of GhDRIN1 has been submitted to NCBI GenBank under the accession number KJ510523. Of total 1, 112-bp long GhDRIN1 genomic sequence comprising of, 634-bp promoter, 88-bp 5′-UTR and 390-bp ORF region. Homology analysis of the protein sequence showed similarity to different uncharacterized/hypothetical proteins. Phylogenetic analysis also showed that it may be a novel protein (Supplementary Fig. 1). Further analyses of the predicted protein using SMART and NCBI-CD programs also resulted in no hits.
Expression analysis of GhDRIN1
Differential expression of GhDRIN1 was studied in cotton during different abiotic and biotic stress conditions. Under drought conditions the expression of this gene in boll tissues gradually increased up to 10 dpa and receded in 20 dpa bolls (Fig. 1a). Under different abiotic stress conditions such as cold, desiccation, heat and H2O2 treatment, the expression of GhDRIN1 showed multifold increase in expression in short time (1 h) as well as long time (12 h). Highest fold change in expression was seen in samples subjected to desiccation and heat. However, when treated with drought inducing agents such as PEG 6000 and mannitol the results were different. GhDRIN1 showed negative log fold change when treated with mannitol whereas it was positive log fold change in case of samples treated with PEG 6000 (Fig. 1b).
Expression analysis of GhDRIN1 transcript in stress-induced cotton samples. a qRT-PCR analysis of drought-induced cotton samples with their respect control samples collected from different boll developmental stages. b qRT-PCR analysis of 7-days-old cotton seedlings treated with various stresses and analyzed after 1 and 12 h time period. c qRT-PCR analysis of bollworm (H. armigera) infected cotton samples with their respective control samples collected from 0, 2, 5 and 10 dpa cotton bolls. d, e qRT-PCR analysis of 7 day-old cotton seedlings infected with conidial suspensions of F. oxysporum f. sp. vasinfectum and C. gloeosporioides fungal culture and collected the samples after 0, 2, 4, 6, 8 and 12 days infection and analyzed, respectively
The boll samples infested with H. armigera and seedlings challenged with conidia of either F. oxysporum f. sp. vasinfectum and C. gloeosporioides showed higher expression of GhDRIN1 (Fig. 1c, d, e). While there was a gradual increase in expression in the case of H. armigera infestation, a rise in eight–tenfold of expression was maintained in the case of fungal challenge. To further gain insights about the expression of GhDRIN1 during biotic stress, its expression in seedlings treated with general biotic stress response inducers, such as MeJA, SA and wounding, was also tested. In these conditions, GhDRIN1 was not upregulated during the early response (1 h), it was upregulated at 12 h by five and eightfold in MeJA and SA treated samples, respectively. However, in wounded samples the expression of GhDRIN1 was upregulated at 1 h as well as at 12 h (Fig. 1b). Expression studies showed that GhDRIN1 upregulated during a variety of abiotic and biotic stress conditions. However, the level and timing of expression seems to change with the type of stress.
GhDRIN1 is targeted to the chloroplast
In silico subcellular localization analysis performed using ProtComp and ChloroP Server indicated that GhDRIN1 is localized in the chloroplasts (Supplementary Table 2). Agroinfiltration of tobacco tissues by Agrobacterium cultures harboring pCAMBIA1302-CaMV35S-GhDRIN1::mGFP (Supplementary Fig. 2a) and control vector pCAMBIA1302-CaMV35S-mGFP (Supplementary Fig. 2b) were carried out. Tissues infiltrated with GhDRIN1::mGFP construct is targeted to the chloroplasts as the GFP fluorescence was completely superimposed with the autofluorescence signals of the chlorophyll, while the cells infiltrated with mGFP control construct showed homogenous and dispersed GFP signals (Fig. 2) confirming the in silico analysis of chloroplast localization of GhDRIN1.
Subcellular localization study of GhDRIN1 in tobacco leaf tissue. Transient expression of CaMV35S-mGFP and CaMV35S-GhDRIN1::mGFP in living tobacco leaf tissue by agroinfiltration method. The infected leaf tissues were analyzed by confocal microscope (Leica) after 4 days (Scale bars in the lower right corners represent 20 µm)
In silico analysis of GhDRIN1 promoter
To understand the regulation of this gene and its response to different stresses, a 722-bp upstream regulatory sequence of GhDRIN1 was obtained using genome walking and analyzed in silico to determine regulatory sequences. A number of regulatory motifs corresponding to several known cis-acting elements related to light, stress, tissue and hormone responsive elements were predicted (Table 1). Light responsive elements, such as AAAC-motif, BOX I and II, G-Box, GTI-motif and Sp1, and other related motifs were identified. Cis-regulatory elements responsive to various stresses such as HSE for heat stress and LTR for low temperature responsiveness were also detected. ARE and GC-motif enhancer-like element involved in anoxic specific response were identified. Hormone responsive elements, such as ABRE, ERE and P-box motifs, are also predicted which respond to ABA, ethylene and gibberellin, respectively. Interestingly, tissue specific motifs, such as AAGAA and RY-elements for seed specific and Skn-1 motif for endosperm specific expression, have been predicted. The presence of different and multiple regulatory sequences corresponding to diverse conditions such as light, stress and hormone responses implies that GhDRIN1 could play a key role during such conditions.
Generation and validation of GhDRIN1 transgenic tobacco plants
The 390-bp coding region of GhDRIN1 was amplified from cDNA that was reverse transcribed from total RNA isolated from 10 dpa drought-induced cotton boll tissue. The amplified RT-PCR product was cloned in pGEM-T Easy vector and verified by sequencing. After sequence verification, the ORF of GhDRIN1 cDNA fragment was cloned in a binary vector pBI121 (replaced by GUS reporter gene) that is driven by constitutive Cauliflower Mosaic Virus (CaMV) 35S promoter and the recombinant binary vector was named pBI121-35S::GhDRIN1 (Supplementary Fig. 2c). The binary vector was mobilized into A. tumefaciens strain LBA4404 by freeze–thaw method. The Agrobacterium-mediated tobacco transformation was performed by leaf disc method. The putative transgenic tobacco plants were initially selected with 200 mg kanamycin/l and the rooted plants were further maintained on 100 mg kanamycin/l.
To confirm the stable integration of the transgene and determination of NPTII gene copy number in transgenic tobacco, Southern blot analysis was carried out with 10 µg genomic DNA from WT and nine T 1 transgenic tobacco plants digested with HindIII, which recognizes a single site within the T-DNA region. The Southern blot results confirmed the stable integration of the transgene. The NPTII gene was integrated as single (T1, T4, T5, T7, T14, T16, T17and T20) and two copies (T19) in the transgenic tobacco plants (Fig. 3a). The expression of GhDRIN1 transcripts in transgenic tobacco was verified by Northern blot analysis which showed varied expression of GhDRIN1 transcript in all the eight single copy southern positive transgenic plants (T1, T4, T5, T7, T14, T16, T17 and T20) whereas no expression was observed in WT as expected (Fig. 3b).
Southern and Northern blot analysis of transgenic tobacco. a Southern blot analysis of GhDRIN1 expressing transgenic T 1 tobacco plants showing the copy number of NPTII. Genomic DNA of transgenic events was digested with HindIII that has no restriction site in the probe and single cutter in the T-DNA region. The blot was hybridized with α [32P]-dCTP labeled with NPTII probe. Molecular weight marker in kb indicated on the left. b RNA gel-blot analysis of WT and GhDRIN1 expressing T 1 tobacco plants. Fifteen micrograms of total RNA was resolved on 1.2 % formaldehyde denaturing agarose gel and transferred to the nylon membrane. The blot was hybridized with α [32P]-dCTP labeled with 390-bp GhDRIN1 cDNA fragment. The ethidium bromide-stained gel demonstrates RNA quantities loaded in each lane (T1, T4, T5 T7, T14, T16, T17 and T20 independent transgenic lines)
Effect of long-term stress on transgenic plants
Fifteen-days-old WT and T 1 transgenic seedlings (T5, T7 and T14) were subcultured in MS medium supplemented with 200 mM each of NaCl (Fig. 4a), mannitol (Fig. 4b) and 10 % (v/v) PEG 6000 (Fig. 4c) for 30 days. The transgenic lines performed much better in terms of growth as well as root initiation when compared to the WT in all the three conditions. The WT plants showed stunted growth and delayed root formation under similar conditions. However, under the stress imposed by 10 % PEG, the transgenic plants showed yellowing of the leaves although no decrease in the growth of the plants was noted.
Effect of long-term stress of WT and T 1 transgenic tobacco. Fifteen-days-old seedling of WT and transgenic tobacco were transferred and grown on MS agar medium supplemented with 200 mM each NaCl (a) mannitol (b) and 10 % PEG 6000 (c) and growth performance were observed after 30 days. d Fresh weight was calculated and plotted. Each bar value represents the mean ± of triplicates (ANOVA test) and means are significantly different from control at P < 0.05 according to Dunnett’s test at each treatment
Fresh weights were measured after 30 days in the WT and transgenic plants grown on amended medium (Supplementary Table 3). Approximately 33, 26 and 22 % of fresh weight was observed in WT plants whereas 84.5, 53.5 and 56.5 % of average fresh weight was noticed in the transgenic plants grown on NaCl, mannitol and PEG 6000, respectively (Fig. 4d). The observation clearly showed that the transgenic plants grown on NaCl medium performed better and accumulated more fresh weight as compared to the transgenic plants grown on mannitol and PEG 6000.
Insect bioassay with GhDRIN1 transgenic leaves
The detached leaves of the transgenic and WT plants were used for insect bioassays. The leaves of the transgenic lines T5, T7 and T14 challenged with H. armigera larvae showed minimal damage to the leaves as compared to WT which was completely damaged after 72 h feeding (Fig. 5). The larvae feed on transgenic leaves was observed less weight as compare to larvae feed on WT leaves. However, we did not observe any mortality of the larvae in the transgenic lines after 72 h experiment.
Insect bioassay performed on WT and GhDRIN1 transgenic tobacco leaves with second/third instar larvae of H. armigera. Leaves were detached from 25-days-old in vitro WT and T 1 transgenic plants (T5, T7 and T14) were placed in Petri dishes with a wet filter paper at the bottom. Five larvae of the second instar stage were released to feed the leaves and maintained the experiment at 24 °C for 72 h and the leaf damage was observed
Effect of water-deficit stress in transgenic plants under glass house condition
The WT and T 1 transgenic lines (T4, T5, T7 and T14) were maintained in a transgenic glass house and water-deficit stress was imposed by withholding water at the flowering stage (Fig. 6a). WT plants completely dried and showed no signs of life after 35 days of withholding water but the GhDRIN1 transgenic plants, although not completely healthy, showed green stems and leaves (Fig. 6b). We also tested the ability of the plants to recover upon rewatering, after 35 days of water-deficit stress treatment. Sixty to ninety percent of the transgenic plants recovered within a week and produced healthy greenish leaves. Flowering was also observed at a later stage whereas more than 80 % of the WT plants did not show any sign of recovery as they wilted completely under water-deficit stress (Fig. 6c, f).
Representative photos for water-deficit stress analysis of WT and transgenic GhDRIN1 tobacco lines. a 30-day-old flowering plants and 0 days water-deficit stress induction (water withholding). b Photo was taken after 35 days water-deficit induction. c Plants recovered after 20 days rewatering. d, e graphs shown the percentage of relative water content and total chlorophyll content analysis at 0 and 15 days after water withholding, respectively. f Percentage of survival rate after recovered from water-deficit stress was calculated and plotted. Each bar value represents the mean ± of triplicates (ANOVA test) and means are significantly different from control at P < 0.05 according to Dunnett’s test at each treatment
The RWC and chlorophyll content were estimated in WT and transgenic plants under water-deficit stress conditions at 0 and 15 days after withholding water. The percentage of RWC was in the range of 45.6–47 % in transgenic plants and 45.4 % in WT at 0 days of water withholding and the percentage of RWC in transgenic plants was in the range of 42–45 % at 15 days water withholding whereas in WT 34.5 % RWC was observed. RWC significantly reduced in WT after 15 days of withholding water and only a slight reduction was observed in transgenic plants (Fig. 6d, Supplementary Table 4). The total chlorophyll content was in the range of 5.3–5.6 mg/g fw in transgenics and 5.5 mg/g fw in WT at 0 days of water withholding and the same was in the range of 5–5.4 mg/g fw in the transgenics and only 4.2 mg/g fw in WT at 15 days after water withholding. Reduction of total chlorophyll was observed in both WT and transgenics at 15 days after water withholding, but transgenics showed less chlorophyll degradation when compared to WT (Fig. 6e, Supplementary Table 5).
Discussion
On an average, 20–40 % of all eukaryotic genomes sequenced to date contain genes that encode proteins of unknown function (Gollery et al. 2006). Over one-quarter of all plant genes encode proteins of unknown function that can be further classified as proteins with obscure features, which lack currently defined motifs or domains and in Arabidopsis more than 70 % of the expressed unknown proteins conferred tolerance to oxidative stress (Luhua et al. 2013) which suggests that the several unknown proteins have an important role in plant biotic and abiotic stress responses and thereby to imparting stress tolerance to the plants. In this present study, such a gene with an unknown function was characterized.
Expression patterns are usually an indicator of gene functions. GhDRIN1 gene, which was originally identified during differential expression studies in our earlier study (Padmalatha et al. 2012), was also found to be over-expressed in cotton bolls infested by bollworm (unpublished data). Quantitative PCR based expression analysis of the seedlings showed that it is also expressed during various abiotic stress treatments. The expression of GhDRIN1 was induced during bollworm infestation and also when challenged with two different fungi. We observed that GhDRIN1 can also be induced by signaling molecules such as SA and MeJA. The expression of GhDRIN1 during different conditions suggests that this gene might play a role in general plant defence mechanisms. To further understand and verify if this gene could confer tolerance to different stresses, ectopic overexpression of GhDRIN1 in transgenic tobacco was carried out.
In line with the expression profile of GhDRIN1, it was able to confer tolerance against drought in vitro and in vivo as well as during salt stress and protect the plants from insect damage. The transgenic lines showed tolerance to drought and also an ability to recover after drought conditions. Importantly, we observed this recovery during flowering stage in tobacco. Cotton is highly susceptible to drought during boll setting stage, and if overexpression of GhDRIN1 in cotton could confer stress tolerance during this stage as is the case with tobacco, it will greatly aid in reducing the yield losses.
Proteins or regulatory factors with unknown function are valuable resources and their functional validation will help us to discover new pathways and mechanisms adapted by the plants to cope with various biotic and abiotic stresses. The present study has shown that GhDRIN1 might be a general stress-induced gene and may act upstream since its overexpression conferred tolerance to different stresses. Moreover, our results suggest that GhDRIN1 may be a promising candidate gene for engineering stress tolerance in agriculturally important crops, particularly cotton.
References
Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M (2010) Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10:49
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Dhandapani G, Kanakachari M, Padmalatha KV, Phanindra MLV, Singh VK, Raghavendrarao S, Jayabalan N, Lakshmi Prabha A, Kumar PA (2014) A gene encoding cold-circadian rhythm-RNA binding-like protein (CCR-Like) from upland cotton (Gossypium hirsutum L.) confers tolerance to abiotic stresses in transgenic tobacco. Plant Mol Biol Rep. doi: 10.1007/s11105-014-0729-x
Dowd C, Wilson IW, McFadden H (2004) Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f. sp. vasinfectum. Mol Plant Microb Interact 17:654–667
Gollery M, Harper J, Cushman J, Mittler T, Girke T, Zhu JK, Bailey-Serres J, Mittler R (2006) What makes species unique? The contribution of proteins with obscure features. Genome Biol 7:757
Gollery M, Harper J, Cushman J, Mittler T, Mittler R (2007) POFs: what we don’t know can hurt us. Trends Plant Sci 12:492–496
Gupta GP, Birah A, Ravi S (2004) Development of artificial diet for mass rearing of American bollworms (Helicoverpa armigera). Indian J Agric Sci 74:548–557
Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61:1041–1052
Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M, Girke T (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol 147:41–57
Luhua S, Ciftci-Yilmaz S, Harper J, Cushman J, Mittler R (2008) Enhanced tolerance to oxidative stress in transgenic arabidopsis plants expressing proteins of unknown function. Plant Physiol 148:280–292
Luhua S, Hegie A, Suzuki N, Shulaev E, Luo X, Cenariu D, Ma V, Kao S, Lim J, Gunay MB, Oosumi T, Lee SC, Harper J, Cushman J, Gollery M, Girke T, Bailey-Serres J, Stevenson RA, Zhu JK, Mittler R (2013) Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol Plant 148:322–333
Padmalatha KV, Dhandapani G, Kanakachari M, Kumar S, Dass A, Patil DP, Rajamani V, Kumar K, Pathak R, Rawat B, Leelavathi S, Reddy PS, Jain N, Powar KN, Hiremath V, Katageri IS, Reddy MK, Solanke AU, Reddy VS, Kumar PA (2012) Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Mol Biol 78:223–246
Wang B, Luttge U, Ratajczak R (2001) Effects of salt treatment and osmotic stress on V-ATPase and V-PPase in leaves of the halophyte Suaeda salsa. J Exp Bot 52:2355–2365
Zhang L, Li FG, Liu CL, Zhang CJ, Zhang XY (2009) Construction and analysis of cotton (Gossypium arboreum L.) drought-related cDNA library. BMC Res Notes 2:120
Acknowledgments
We are grateful to National Agricultural Innovation Project (NAIP/C4/C10103), Indian Council for Agricultural Research, New Delhi, India for the financial assistance. We thank Prof. I. S. Katageri, Agricultural Research Station, University of Agricultural Sciences, Dharwad, Karnataka for providing the drought-induced and bollworm infested cotton boll samples used in the study. We also thank to Dr. Shelly Praveen and Mr. Vipin Kumar, Division of Plant Pathology, Indian Agricultural Research Institute (IARI), New Delhi for facilitating confocal microscopy studies.
Supporting information
Supplementary Table 1—List of primers used in this study.
Supplementary Table 2—Prediction for subcellular localization of GhDRIN1 protein.
Supplementary Table 3—Percentage of fresh weight of WT and GhDRIN1 T 1 transgenics under long-term stress amended MS medium supplemented with 200 mM each of NaCl, mannitol and 10 % PEG.
Supplementary Table 4—Analysis of relative water content for WT and T 1 transgenic tobacco plants under transgenic glass house condition for 0 and 15 days after water withholding.
Supplementary Table 5—Analysis of total chlorophyll content in WT and T 1 transgenic tobacco plants under transgenic green house condition for 0 and 15 days after water withholding.
Supplementary Fig. 1—The phylogenetic tree showing the relationship between Gossypium hirsutum drought-induced protein with closely related other plant proteins. Phylogram was generated based on protein sequences using neighbor joining algorithm of MEGA (Molecular Evolutionary Genetics Analysis) software, version 6.0. Numbers above or below branches indicate bootstrap values from 1000 replicates. The plant name is followed by protein ID.
Supplementary Fig. 2—T-DNA region of binary vectors used for subcellular localization and plant transformation study. a GhDRIN1 cDNA fragment was cloned upstream of mGFP gene under BglII-SpeI site in the T-DNA region of pCAMBIA1302 and the derived vector named as pCAMBIA1302-CaMV35S-GhDRIN1::mGFP. b T-DNA regions of pCAMBIA1302 binary vector carrying CaMV35S-mGFP cassette in right border (RB) and HPTII as plant selection marker in left border. c pBI121 has the NPTII gene as plant selectable marker in right border (RB) and cotton GhDRIN1 cDNA fragment was cloned in the place of GUS reporter gene in left border (LB) driven by CaMV 35S promoter and NOS-terminator. The right border (RB) junction fragment of the T-DNA is >3.3 (the distance between HindIII and RB) for NPTII used as the probe.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dhandapani, G., Lakshmi Prabha, A., Kanakachari, M. et al. GhDRIN1, a novel drought-induced gene of upland cotton (Gossypium hirsutum L.) confers abiotic and biotic stress tolerance in transgenic tobacco. Biotechnol Lett 37, 907–919 (2015). https://doi.org/10.1007/s10529-014-1733-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-014-1733-9