Abstract
Photoperiod is one of the important environmental triggers of flowering, and the CONSTANS (CO) gene is a key regulator of this pathway in the model plant Arabidopsis thaliana. CO-like genes are present in many plant species. We describe here the isolation of the CO homolog Glycine max CONSTANS-like 9 (GmCOL9) from the soybean cultivar Kennong18. Sequence comparisons showed that the closest A. thaliana gene to GmCOL9 is COL13. The expression of GmCOL9 was investigated using quantitative real-time reverse transcriptase polymerase chain reaction, which showed that GmCOL9 was not regulated by a circadian mechanism, since its expression oscillated at a cycle of 12 h instead of 24 h. GmCOL9 expression was influenced by the developmental stage of the plant and varied between tissue/organs. The expression of GmCOL9 was concentrated in the unifoliolate leaves during the vegetative phase of growth and in the stem during the reproductive phase. However, traces of expression were also present throughout the plant, indicating that GmCOL9 may be a multifunctional gene in the context of the development of the soybean plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The successful completion of a plant’s life cycle depends heavily on the environmental conditions it experiences. Of the many distinct components of the growing environment, photoperiod is especially important, as it represents the trigger for the switch from vegetative to reproductive growth in many plant species. Plants that respond to a lengthening day length are referred to as long-day plants and those to a shortening one as short-day ones. The sensing of photoperiod and the subsequent molecular events which respond to this stimulus forms the so-called photoperiod pathway. A key component in this pathway is the gene CONSTANS (CO; Putterill et al. 1995; Mouradov et al. 2002), which has orthologs in a wide range of plant species (Yano et al. 2000; Liu et al. 2001; Griffiths et al. 2003; Miller et al. 2008; Holefors et al. 2009; Serrano et al. 2009). CO is a transcription factor containing two B boxes and one CCT (CO, CO-like, TOC1) domain (Robson et al. 2001). The CO gene family has been classified into three recognizable groups on the basis of B box content. Group I members have both a B1 and a B2 box, group II members a B1 and a variant B2 box, while group III members possess only one B box; all, however, retain the CCT domain(Robson et al. 2001; Griffiths et al. 2003). The expression profiles of some CO orthologs are also well conserved, and these CO orthologs can complement the Arabidopsis thaliana co mutant. For example, when the Pharbitis nil (short-day species) gene PnCO was over-expressed in the background of the A. thaliana co mutant, flowering was accelerated independently of photoperiod (Liu et al. 2001; Hayama et al. 2007); similarly, BvCOL1 (isolated from the long-day species Beta vulgaris) complements the late flowering phenotype of the A. thaliana co-2 mutant under various photoperiodic conditions (Chia et al. 2008). Thus, CO function seems to be well conserved across the plant kingdom.
However, CO orthologs have different functions in different species, and CO-like genes have differently functions compared with CO flowering activity. Whereas AtCOL3 is active during root development (Datta et al. 2006), AtCOL9 is involved in the regulation of flowering time via the suppression of CO (Cheng and Wang 2005). Under non-inductive long-day conditions, Hd1 (a rice ortholog of CO) delays flowering by inhibiting the action of Hd3a but accelerates it under inductive short-day conditions (Hayama et al. 2003). The CO/FT regulatory module in Populus trichocarpa controls both flowering and growth cessation and bud set in the fall (Bohlenius et al. 2006). The over-expression of CO impairs tuber formation in transgenic potato grown under short-day conditions (Martinez-Garcia et al. 2002). Thus, the CO gene family clearly has a wide-ranging influence over plant development.
Here, we report the isolation of a CO-like gene from soybean (Glycine max L.), named GmCOL9 (CONSTANS-like 9), and present an analysis of its structure and expression profiles in circadian, developmental, and tissue-/organ-specific patterns. Our results suggested that GmCOL9 may be involved in several aspects of the development of the soybean plant.
Materials and Methods
Plant Materials
The soybean cultivar Kennong18 (G. max L. KN18) was employed for all experiments. Plants were grown in a growth room under either short-day conditions (8:16 h light/dark) or long-day conditions (16:8 h light/dark) at 28°C under a light fluency of 100–150 μmol m−2 S−1. Seedlings were harvested before the expansion of the unifoliolate leaves. Various tissues/organs including roots, hypocotyls, epicotyls, cotyledons, unifoliolates, shoot apex (including the apical meristem and immature leaves), stems, leaves on lateral branches, and the whole aerial organs of plants were individually sampled when the unifoliolate leaves, the first trifoliolates, the second trifoliolates, the third trifoliolates, or the fourth trifoliolates had become fully expanded or when plants flowering. Seeds and pods without seeds were sampled at 7, 14, and 21 days after flowering, as well as at maturity. To determine the influence on gene expression of photoperiod, fully expanded unifoliolate leaves were sampled at ZTL0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, and 96, which spanned two successive 24-h light/dark cycles followed by two successive 24-h periods of continuous light or darkness. All samples were immediately frozen in liquid nitrogen and stored at −80°C until required.
RNA Isolation and cDNA Synthesis
Total RNA was extracted by the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Total RNA from the stems was isolated by the cetyltrimethylammonium bromide method (Suzuki et al. 2008). All RNA preparations taken forward had an A260/A280 ratio of 1.8–2.0 and an A260/A230 ratio of >2.0. The integrity of the RNA was checked electrophoretically. Prior to cDNA synthesis, the RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA), and first-strand cDNA synthesis was carried out using 4 μg RNA with the help of a RevertAid first-strand cDNA synthesis kit (Fermentas, St. Leon-Roth, Germany) provided with oligo-dT primers.
Gene Isolation and In Silico Analysis
The Glyma19g05170 sequence, which included both the untranslated and the coding (cds) regions, was first cloned using reverse transcriptase polymerase chain reaction (RT-PCR), based on the primers GmCOL9F-F and GmCOL9F-R (Table 1). Three independent fragments were sequenced (SunBio, China), and the resulting consensus sequence was used as the template to amplify the cds using the primer pair GmCOL9C-F and GmCOL9C-R (Table 1). This amplicon was inserted into the pGWC vector (Chen et al. 2006), resulting in the construct pGWC-GmCOL9, which was verified by sequencing. The alignment was performed using Molecular Evolutionary Genetics Analysis (MEGA) v4.0. Phylogenetic analyses were conducted using v4 of MEGA software (Tamura et al. 2007). To generate a phylogenetic tree, predicted full-length proteins were aligned by ClustalW software using default parameters. A neighbor-joining method was used to align and construct phylogenetic trees. Bootstraps with 1,000 replicates were performed to provide probabilistic support to tree nodes.
Quantitative Real-Time RT-PCR
Expression profiles were evaluated by quantitative real-time RT-PCR, carried out on an ABI StepOne Detection System (Applied Biosystems, USA), based on SYBR Premix Ex Taq polymerase (TaKaRa, Tokyo, Japan). Each 15-μl reaction consisted 4 μl template, 7.5 μl 2× SYBR Premix, 200 nM of each primer (qGmCOL9-F and qGmCOL9-R, see Table 1), and 0.3 μl ROX. Two independent reference genes were included to improve the level of reliability (Hu et al. 2009)—for the developmental samples, these were SKIP16 and UKN1; for tissues/organs, these were ACT11 and UKN1; and for photoperiod comparisons, these were ACT11 and TUA5. The relevant primer sequences are listed in Table 1. The data were analyzed by StepOne software v2.0. Each experiment was repeated at least three times.
Sub-cellular Localization of GmCOL9 Proteins
The coding sequence of GmCOL9 was introduced in pGWC (Chen et al. 2006) and subsequently cloned into destination vector pENSG-YFP (provided by Dr. Jane Parker, Max Planck Institute, Cologne, Germany) by LR Gateway recombination (Invitrogen). The resulting construct pENSG-YFP-GmCOL9, which includes the CaMV 35S promoter, was biolistically co-transformed into onion epidermis cells with the vector pENSG-CFP-AHL22, which served as a maker of nuclear proteins (Xiao et al. 2009). Transformation was achieved with the PDS 1000/He device (Bio-Rad), utilizing the following parameters: target distance 6 cm, vacuum 25″ Hg, and rupture disk pressure 1,100 psi. Yellow fluorescent protein (YFP) was monitored by confocal microscopy (LEICA TCS SP2, Germany).
Results
Cloning the Gene of GmCOL9
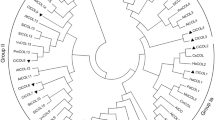
A search of the soybean genome sequence (http://www.phytozome.net/), based on the CO peptide sequence, produced 23 hits. The sequence of the locus Glyma19g05170 was related more closely to AtCOL13 than to any other AtCO family member (Fig. 1). The Glyma19g05170 locus was re-sequenced from cv. Kennong18 to produce full-length cds of 1,056 bp, which translated into a 352 residue polypeptide. Its sequence differed from that of Glyma19g05170 with respect to three base positions: G287 to A, A882 to G, and T921 to C, but only the first of these variants resulted in an amino acid change (glycine to aspartic acid; such an amino acid change did not fall in B boxes or CCT domain). Such single-nucleotide polymorphisms may result from the variation of different cultivars (KN18 in this study and Williams 82 in Phytozome). As do all CO group III members (Robson et al. 2001), the GmCOL9 protein contained two B boxes and a CCT domain (Fig. 2). The GmCOL9 B box sequences included both the conserved CX2CX8CX7CX2C motif, the critical C and H residues, and the consensus spacing defining B box function (Robson et al. 2001; Griffiths et al. 2003). The C-terminus of the GmCOL9 protein contained a conserved CCT domain NF-YA1/linker/NF-YA2 structure, and all the key residues were present (Fig. 2). A 24 residue stretch followed the B2 box, but this lacked any functional motif except for an SVAD phosphorylation site.
A sequence-based phylogeny of GmCOL9 and the A. thaliana CO family. GmCOL9 is related to the A. thaliana CO family group II, and its most closely related member is AtCOL13. The classification of CO family in Arabidopsis is according to Robson et al. 2001
Conserved domains alignment of GmCOL9 and COL13 from Arabidopsis. Shown are the conserved cysteine (black arrowheads) and histidine (black stars) residues, along with the consensus spacing (Xn) defining the two B box domains (B box1 shown by a single line, B box2 by a double line). The CCT sub-domains NF-YA1 and NF-YA2 and their linker, as well as the key residues within it, are marked by gray arrowheads
Circadian Expression Pattern
The expression profile of GmCOL9 in response to variation in the photoperiod was obtained from measurements of mRNA abundance in unifoliolates over the course of two successive 24-h cycles of either long or short days, followed by 48 h of continuous light or darkness. Under the short-day conditions, expression followed an oscillation period of 12 h, peaking first at dawn (ZTL0) and then again 4 h after the beginning of the dark period (ZTL12; Fig. 3a, b). Such a period was damped in subsequent continuous light (LL) but increased the amplitude in subsequent continuous dark (DD). And GmCOL9 expression in LL (Fig. 3a) and DD (Fig. 3b) was random. The expression pattern of GmCOL9 in long-day conditions was similar to that in short-day conditions, except that the peaks appeared 4 h later. And this pattern was also not maintained once the plants were transferred into continuous light or continuous darkness. In fact, mRNA abundance was generally greater in the dark (Fig. 3d) than in the light (Fig. 3c). The results suggested that GmCOL9 did not show a circadian expression pattern.
Developmental Expression Pattern
The effect of developmental stage on GmCOL9 expression was followed from comparisons between leaves harvested from plants at several range of developmental stages (Fig. 4). GmCOL9 transcript accumulated at higher levels the younger the plants (Fig. 4a, b). Comparing the expression levels in leaves at distinct developmental stages, higher level was observed in the unifoliolates, while the fourth trifoliolates displayed lowest levels of GmCOL9 mRNA. At the stage when the first trifoliolates were fully opened, GmCOL9 was mainly expressed in the unifoliolates and the first trifoliolates. Once the second trifoliolates were fully opened, most of the transcript was present in the unifoliolates, and by the time that the third trifoliolates were fully opened, most of the transcript was present in the unifoliolates and the second trifoliolates. In the developing seed, GmCOL9 expression was at a low level, but the level of expression rose as the seeds approached maturity (Fig. 4c).
The expression profile of GmCOL9 in leaves (a), aerial parts of the plant (b), and seeds (c). a The left-hand Y-axis shows unifoliolates (black column), and the right-hand Y-axis various trifoliolates (gray columns). U and T1–T4 indicated different leaves (organs) or the developmental stages when leaves opened fully (stages). U fully opened unifoliolates, T1 fully opened first trifoliolates, T2 fully opened second trifoliolates, T3 fully opened third trifoliolates, T4 fully opened fourth trifoliolates, F flowering. 7DAF, 14DAF, and 21DAF indicate, respectively, 7, 14, and 21 days after flowering. M seed maturation. Error bars denote SD
Tissue/Organ-Specific Expression Pattern
Although GmCOL9 transcript was detectable throughout the plant, the level of expression varied among tissues/organs (Fig. 5). The level in the stem was up to 100-fold that in other parts of the plant. Transcript abundance was also substantial in the unifoliolates. The expression of GmCOL9 was detected in roots although in lower levels than that in leaves and cotyledons. Lateral leaves, flower buds, pods excluding the seeds, shoot meristems, epicotyls, and hypocotyls also had different level of GmCOL9.
Tissue/organ expression profiles. SL seedling, R root, HH hypocotyl, EH epicotyl, C cotyledon, U unifoliolate leaf, SAM shoot apex (including the apical meristem and immature leaves), St stem, L lateral leaf, T1 to T4 first to fourth trifoliolates, F flower buds, P1 to P3 pods (excluding seed) at 7, 14, and 21 days after flowering. Stages defined as follows: unifoliolates unifoliolates fully opened, flowering onset of flowering, seed set initiation of seed growth. The right-hand Y-axis refers only to St at flowering (gray column), while the left-hand Y-axis applies to the other samples (black columns). Error bars denote SD
Sub-cellular Localization of GmCOL9 Protein
To study the sub-cellular localization of GmCOL9 protein, an expression vector containing yellow fluorescence protein-GmCOL9 (YFP-GmCOL9) fusion gene driven by 35S promoter was constructed and introduced into onion epidermis cells by biolistic transformation. As shown in Fig. 6, YFP-GmCOL9 was co-localized with the nuclear protein AHL22 (Xiao et al. 2009) in nuclei, not in the cytoplasm.
GmCOL9 protein co-localized with a nuclear protein AHL22 in nucleus of onion cells. YFP was fused to the N-terminus of GmCOL9, and CFP was also at the N-terminus of AHL22. Both the gene fusions were driven by a constitutive promoter. Two constructs were co-bombarded into onion epidermis cells and fluorescence visualized by confocal microscopy. a Yellow fluorescent signal, b cyan fluorescent signal, c bright field, d merged of a and c, e merged of b and c, f merged of d and e. Bar, 50 μm
Discussion
CO is a key gene in photoperiod pathway of flowering regulation, and its orthologs were found in various plants (review by Khanna et al. 2009) and green alga (Serrano et al. 2009). These orthologs exhibit high sequence and functional conservation. But the members in CO family in various plants may have different variations in sequences and functions. In Arabidopsis, there are 17 members in CO family, which can be further classed into three subgroups. Group I members contain two B boxes, group II has only one B box, and group III members have a conserved B box and a variant of B box. The proteins in all of three groups contain a CCT (CO, CO-like, TOC) domain at C-terminus (Robson et al. 2001). Based on the characters of all domains, GmCOL9 belongs to group III and is the closest to COL13 in Arabidopsis, indicating that they may have similar functions. The NF-YA1 sub-domain forms a helix proposed to interact with the HAP3/HAP5 dimer, whereas NF-YA2 is interacted with the DNA of the CCAAT box (Romier et al. 2003; Wenkel et al. 2006). GmCOL9 shares key residues with AtCOL13 in this sub-domain, as well as having a similar B box content. Transient expression analyses in onion epidermal cells have demonstrated the nuclear localization of GmCOL9 protein, in agreement with its role as a transcriptional regulator.
GmCOL9 is expressed in a non-circadian manner of 24 h, but an oscillation period of 12 h regardless of photoperiod conditions. However, the pattern was largely disrupted when plants were exposed to either continuous light or continuous darkness. And such a pattern was damped in subsequent continuous light conditions or the amplitude was increased in continuous dark. As Aschoff’s rule says, in animals, light enhances activity of diurnal model and suppresses the activity of the nocturnal model (Carpenter and Grossberg 1984). Thus, different from CO in Arabidopsis which oscillates following a typical 24-h circadian pattern (Putterill et al. 1995), the regulation of GmCOL9 expression appears to be regulated by a non-circadian mechanism, but the light/dark cycle could be an effecter for GmCOL9 expression. A nocturnal model may be involved in the control of GmCOL9 expression.
CO function is largely dependent on its regulation on FT expression (An et al. 2004); thus, the CO protein accumulation in cells is more important than its transcript abundance (Valverde et al. 2004; Michaels 2009; Fornara et al. 2010). However, CO protein also shows a circadian pattern similar to its transcripts (Valverde et al. 2004). Therefore, CO mRNA change indicates its function as in Arabidopsis (Valverde et al. 2004) and other plants such as P. nil (Hayama et al. 2007). For lack of specific antibody, we tried to analyze the function of GmCOL9 by its transcript profiles. As GmCOL9 was constitutively expressed throughout the plant, it is possible that it participates in a range of functions concerned with plant development. It is of interest that transcript abundance varied substantially between different parts of the plant, with the majority of the message being concentrated in the unifoliolates and stems during vegetative and reproductive stages, respectively. It is possible that GmCOL9 functions mainly in the unifoliolates during the vegetative phase of growth and shifts its activity to the stem when the plants enter their reproductive phase. Its expression in the hypocotyl, epicotyl, and shoot meristem indicates a possible role in morphogenesis. GmCOL9 may affect root development due to its expression in both before and after flowering. Its function on seed maturation other than in seed developmental progress may be another important role for GmCOL9. In summary, GmCOL9 may have its functions in multiple aspects.
References
An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131:3615–3626
Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312:1040–1043
Carpenter GA, Grossberg S (1984) A neural theory of circadian rhythms: Aschoff’s rule in diurnal and nocturnal mammals. Am J Physiol 247:R1067–R1082
Chen QJ, Zhou HM, Chen J, Wang XC (2006) Using a modified TA cloning method to create entry clones. Anal Biochem 358:120–125
Cheng XF, Wang ZY (2005) Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J 43:758–768
Chia TY, Muller A, Jung C, Mutasa-Gottgens ES (2008) Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J Exp Bot 59:2735–2748
Datta S, Hettiarachchi GH, Deng XW, Holm M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18:70–84
Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141:550, 550 e551–552
Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131:1855–1867
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422:719–722
Hayama R, Agashe B, Luley E, King R, Coupland G (2007) A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 19:2988–3000
Holefors A, Opseth L, Ree Rosnes AK, Ripel L, Snipen L, Fossdal CG, Olsen JE (2009) Identification of PaCOL1 and PaCOL2, two CONSTANS-like genes showing decreased transcript levels preceding short day induced growth cessation in Norway spruce. Plant Physiol Biochem 47:105–115
Hu R, Fan C, Li H, Zhang Q, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93
Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21:3416–3420
Liu J, Yu J, McIntosh L, Kende H, Zeevaart JA (2001) Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol 125:1821–1830
Martinez-Garcia JF, Virgos-Soler A, Prat S (2002) Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS. Proc Natl Acad Sci USA 99:15211–15216
Michaels SD (2009) Flowering time regulation produces much fruit. Curr Opin Plant Biol 12:75–80
Miller TA, Muslin EH, Dorweiler JE (2008) A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227:1377–1388
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14(Suppl):S111–S130
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857
Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28:619–631
Romier C, Cocchiarella F, Mantovani R, Moras D (2003) The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem 278:1336–1345
Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F (2009) Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol 19:359–368
Suzuki Y, Mae T, Makino A (2008) RNA extraction from various recalcitrant plant tissues with a cetyltrimethylammonium bromide-containing buffer followed by an acid guanidium thiocyanate–phenol–chloroform treatment. Biosci Biotechnol Biochem 72:1951–1953
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303:1003–1006
Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984
Xiao C, Chen F, Yu X, Lin C, Fu YF (2009) Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol 71:39–50
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2484
Acknowledgments
Thanks to Dr. Jane Parker (Max Planck Institute, Cologne, Germany) for kindly providing pENSG-YFP and pENSG-CFP vectors. This work was supported in part by the Key Technology R&D Program (2007BAD59B02), Transgenic program (nos. 2008ZX08009-001, 2008ZX08004-005, and 2009ZX08009-133B), the Chinese National Key Basic Research “973” Program (2010CB125906), and the Chinese National “863” Program (nos. 2006AA10A111 and 2007AA10Z119).
Author information
Authors and Affiliations
Corresponding author
Additional information
Guowen Huang and Jinghua Ma contributed equally to the work.
Rights and permissions
About this article
Cite this article
Huang, G., Ma, J., Han, Y. et al. Cloning and Expression Analysis of the Soybean CO-Like Gene GmCOL9 . Plant Mol Biol Rep 29, 352–359 (2011). https://doi.org/10.1007/s11105-010-0240-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0240-y