Abstract
The vacuolar H+-ATPase plays a crucial role in secondary transport and in plant response to environmental stress. In this study, a vacuolar H+-ATPase (MxVHA-c) gene, consisting of an ORF of 498 base pairs and 165 amino acid residues, has been cloned from the iron-efficient genotype of Malus xiaojinensis. Subsequently, this gene has been targeted to the tonoplast using transient expression analysis. Quantitative real-time (qRT) PCR results reveal that the MxVHA-c gene is expressed in both roots and leaves of Fe-deficient plants; however, it is sensitive to iron stress in roots. This suggests that MxVHA-c expression in roots may mediate iron-dependent responses. MxVHA-c expression is up-regulated following exogenous treatment with abscisic acid (ABA) and down-regulated following treatment with CaCl2. Overexpression of the MxVHA-c gene in yeast strains has revealed that MxVHA-c transiently alleviated cadmium toxicity via the Cd2+/H+ antiport protein. H+-ATPase activity is slightly increased in yeast overexpressing the MxVHA-c gene compared to that in yeast transformed with an empty vector. In addition, this transgenic yeast strain can grow in a liquid medium containing 40 μM ferrozine. These findings may provide useful information in elucidating molecular mechanisms that mediate resistance to iron deficiency.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron (Fe) is an important nutrient element and is involved in many physiological and biochemical processes. Iron deficiency causes chlorosis and affects the production of crops (Briat et al. 1995; Mori 1999; Marschner et al. 1986). Plants can absorb iron from soil and also regulate iron balance in cells to satisfy the need of iron element. The vacuole is a primary storage site that mediates the distribution and transportation of nutrient in the plants. Following environmental damage in plants, vacuolar H+-ATPase (V-H+-ATPase) pumps H+ into the vacuole to establish an electrochemical potential gradient between the vacuole and cytoplasm, which may promote ion transportation. Furthermore, V-H+-ATPase can maintain ion balance under stress (Schumacher and Krebs 2010).
Malus xiaojinensis is an iron-efficient plant species that has been selected from more than 40 plant species and ecotypes in the genus Malus for investigation in this study. Understanding the transport of nutrients in fruit trees, from roots to scions, is very important (Han et al. 1994a; Xu et al. 2011). Thus, cloning and analysis of V-H+-ATPase genes from plants, especially from woody stock plants, may aid in providing insights into our understanding of balance of ions, such as that of iron, at the cellular level.
V-H+-ATPase is a functional complex and multi-subunit enzyme consisting of two sub-complexes as follows: a membrane-external V1 subunit with eight sub-complexes (A, B, C, D, E, F, G and H) and a membrane-integral V0 subunit with six sub-complexes (a, c, c′, c″, d and e) (Xiao et al. 2008; Ratajczak 2000; Dettmer et al. 2010). V-H+-ATPase genes have been cloned from a broad range of plant species: monocots, dicots, trees as well as fruits such as wheat (Zhao et al. 2009), Arabidopsis (Kluge et al. 1999), tobacco (Rouquie et al. 1998), cotton (Wan and Wilkins 1994) and Fuji (Yao et al. 2009). V-H+-ATPase subunits exhibit tissue specificity. VHA-c1 is expressed ubiquitously, and VHA-c3 expression is limited to roots caps. VHA-E2 is a pollen-specific gene (Hirata et al. 2003; Padmanaban et al. 2004; Gaxiola et al. 2007). Disruption of each V-H+-ATPase subunit (except for VPH1 or STV1) results in an identical phenotype that is characterized by the inability of yeast cells to grow at a pH higher than 7 and sensitivity to calcium concentrations in the medium (Aviezer-Hagai et al. 2000; Drory et al. 2004). In Arabidopsis, VHA-c is likely to be involved in transporting protons, and the rotation of a ring of six c subunits is necessary for driving proton transport.
V-H+-ATPase is not only a simple proton-pump but also has other functions. The Na+/H+ antiport protein in the tonoplast transports Na+ into the vacuole based on the gradient by V-H+-ATPase genes under salt stress (Zhang et al. 2009). V-H+-ATPase genes are involved in glucose signaling and in regulating the vacuolar pH value, which is essential for Arabidopsis root gravity perception (Fasano et al. 2001). VHA-c has a major role in transporting ions. In addition, it is involved in many stress responses such as salt stress, low temperature, heat and so on. The enhancement in the expression of V-H+-ATPase subunits has been demonstrated under salt- and abscisic acid (ABA)-induced stress (Kasai et al. 1994). Exogenous hormones can also induce the expression of the VHA-c gene (Zhang et al. 2006; Chinnusamy et al. 2006). VHA-c and H+-PPase are mainly used to transport protons across the tonoplast. VHA-c enhances the SOD and POD activities to improve the salt tolerance in transgenic tobacco (Xu et al. 2010).
Although V-H+-ATPase genes have been cloned from many species, there is little research analyzing these genes in woody plants, especially in apple. So we cloned a vacuolar H+-ATPase subunit c from M. xiaojinensis and analyzed its expression patterns in roots and leaves in response to iron deficiency, ABA and CaCl2. The sub-cellular location of MxVHA-c protein has been detected by transient expression analysis. Furthermore, the vacuolar H+-ATPase activity and the cell growth of yeast with overexpressed MxVHA-c gene under ion stresses were investigated. Therefore, the function of MxVHA-c gene in ion absorption and bivalent ion transportation was confirmed in yeast.
Materials and Methods
Plant Material and Growth Conditions
M. xiaojinensis seedlings were grown in a container with Murashige and Skoog (MS) medium containing 0.5 mg L−1 Indole-3-butytric acid (IBA). After 30 days, when white roots were approximately 10 cm, the roots were rinsed in distilled water three times. The seedlings were then transferred to 1/2 Hoagland and Hoagland nutrient solution for 15 days, respectively. The composition of Hoagland nutrient solution was: 40 μM Fe(III)–EDTA, 0.5 mM KNO3, 0.5 mM NH4H2PO4, 1 mM Ca(NO3)2, 0.5 mM MgSO4·7H2O, 0.5 mM CaCl2, 0.3 mM Mg(NO3)2·6H2O, 23 μM H3BO3, 0.4 μM ZnSO4·7H2O, 0.15 μM CuSO4·5H2O, 0.05 μM H2MoO4·H2O and 3 μM MnCl2. The pH was adjusted to 6.3 with 0.1 N KOH. And then, the solutions were changed to iron-insufficient (4 μM Fe–EDTA) (Han et al. 1994b), ABA (50 mg L−1) and CaCl2 (50 mg L−1) solutions. Roots and mature leaves were harvested being cultivated plants in iron-insufficient solutions for 0 h, 12 h, 1 day, 3 days, 6 days and 9 days, respectively. The same samples were harvested after being exposed to ABA and CaCl2 for 0, 12 and 24 h, respectively.
Cloning of MxVHA-c cDNAs
Total RNA was isolated from the roots and mature leaves using CTAB methods with some modifications (Gasic et al. 2004). Approximately 1 μg of RNA was digested by DNase (TaKaRa Biotechnology Co. Ltd., Dalian, China) and reverse-transcribed using an oligo-dT primer and reverse transcriptase (TaKaRa Biotechnology Co. Ltd., Dalian, China) in a total volume of 20 μL. The initial complementary DNAs (cDNAs) were used to clone MxVHA-c cDNAs. Based on the gene sequence of VHA and the genome of the Golden Delicious (http://genomics.research.iasma.it/) (Velasco et al. 2010), the primers were designed using primer 5.0. The primers were 5′-AAAGAATTCATGTCTTCTTCAACCTTC-3′ with EcoRI site and 5′-AAAGTCGACCCCTCAGCTCTTGACTGACC-3′ with SalI site. Amplification of the cDNA clone was performed at 94 °C for 5 min, 29 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, and the final extension was performed at 72 °C for 10 min. The amplified PCR products were purified and subcloned into the Peasy-T1 vector (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. Three clones were sequenced. Alignment of sequences was performed with DNAMAN software. Potential transmembrane segments were identified using TMHMM-2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Real-Time PCR
The cDNA reaction mixture was diluted with 20 μL distilled water for quantitative real-time (qRT) PCR, which was performed using the SYBR Premix Ex Taq (TaKaRa Biotechnology Co. Ltd., Dalian, China). Primers were designed by using Primer Premier 5 (PREMIER Biosoft International, Palo Alto, CA) to give an amplicon length of 100–150 bp. The primers were 5′-GGTAGTTATGGCGGGAGTGTGGGT-3′ and 5′-GGTAATAGGACTTAGCCAGCAGCCTTGGGGTT-3′. Gene specificity of primers was ensured using qRT-PCR and by analysis of the melting curves of the products. The Malus domestica 18S gene was used as a reference gene (GenBank: DQ341382), and the primers were 5′-ACACGGGGAGGTAGTGACAA-3′ and 5′-CCTCCAATGGATCCTCGTTA-3′. Each reaction was carried out in triplicate. The relative expression of the target gene in the treatment was calculated using quantitative real-time PCR and the 2−△△C T method (Livak and Schmittgen 2001; Qi et al. 2010; Tai et al. 2009).
Sub-cellular Localization of MxVHA-c Protein in Onion Epidermal Cells
The MxVHA-c gene and pEZS-NL plasmid (transient expression carrier) was digested using EcoRI and SalI and ligated with T4 DNA ligase (TransGen Biotech, Beijing, China) to form the pEZS-NL+MxVHA-c plasmid. The recombinant plasmid was transformed into DH-5α for amplification. Recombined plasmid DNA was extracted. The recombinant plasmid DNA and pRTL2-GFP plasmid DNA (control) were integrated into onion epidermal cells using a particle bombardment device (Biolistic PDS-1000/He, Bio-Rad) according to the manufacturer’s instructions. After 12 h, the fluorescence was observed using a florescence microscope (Nikon Eclipse TE2000-E).
Isolation of the Vacuolar Membrane Vesicles and Enzyme Activity Analysis
The pEZS-NL+MxVHA-c and pEZS-NL plasmid DNAs were digested with EcoRI and XbaI to obtain MxVHA-c+eGFP and eGFP fragments. These two fragments were ligated to the pYES2.0 plasmid respectively which contained Ura selection maker driven by GAL promoter. Therefore, the recombinant yeast expression vector pYES2.0+MxVHA-c+eGFP plasmid was created, and pYES2.0+eGFP plasmid was used as a control. The recombinant plasmids were transformed into the wild-type yeast strain BJ2168 with pYES2 vector kit instructions (Invitrogen). Yeast growth (YPD) medium and selection (SD-Ura−) medium were prepared. A single colony from the SD-Ura− plate was inoculated into 2 mL of SD-Ura− liquid medium and incubated at 30 °C with shaking at 200 rpm for 24 h. Vacuolar membrane vesicles were isolated using the supercompetent cell membrane vesicle preparation kit (GENMED SCIENTIFICS INC., USA). The ability of H+ transport was measured by green fluorescence indicating H+ transportation using an aspartame assay kit (GENMED SCIENTIFICS INC., USA).
The Function of MxVHA-c in Fe-Deficient Yeast
The MxVHA-c overexpressing yeast strains and the empty vector-expressing yeast strains were cultured overnight in SD-Ura− liquid medium. When the OD value reached 1, 40 μM ferrozine (to induce Fe deficiency) was added to the medium. After 12, 24 and 48 h, the OD values were determined.
The Tolerance of Cd 2+
The yeast strain expressing pYES2.0+MxVHA-c+eGFP and the strain transformed with the empty vector were grown in 20 mL SD-Ura− liquid medium until the OD reached 0.1, respectively. The cells were cultured with 10 μM CdCl2 at 30 °C with shaking at 200 rpm (Gao et al. 2011). The OD values were measured at 0, 24, 48, 72 and 96 h since CdCl2 addition.
Results
Cloning and Sequence Analyses of the MxVHA-c Gene from Malus xiaojinensis
A complete open reading frame of 498 base pairs was cloned from M. xiaojinensis using PCR. The predicted protein encoded by MxVHA-c gene consisted of 165 amino acids with a predicted molecular weight of 16.6 kDa.
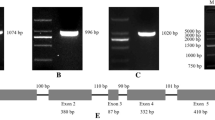
To investigate the homology and genetic relationship with other plants, the alignment of the deduced amino acid was performed. The MxVHA-c gene exhibited similarities with the VHA gene from Arabidopsis thaliana (GenBank ID: NP177693.1), Citrus unshiu (GenBank ID: BAA75516.1), Nicotiana tabacum (GenBank ID: CAA65063.1), Plantago major (GenBank ID: CAH58637.1) and Golden Delicious with the sequence identity up to 98 %. VHA gene families in different species may have similar structures. However, difference in a single amino acid may lead to species-specific distinctions (Fig. 1a, b). It was shown that this protein has four potential transmembrane domains (Fig. 1c, Table 1). Using the genome of domesticated apple as a comparison (Velasco et al. 2010), the results ascertained that MxVHA-c gene included three exons and two introns. This result was consistent with the structure of the PgVHA-c1 gene (Tyagi et al. 2005).
Identification and sequence alignment of deduced amino acids of MxVHA-c with other related VHA genes. a Compared protein sequences are from Arabidopsis thaliana (AVA-p4), Citrus unshiu (CitVATP-c), Nicotiana tabacum (NtVATP-c), Plantago major (PmVATPc-1) and Golden Delicious (MdVHA-c). Identical domains are marked by black color. The different amino acids are labeled by white color. b Evolutionary tree analysis of MxVHA-c with VHA genes in others species. The identity is confirmed by percentage. c The transmembrane domains analysis of MxVHA-c protein by TMHMM-2.0
Expression Analysis of MxVHA-c Gene
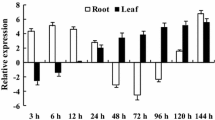
The expression analysis of MxVHA-c gene during iron deficiency was investigated using qRT-PCR (Fig. 2). MxVHA-c expression was detected in roots and mature leaves. After iron deficiency for 12 h, MxVHA-c expression in roots increased by 4.0-fold. The transcript correlated with the length of iron deficiency. In leaves, the expression of MxVHA-c gene increased at the initial stages of iron deficiency, reached peak at 3 days and subsequently decreased. The transcript levels of the MxVHA-c gene were 1.2-fold and 1.8-fold up-regulated in leaves and roots under ABA stress. MxVHA-c expression was down-regulated in response to CaCl2.
Quantitative real-time PCR analysis of MxVHA-c expression in leaves and roots. With 18 s as control, the relative expression of this gene was calculated by 2−△△C T method. Values are means of three biological replicates. Standard error is also labeled. a Iron deficiency stress. b 50 mg L−1 IAA. c 50 mg L−1 CaCl2
Sub-cellular Localization of the MxVHA-c Gene
The MxVHA-c gene encoded a membrane protein. The sub-cellular localization of MxVHA-c was visualized using a transiently expressed eGFP fusion protein. We detected GFP-tagged MxVHA-c protein around tonoplast on onion epidermal cells (Fig. 3d), while the pRTL2-GFP was located around all the cells (Fig. 3b). There was no GFP fluorescence in other sites of cells.
Sub-cellular localization of MxVHA-c protein in onion epidermal cells using eGFP by common florescence microscope. Cells were observed after 12 h of infection. Images a and c are pictured in the dark field, while b and d are bright field images. Panels a and b represent fusion protein (GFP) in the pRTL2-GFP, while c and d represent MxVHA-c fused to GFP in the pEZS-NL
Vacuolar H+-ATPase Activity Analysis
H+ transport ability reflected vacuolar H+-ATPase activity. The relative fluorescence unit (RFU) in transgenic yeast was lower than that in yeast with pYES2.0+eGFP (Fig. 4). This result illustrates that MxVHA-c gene enhances H+ transport from the cytoplasm into vacuole, indicating that the MxVHA-c gene encoded a vacuolar H+-ATPase of M. xiaojinensis.
The Growth of Transgenic Yeast in the Iron-Deficient Medium
MxVHA-c overexpression yeast strains exhibited normal growth in iron-deficient medium. The OD value of transgenic yeast was more than 1.5 times (1.6 ± 0.02) greater than that of the control (1.096 ± 0.002) at 12 h. After 12 h, the differences between the two values were less dramatic. These results indicate that the kinetics of MxVHA-c mediating Fe2+ transport is time-dependent with a peak at 12 h (Fig. 5).
Yeast strains which overexpressed MxVHA-c gene can grow in the iron-deficient medium. Cells were cultured overnight in the SD-Ura− medium at 30 °C with shaking at 200 rpm. Until the OD value was up to 1, then 40 μM ferrozine was added in the medium; after that the OD value was determined after 12, 24 and 48 h. Three replicates can be done every time. The average value and standard error are also printed
Cd 2+ Tolerance in the Transgenic Yeast
To determine whether the transgenic yeast can enhance the tolerance of heavy metals, we measured the cell growth value. During cultivation, 10 μM CdCl2 inhibited the growth of wild-type yeast that was transformed with the empty vector compared to that of yeast transformed with the MxVHA-c gene. The OD value of yeast with empty vector decreased, whereas the OD value of yeast transformed with MxVHA-c gene increased within 24 h. After 24 h, the growth decreased (Fig. 6). This observation suggested that cadmium toxicity was transiently alleviated in the transgenic yeast strain. However, transgenic yeast cannot maintain normal growth after 2 days.
Yeast strains which overexpressed MxVHA-c gene can transiently alleviate cadmium toxicity. Pre-cultured cells were cultured in the SD-Ura− medium with 10 μM CdCl2 at 30 °C with shaking at 200 rpm which started from an OD of 0.1. After 24, 48, 72 and 96 h of culture, cell growth can be got by OD600. Three replicates can be done every time. The average value and standard error are also printed
Discussion
Plants must make appropriate physiological and biochemical changes in order to deal with environmental stresses (Fu et al. 2010). The H+-ATPase gene plays a crucial role in these processes. In this study, we reported a V-H+-ATPase subunit c gene consisting of 498 base pairs fragment in the M. xiaojinensis. This gene has a high homology with VHA genes in other species. The proton-pumping activity may be regulated by complex formation with the V0 subcomponent, which is located within the membrane. One of V-H+-ATPase’s functions in plants is to create the electrochemical gradient across the tonoplast, which maintains a stable environment despite stress. Together with other V-H+-ATPase genes, VHA genes may influence the pH homeostasis in the cytoplasm (Nishi and Forgac 2002). The V-H+-ATPase subunit c is essential for transporting protons and regulating V-H+-ATPase activity (Sze et al. 2002). The vacuole is an important storage organ for nutritional elements, including Fe. The MxVHA-c gene expression was up-regulated in Fe-deficient plant roots and leaves. The yeast strain transformed with the MxVHA-c gene displayed an improved survival rate under 40 μM ferrozine, relative to the strain transformed with the empty vector. This result indicates that MxVHA-c regulates ion transport and balance in transformed cells. Iron balance in the cells requires the coordination of many genes. AtNRAMP3 and AtNRAMP4, which encode proteins located on the surface of the tonoplast, are expressed earlier than IRT1 (iron-transport protein 1). These proteins transport Fe2+ to the cytoplasm to meet the demands of plants (Lanquar et al. 2005). The VIT1 (vacuolar membrane transport) gene regulates iron balance between vacuole and cytoplasm, and vit1 mutants cannot grow in alkaline soil (Kim et al. 2006). In our study, MxVHA-c gene was expressed in Fe-deficient plant roots and leaves. In addition, MxVHA-c mediates H+ balance in the vacuole. From above analysis, we can deduce that MxVHA-c gene may also interact with other genes to regulate iron homeostasis in M. xiaojinensis.
In Fe-deficient plants, roots initially induce signals in response to Fe deficiency. Our results are consistent with this finding. QRT-PCR indicates that MxVHA-c gene is expressed in roots and leaves. MxVHA-c expression peaks at 12 h in the roots. The increasing of H+ in the vacuole causes acidification, enhancing Fe2+ removal. The signals that are involved in this process have not been well characterized. Iron content in the surrounding soil of the roots induces changes in local signals in the plants (Curie and Briat 2003; Yang et al. 2010). V-H+-ATPase activity and endogenous ABA content increase under salt stress. In addition to this, exogenous ABA enhances V-H+-ATPase activity (Zhao et al. 2009). The transcriptional levels will be up-regulated under ABA and CaCl2 treatments (Tyagi et al. 2005). Exogenous ABA enhanced the expression of MxVHA-c gene. Ca2+ signaling has different effects on V-H+-ATPase genes in different plants (Tyagi et al. 2005). CaCl2 suppressed MxVHA-c expression in M. xiaojinensis. Because Ca2+ and ABA may influence MxVHA-c gene expression, the properties of MxVHA-c gene in signal pathway require further investigation.
Yeast is a useful model organism that has been extensively studied, and it has a mechanism of iron absorption that is similar to that of plants (Zhang et al. 2009). Yeast stores many ions in the vacuole. Overexpressing of MxVHA-c gene in yeast induces elevation of V-H+-ATPase activity compared to yeast transformed with an empty vector. Yeast transformed with MxVHA-c gene reveals lower levels of RFU. This result indicates that H+ transportation from the cytoplasm into vacuole influences ion distribution between cytoplasm and vacuole. This phenomenon is in accordance with previous studies. Transferability of Cd2+ in plants is regulated by pH. In oat roots, Cd2+ transportation from cytoplasm to vacuole across the tonoplast is demonstrated through Cd2+/H+ antiport protein (Prasad 1995). And also the absorption rate of Cd2+ in the Fe-deficient plant roots is seven times greater than that in the Fe-sufficient plant roots (Cohen et al. 1998; Kabała et al. 2008). The increasing of Cd2+ absorption may cause metal ion poisoning. The ThVHA-c1 can greatly improve CdCl2 tolerance in transgenic yeast (Gao et al. 2011). The transgenic yeast had a relatively high survival rate under 10 μM CdCl2. This phenomenon illustrated that overexpression of MxVHA-c gene inhibited cadmium toxicity. Therefore, we hypothesize that Cd2+/H+ antiport protein is present in the tonoplast to transport excess Cd2+ from cytoplasm into vacuole in M. xiaojinensis. However, the transporting activity of the Cd2+/H+ antiport protein is limited. Similar to the cellular response to salt stress, this process may also be regulated by SOS signals (Zhao et al. 2009).
Abbreviations
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- VHA :
-
Vacuolar H+-ATPase
- YPD:
-
Yeast growth medium
- SD:
-
Selection medium
- RFU:
-
Relative fluorescence unit
- eGFP:
-
Enhanced green fluorescent protein
References
Aviezer-Hagai K, Nelson H, Nelson N (2000) Cloning and expression of cDNAs encoding plant V-ATPase subunits in the corresponding yeast null mutants. Biochim Biophys Acta 1459:489–498
Briat JF, Fobis-Loisy I, Grignon N, Lobreaux S, Pascal N, Savino G, Thoiron S, von Wiren N, Van Wuytswinkel O (1995) Cellular and molecular aspects of iron metabolism in plants. Biol Cell 84:69–81
Chinnusamy V, Zhu JH, Zhu JK (2006) Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng 27:141–177
Cohen CK, Fox TC, Garvin DF, Kochian LV (1998) The role of iron-deficiency stress responses in stimulating heavy-metal transport in plants. Plant Physiol 116:1063–1072
Curie C, Briat JF (2003) Iron transport and signaling in plants. Annu Rev Plant Biol 54:183–206
Dettmer J, Liu TY, Schumacher K (2010) Functional analysis of Arabidopsis V-ATPase subunit VHA-E isoforms. Eur J Cell Biol 89:152–156
Drory O, Frolow F, Nelson N (2004) Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep 5:1148–1152
Fasano JM, Swanson SJ, Blancaflor EB, Dowd PE, Kao TH, Gilroy S (2001) Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell 13:907–921
Fu WD, Shuai L, Yao JT, Yu SH, Liu FL, Duan DL (2010) Molecular cloning and analysis of a cytosolic Hsp70 gene from Enteromorpha prolifera (Ulvophyceae, Chlorophyta). Plant Mol Biol Report 28:430–437
Gao CQ, Wang YC, Jiang B, Liu GF, Yu LL, Wei ZG, Yang CP (2011) A novel vacuolar membrane H+-ATPase c subunit gene (ThVHAc1) from Tamarix hispida confers tolerance to several abiotic stresses in Saccharomyces cerevisiae. Mol Biol Rep 38:957–963
Gasic K, Hernandez A, Korban SS (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Report 22:437a–437g
Gaxiola RA, Palmgren MG, Schumacher K (2007) Plant proton pumps. FEBS Lett 581:2204–2214
Han ZH, Shen T, Korcak RF, Baligar VC (1994a) Screening for iron-efficient species in the genus Malus. J Plant Nutr 17:579–592
Han ZH, Wang Q, Shen T (1994b) Comparison of some physiological and biochemical characteristics between iron-efficient and iron-inefficient species in genus Malus. J Plant Nutr 17:1257–1264
Hirata T, Iwamoto-Kihara A, Sun-Wada GH, Okajima T, Wada Y, Futai M (2003) Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits. J Biol Chem 278:23714–23719
Kabała K, Janicka-Russak M, Burzynski M, Kłobus G (2008) Comparison of heavy metal effect on the proton pumps of plasma membrane and tonoplast in cucumber root cells. J Plant Physiol 165:278–288
Kasai M, Yamamoto Y, Matsumoto H (1994) In vivo treatment barley roots with vanadate increases vacuolar H+-translocating ATPase activity of the tonoplast-enriched membrane vesicles and the level of endogenous ABA. Plant Cell Physiol 35:291–295
Kim SA, Punshon T, Lanzirotti A, Li LT, Alonso JM, Ecker JR, Kaplan J, Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314:1295–1298
Kluge C, Golldack D, Dietz KJ (1999) Subunit D of the vacuolar H+-ATPase of Arabidopsis thaliana. Biochim Biophys Acta 1419:105–110
Lanquar V, Lelievre F, Bolte S, Hames C, Alcon C, Neumann D, Vansuyt G, Curie C, Schroder A, Kramer U, Barbier-Brygoo H, Thomine S (2005) Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J 24:4041–4051
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 25:402–408
Marschner H, Romheld V, Kissel M (1986) Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr 9:695–713
Mori S (1999) Iron acquisition by plants. Curr Opin Plant Biol 2:250–253
Nishi T, Forgac M (2002) The vacuolar (H+)-ATPases—nature’s most versatile proton pumps. Nat Rev Mol Cell Biol 3:94–103
Padmanaban S, Lin XY, Perera I, Kawamura Y, Sze H (2004) Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi. Plant Physiol 134:1514–1526
Prasad MNV (1995) Cadmium toxicity and tolerance in vascular plants. Environ Exp Bot 35:525–545
Qi JN, Yu SC, Zhang FL, Shen XQ, Zhao XY, Yu YJ, Zhang DS (2010) Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol Biol Report 28:597–604
Ratajczak R (2000) Structure, function and regulation of the plant vacuolar H+-translocating ATPase. Biochim Biophys Acta 1465:17–36
Rouquie D, Tournaire-Roux C, Szponarski W, Rossignol M, Doumas P (1998) Cloning of the V-ATPase subunit G in plant: functional expression and sub-cellular localization. FEBS Lett 437:287–292
Schumacher K, Krebs M (2010) The V-ATPase: small cargo, large effects. Curr Opin Plant Biol 13:724–730
Sze H, Schumacher K, Müller ML, Padmanaban S, Taiz L (2002) A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends Plant Sci 7:157–161
Tai HH, Conn G, Davidson C, Bud Platt HW (2009) Arbitrary multi-gene reference for normalization of real-time PCR gene expression data. Plant Mol Biol Report 27:315–320
Tyagi W, Rajagopal D, Singla-Pareek SL, Reddy MK, Sopory SK (2005) Cloning and regulation of a stress-regulated Pennisetum glaucum vacuolar ATPase c gene and characterization of its promoter that is expressed in shoot hairs and floral organs. Plant Cell Physiol 46:1411–1422
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Wan CY, Wilkins TA (1994) Isolation of multiple cDNAs encoding the vacuolar H+-ATPase subunit B from developing cotton (Cossypium hirsutum 1.) ovules. Plant Physiol 106:393–394
Xiao ZY, Tan KL, Hu MY, Liao P, Chen KJ, Luo M (2008) Cloning and expression analysis of GhDET3, a vacuolar H+-ATPase subunit C gene, from cotton. J Genet Genomics 35:307–312
Xu CX, Zheng L, Gao CQ, Wang C, Liu GF, Jiang J, Wang YC (2010) Overexpression of a vacuolar H+-ATPase c subunit gene mediates physiological changes leading to enhanced salt tolerance in transgenic tobacco. Plant Mol Biol Report 29:424–430
Xu HM, Wang Y, Chen F, Zhang XZ, Han ZH (2011) Isolation and characterization of the iron-regulated MxbHLH01 gene in Malus xiaojinensis. Plant Mol Biol Report 29:936–942
Yang TJW, Lin WD, Schmidt W (2010) Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol 152:2130–2141
Yao YX, Li M, Liu Z, You C-X, Wang D-M, Zhai H, Hao Y-J (2009) Molecular cloning of three malic acid related genes MdPEPC, MdVHA-A, MdcyME and their expression analysis in apple fruits. Sci Hortic 122:404–408
Zhang JH, Liu YP, Pan QH, Zhan JC, Wang XQ, Huang WD (2006) Changes in membrane-associated H+-ATPase activities and amounts in young grape plants during the cross adaptation to temperature stresses. Plant Sci 170:768–777
Zhang QX, Xu XF, Wang Y, Li TZ, Han ZH (2009) Intracellular localization of Na+/H+ antiporter from Malus zumi (MzNHX1). Afr J Biotechnol 8:6784–6786
Zhao Q, Zhao YJ, Zhao BC, Ge RC, Li M, Shen YZ, Huang ZJ (2009) Cloning and functional analysis of wheat V-H+-ATPase subunit genes. Plant Mol Biol 69:33–46
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (30971982), The National Transgenic Special Project (2009ZX08009-122B), and The Key Laboratory of Beijing Municipality of Stress Physiology and Molecular Biology for Fruit Trees.
Author information
Authors and Affiliations
Corresponding author
Additional information
Qian Zhang and Yi Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, Q., Wang, Y., Zhang, X.Z. et al. Cloning and Characterization of MxVHA-c, a Vacuolar H+-ATPase Subunit C Gene Related to Fe Efficiency from Malus xiaojinensis . Plant Mol Biol Rep 30, 1149–1157 (2012). https://doi.org/10.1007/s11105-012-0426-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-012-0426-6