Abstract
The Arabidopsis genome encodes 29 AHL (AT-hook motif nuclear localized) proteins, but the function for most of them remains unknown. We report here a study of the AHL22 gene, which was originally identified as a gain-of-function allele that enhanced the phenotype of the cry1 cry2 mutant. AHL22 is a nuclear protein with the binding activity for an AT-rich DNA sequence. AHL22 overexpression delayed flowering and caused a constitutive photomorphogenic phenotype. The loss-of-function AHL22 mutant showed no clear phenotype on flowering, but slightly longer hypocotyls. However, silencing four AHL genes (AHL22, AHL18, AHL27, and AHL29) resulted in early flowering and enhanced ahl22-1 mutant phenotype on the growth of hypocotyls, suggesting genetic redundancy of AHL22 with other AHL genes on these plant developmental events. Further analysis showed that AHL22 controlled flowering and hypocotyl elongation might result from primarily the regulation of FT and PIF4 expression, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of a plant is heavily dependent on its surroundings, and plants respond to environmental and endogenous signals by generating cellular signals which trigger cell- and development-specific patterns of gene expression. This response depends, as in other eukaryotes, on the organization of genomic DNA into chromatin, a process which comprises the first step in gene transcription (Bastow and Dean 2003; Sung and Amasino 2004). The AT-hook is a small DNA-binding motif, and has been shown to be present in a broad range of DNA-binding proteins, including high mobility group (HMG) proteins, homeodomains, bromodomains, and both PHD and zinc finger proteins. This group of proteins are mechanistically involved in both local and global changes in chromatin structure, and act to increase the structural flexibility of DNA via the promotion of the assembly of the nucleoprotein complex, which controls transcription (Harrer et al. 2004; Grasser et al. 2007a). The AT-hook motif proteins regulate gene expression through the interaction of the motif with the narrow minor groove of AT-rich DNA sequences. The HMGA and HMGB proteins appear to bind chromatin only transiently before moving on to the next site, and are implicated in a continuous genome scan for targets (Harrer et al. 2004; Launholt et al. 2006).
The AT-hook motif is characterised with the peptide sequence RGRP, with the flanking sequences of this motif determining its DNA-binding affinity, and thereby its function of the protein. Aravind and Landsman (1998) have recognized three types of such proteins. Type I proteins have the highest affinity to DNA, as the C-terminal of their core RGRP carries an additional module composed of a number of basic residues, usually along with a glycine, which together form a supporting polar network and provide additional contacts with the target DNA. In addition to the AT-hook motif, plant members of this protein family also carry one PPC (plant and prokaryotic conserved) domain, the hydrophobic regions of which are essential for their nuclear localization (Fujimoto et al. 2004; Matsushita et al. 2007; Street et al. 2008). The AT-hook motif itself also contributes to nuclear localization, probably as a result of methylation of the arginine residue within the motif (Sgarra et al. 2006; Cattaruzzi et al. 2007).
There are many AT-hook proteins (containing AT-hook motif proteins) in plants identified from a range of species, including Arabidopsis thaliana (At), rice (Oryza sativa), maize (Zea mays), pea (Pisum sativum) and the Madagascar periwinkle (Catharanthus roseus). In At, they are referred to as AHLs (AT-hook motif nuclear localized proteins) (Fujimoto et al. 2004; Grasser et al. 2007b; Vom Endt et al. 2007). Their expression has been detected in most plant organs (Matsushita et al. 2007; Street et al. 2008). When transgenic GUS activity was driven by AT-hook promoters of AHL27 and AHL29, expression was concentrated in the vascular system, and was not modulated by light (Street et al. 2008). AT-hook proteins have been shown to be involved in the maintenance of meristem identity (Su et al. 2006), leaf longevity (Lim et al. 2007), photomorphogenesis and the development of flowers and leaves (Street et al. 2008), the jasmonate signal pathway (Vom Endt et al. 2007) and gibberellin homeostasis (Matsushita et al. 2007). These observations suggest that the AT-hook genes play multiple roles in plant development, although single AHL mutants, such as the loss-of-function mutant of AHL27 (At1g20900), have apparently no observable phenotype with respect to either seedling or adult plant morphology. A double mutant of AHL27 and AHL29 (At1g76500), two genes which are closely related to each other, flowered at the same time as the wild type. However, under low levels of illumination, seedlings of the double mutant produced longer hypocotyls than either the wild type or either of the single mutants. These observations imply a level of redundancy among the AHL genes, and that the various functions of a particular AHL protein are arrived at via distinct mechanisms. In contrast to the loss-of-function mutants, dramatic phenotypic changes are induced when various AHL genes are transgenically over-expressed (Su et al. 2006; Lim et al. 2007; Street et al. 2008).
The plant cryptochromes encoded by CRY1 (Ahmad and Cashmore 1993; Lin et al. 1996) and CRY2 (Lin et al. 1998) are the major receptors of blue light, and are involved in the control of numerous developmental events, including the determination of flowering time and hypocotyl growth. Compared to the wild type, the flowering of the cry2 mutant is much delayed under long, but not under short day conditions (Guo et al. 1998; El-Din El-Assal et al. 2003), while mutations at CRY1 largely affect hypocotyl growth (Ahmad and Cashmore 1993; Lin et al. 1996). The double mutant cry1 cry2 flowers later and has longer hypocotyls than either of the single mutants (Mockler et al. 1999).
Activation tagging is a gain-of-function mutagenesis method which permits the identification of genes involved in a given process, and the phenotypes conferred by the over-expression of these genes can be informative as to the function of the tagged genes (Weigel et al. 2000). We describe here the screening of an activation tagged library in background of the cry1 cry2 double mutant, which led to the identification of a late-flowering individual. We show that this phenotype is the outcome of AHL22 (At2g45430, an AT-hook gene) over-expression, and that AHL22 affects on hypocotyl growth either.
Methods
Gene cloning and plasmid construction
Activation-tagging mutagenesis was performed in a cry1 cry2 mutant background as (Weigel et al. 2000; Zhao et al. 2007) did. The mutants with flowering phenotype were screened, and one of later flowering mutant, ecc (enhancer of cry1 cry2) 2, was identified. With the results of Tail-PCR (Weigel and Glazebrook 2002), ecc2 carried a T-DNA which inserted in the upstream of At2g45430. Using genomic sequences from the TAIR8 database (http://www.arabidopsis.org/), the full-length AHL22 open reading frame was amplified by PCR with primers (see Table S1) containing appropriate restriction sites, and the amplicon was cloned into the 35SpBARN (LeClere and Bartel 2001) and pEGAD vectors (Cutler et al. 2000) to obtain the binary vectors p35S::AHL22 and p35S::GFP:AHL22. The open reading frame of AHL18 (At3g60870) was cloned into pLeela vector using Gateway system (Invitrogen) to obtain the binary vector p35S::AHL18. The special fragment sequences of AHL22 and AHL18 were combined using overlap PCR (Lu 2005), and cloned into pJawohl8 to obtain the AHL22-AHL18 double RNAi construct. With the same approach, the quadruple RNAi vector of AHL22, AHL18, AHL27 and AHL29 was constructed. Sequence alignment was performed using MEGA v3.1.
Plant materials and growth conditions
Arabidopsis thaliana seeds were pretreated at 4°C for 3 days and grown at a constant temperature of 22°C under various lighting regimes. For hypocotyl measurement, the long day regime was 16 h light 80 μmol m−2 s−1 and 8 h dark, the short day regime 8 h light 100 μmol m−2 s−1 and 16 h dark, continuous blue light 6 μmol m−2 s−1, continuous red light 30 μmol m−2 s−1, continuous far-red light 0.6 μmol m−2 s−1. At plants were agroinfected with Agrobacterium tumefaciens strain GV3101 MP90 and pGV3101 MP90RK, using the floral dipping method (Clough and Bent 1998). Flowering time data were measured on the basis of the number of rosette and cauline leaves formed by the primary meristem and days to flower, and represent the mean of at least 16 per each line. T-DNA insertion lines of Salk_018866 and Salk_143279 were obtained from ABRC. PCR based genotyping used primers recommended by http://signal.salk.edu/tdnaprimers.2.html.
Electrophoretic mobility shift assay
The open reading fragments encoding the AHL22 and AHL27 full-length proteins were cloned into the entry clone pDONR201 (Invitrogen), and recombined into the destination vector pDEST17 (Invitrogen). Expression of the His-AHL22 and His-AHL27 fusion proteins was induced by 1 mM IPTG (Sigma) in E.coli strain BL21-star. After four additional hours at 28°C, the bacterial cells were harvested by centrifugation at 10,000g for 15 min at 4°C, and resuspended in 20 ml of EXB buffer [50 mM Sodium Phosphate (pH 8.0), 300 mM NaCl, 1 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM Imidazole, 8 M Urea]. The cells were disrupted by sonication and the supernatants were obtained by centrifugation at 16,000g for 30 min at 4°C. The soluble fraction was incubated with Ni-NTA Agarose Beads (Qiagen) with shaking for 1 h, and then applied into a purification column and washed two times using the EXB buffer. Finally the His-tagged protein was eluted by five bed-volumes of ELB buffer [50 mM Sodium Phosphate (pH 7.0), 300 mM NaCl, 200 mM Imidazole, 8 M Urea].
An electrophoretic mobility shift assay was performed using LightShift Chemi-luminescent EMSA kits (Pierce). The 39-bp synthetic oligonucleotide (5′-TAACACATATTTTGATAAATTTATTACTAAAACTATTTT-3′)(bold letters indicate A/T rich motif; Lim et al. 2007) was used as a probe. Competition experiments using the wild-type competitor and mutated competitors (5′-TAACACACTGCAGGATAAATTTATTACTAAAACTATTTT-3′) (bold letters indicate mutated sites) were also performed. The probes were labeled with biotin 3′-end DNA labeling kit (Pierce) according to the instruction. Briefly, binding reactions containing 10 μg of His-tagged proteins and 1 nmol of oligonucleotide were performed for 30 min in binding buffer [2.5% glycerol, 0.05% Nonidet P-40, 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, 10 mM Tris, pH 7.6, and 50 ng of poly(dI-dC)]. Protein-nucleic acid complexes were resolved using a nondenaturating polyacrylamide gel consisting of 6% acrylamide, and transferred to a nylon membrane. The membrane was incubated in blocking solution followed by incubation with streptavidin-peroxidase. After extensive washing, signal was detected with chemiluminescence solution.
Measurement of hypocotyl length
Arabidopsis thaliana seeds were surface-sterilized and sown on MS medium (0.5 g l−1 MES pH 5.7 and 0.8% w/v agar). The seeds were held at 4°C for 3 days, exposed to white light for 4 h to stimulate germination, and then cultured for 6 or 8 days at 22°C under various light and photoperiod conditions to measure the hypocotyl length. The length of hypocotyl was measured to an accuracy of 0.5 mm (Lin et al. 1998; Mockler et al. 1999).
GUS staining
A 2.3 kbp fragment upstream of the AHL22 coding sequence was cloned from the genomic DNA of the Columbia ecotype, and fused to the GUS report gene to form a binary vector using the GATEWAY cloning system (Invitrogen). The GUS fusion gene was introduced into Columbia, and T2 lines were used for analysis. For GUS staining, plant tissue was immersed in a buffer composed of 50 mM sodium phosphate pH 7.0, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, 0.2% v/v Triton X-100 and 2 mM 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid, cyclohexylammonium salt and held at 37°C overnight. The material was decoloured in ethanol:acetic acid (3:1, v/v) and analysed by light microscopy (Nikon SMZ1000, Japan).
Subcellular localization of GFP-AHL22 in Arabidopsis thaliana
GFP (green fluorescent protein) was monitored in guard cells, hypocotyl and root cells of 3-day-old 35S::GFP:AHL22 seedlings grown on MS medium by confocal microscopy (LEICA TCS SP2, Germany). GFP fluorescence was induced by exposure to blue light.
Semi-quantitative RT-PCR and quantitative real-time RT-PCR
Total RNA was extracted from 10-day-old seedlings and different organs using Trizol reagent (Invitrogen). The samples were treated with RQ1 RNase-free DNase (Promega) to remove any contamination with genomic DNA. RNA quality was verified by agarose gel electrophoresis, and first-strand synthesis was performed with an oligo(dT17) primer (Fermentas), following the manufacturer’s instructions. The constitutively expressed ACT2 was used as an internal control. Quantitative real-time RT-PCR was performed following the StepOne Real-Time PCR system (Applied Biosystems). Data were analysed using StepOne software (ABI, Applied Biosystems) and transcript levels were calculated relative to the transcript level of ACT2. The sequences of the primers used for semi-quantitative RT-PCR and quantitative real-time RT-PCR are listed in Table S1.
Results
The over-expression of AHL22 delays flowering in long days and short days
One activation-tagging mutant, ecc (enhancer of cry1 cry2) 2 with late-flowering phenotype, was identified from the background of cry1 cry2 double mutant. After genotyping (Weigel and Glazebrook 2002), it was confirmed that the phenotype of ecc2 may result from overexpression of an AT-hook gene, AHL22 (At2g45430). But the ecc2 plants were sterile and no seeds were got from them. So, AHL22 was cloned from the wild type plants (ecotype Columbia) using RT-PCR, with primers based on the genomic sequence of At2g45430. The protein sequence shows that AHL22 is an intron-less gene encoding a 317 residue peptide. Its protein has a single AT-hook motif and a conserved PPC domain (Fig. S1A). Its characters of motif and its flanking sequence is typical of type I proteins, as described by Aravind and Landsman (1998).
Then AHL22 gene driven by the cauliflower mosaic virus (CaMV) 35S promoter was expressed in the background of wild type COL. In all, 36 independent transgenic lines were obtained and classified on the basis of phenotype (flowering time and fertility) into three types (Fig. S1B). Type I plants (6 out of 36) took 150 days to reach flowering under long days, and were branchy and sterile (Fig. S1C); in these plants the level of AHL22 mRNA was maintained at a high level (Fig. S1D). Type II plants (6 out of 36) flowered later than the wild type, but earlier than type I, and were partially sterile—in these, the level of AHL22 mRNA was moderate (Fig. S1D). Although AHL22 transcript in type III plants (24 out of 36) was more abundant than in the wild type (Fig. S1D), the flowering time and fertility of these plants were indistinguishable from that of the wild type. Thus the expression level of AHL22 was correlated with phenotype. Type I plants resembled the ecc2 mutant, suggesting that the over-expression of AHL22 was responsible for the ecc2 mutant phenotype. In a more detailed study of the function of AHL22 on plant development, the type II plants were used—these were hereafter referred to as over-expressing-AHL22 (AHL22ox) plants. AHL22ox plants flowered later than the wild type under both long day and short day conditions, and the total number of leaves was more than triple in short days than that in long days (Fig. 1A, Ea), suggesting that AHL22ox was still sensitive to photoperiod.
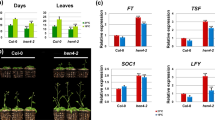
The flowering phenotypes of AHL22ox and mutants. A Flowering phenotype of AHL22ox plants was delayed under both long day (LD) and short day (SD) conditions. B The ahl22-1 mutant flowered at the same time as the wild type plants (WT) under both long and short day conditions. C The flowering time of AHL18 AHL22 double RNAi plants under both long and short day conditions (15 and 18: two independent transgenic lines). D The flowering time of AHL18-AHL22-AHL27-AHL29 quadruple RNAi plants under both long and short day conditions (21 and 23: two independent transgenic lines). The right panel of A, B, C, and D shows the number of rosette and cauline leaves and days at flowering. Error bars indicate standard deviation, every phenotype has at least 16 plants. Data in (A, B, C, D) were analyzed statistically using the t-test and a double asterisk indicates significance at P < 0.01 level. E mRNA expression level of AHL22, AHL18, AHL27 and AHL29 in different plants determined by quantitative real-time PCR. Error bars represent the standard deviation (n = 3). The labels of plants are the same as in A, B, C, and D
AHL22 involves in flowering regulation by redundant function with other AHL genes
To analyze the function of AHL22, we ordered two mutant lines of AHL22 gene of Salk_018866 (ahl22-1) and Salk-143279 (ahl22-2) from ABRC (http://www.arabidopsis.org/). There was no transcripts amplified from ahl22-1 allele, but AHL22 mRNA was clearly detected in ahl22-2 line (Fig. 1Eb, S2), suggesting that ahl22-1 was a null mutant. The ahl22-1 mutant, however, reaches flowering at nearly the same time as do the wild type plants (Fig. 1B), suggesting functional redundancy exists between AHL22 and other genes. Another AHL gene, AHL18, may be the candidate, since AHL18 is in the same phylogenetic clade of AHL22 (Fujimoto et al. 2004). Therefore, RNAi plants silencing both AHL18 and AHL22 genes were constructed, and plants showed a decreased expression of AHL22 and AHL18 (Fig. 1Ec). Unexpectedly, no clear flowering phenotype was observed in these plants (Fig. 1C). However, plants of overexpression of AHL18 (AHL18ox) had similar phenotype (late flowering) to that of AHL22ox (Fig. S3). The results suggested that there were more genes involved in the network of flowering regulation of AHL22.
Because overexpression of AHL27 is known to delay flowering time (Weigel et al. 2000) and AHL29 is the gene closest to AHL27, AHL27 and AHL29 may be the candidates with the redundant function of flowering regulation with AHL22, even though a double mutant ahl27 ahl29 flowered at the same time as the wild type (Street et al. 2008). The proteins of four AHLs share highly conserved sequences (Fig. S1A). Therefore, we constructed AHL18-AHL22-AHL27-AHL29 quadruple genes silencing plants. Interestingly, the quadruple silencing plants displayed obviously early flowering phenotype (Fig. 1D), even though the transcripts of all of these four genes were not completely knocked out (Fig. 1Ed). The results suggest that these four AHLs have a redundant role of flowering regulation even though AHL22 has a very low level of transcripts in leaves as indicated below.
The over-expression of AHL22 inhibits hypocotyl growth in a light-independent manner
Hypocotyl elongation, cotyledon opening, hook opening and plant greening are important events in photomorphogenesis. In AHL22ox plants, photomorphogenesis is constitutively expressed (Fig. 2A). Regardless of growth conditions (dark, different photoperiods and different lights), AHL22ox plants produced short hypocotyls, and displayed an open hook and open cotyledons. This combination of traits is reminiscent of the phenotype of constitutive photomorphogenic (cop) (Osterlund et al. 2000; Saijo et al. 2003), long hypocotyl5 (hy5) (Ulm et al. 2004). Compared to the wild type, AHL22ox photomorphogenesis suggests a response to variation in photoperiod and light quality. Red light and short days had a stronger effect than either blue light or long days (Fig. 2B), indicating that photoreceptors and the circadian clock could be involved in the process regulated by AHL22. Furthermore, AHL22ox displayed the same phenotype even in the absence of light. However, the ahl22-1 mutant had slightly longer hypocotyls than the wild type in dark, long day and short day conditions (Fig. 2C, D), and the quadruple RNAi mutant had much longer hypocotyls than the wild type in different light conditions (Fig. 2E, F). The results suggest the function of AHL22 in both photomorphogensis and skotomorphogenesis.
AHL22 affects hypocotyl elongation under various light conditions. A The hypocotyls of 6-day-old the wild type plants (WT) and AHL22ox seedlings under continuous darkness (Dark), long day (LD), short day (SD), continuous blue light (Blue), continuous red light (Red) and continuous far-red light (Far-red). B Quantitative analysis of the samples in A. The digits at the top of the empty bar show the ratio of hypocotyl length of AHL22ox to wild type under the same light regime (n > 20). C The hypocotyls of 8-day-old the wild type plants (WT) and ahl22-1 mutant grown under continuous darkness, long days and short days. D Quantitative analysis of the samples in C (n > 20). E Hypocotyl phenotypes of 6-day-old AHL18-AHL22-AHL27-AHL29 quadruple RNAi plants in different conditions. F Quantitative analysis of the samples in E (n > 30). Data in (B, D, F) were analysed statistically using the t-test. A single asterisk indicates significance at P < 0.05 and a double asterisk indicates significance at P < 0.01
The effect of AHL22 over-expression on the expression of genes related to flowering and photomorphogenesis
To study the mechanism of AHL22 on flowering and hypocotyl growth, the expression of genes involved in the determination of flowering time and the development of the hypocotyl was monitored. AHL22ox plants showed lower levels of CONSTANS (CO) and FLOWERING LOCUS T (FT) transcripts than the wild type plants, while the expression levels of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), FLOWERING LOCUS C (FLC) and LEAFY (LFY) were not measurably affected (Fig. 3). The reduced expression of CO and FT was independent of sampling time during the diurnal cycle, suggesting that both may be implicated in AHL22 signalling. The flower identity gene SEPALLATA3 (SEP3) is known to interact with AP1/FUL, and its over-expression to hasten flowering (Pelaz et al. 2001; Ferrario et al. 2003). In AHL22ox plants, the expression of SEP3 was reduced (Fig. 3), indicating that the function of AHL22 may require the participation of SEP3. On the contrary, the mRNA expression level of FT was upregulated in the AHL18-AHL22-AHL27-AHL29 quadruple RNAi seedlings (Fig. S4).
The effect of AHL22 over-expression on the transcription of CO, FT, SOC1, FLC, LFY and SEP3. Ten-day-old the wild type and AHL22ox seedlings were harvested every 4 h during a long day, and mRNA expression level was determined by quantitative real-time RT-PCR. Error bars represent the standard deviation (n = 3). ZT Zeitgeber
Among the genes related to hypocotyl growth, the expression of various members of the COP (Osterlund et al. 2000; Saijo et al. 2003) and PIF (Ni et al. 1998; Huq and Quail 2002) families, CRY1 and CRY2 (Ahmad and Cashmore 1993; Lin et al. 1996), phyA and phyB (Nagy and Schafer 2002) and HY5 (Ulm et al. 2004) was determined (Fig. 4). The only ones of these genes showing a significantly reduced level of expression were PIF3 and PIF4. Although the AHL22ox photomorphogenesis phenotype is reminiscent of that of various cop mutants, the only family member showing an expression level difference between the wild type and AHL22ox is COP1. However, this change is thought to be unrelated to the AHL22ox phenotype, since AHL22 over-expression increased, rather than decreased COP1 transcript abundance. The expression of neither HY5 nor any of the photoreceptors was substantially altered. As expected, the mRNA expression level of PIF4 was upregulated in the AHL18-AHL22-AHL27-AHL29 quadruple RNAi seedlings (Fig. S4).
Shorter hypocotyls in AHL22ox plants may be caused by induction of PIF3 and PIF4 expression. The diurnal variation in transcript level of PIF3, PIF4, COP1, COP8, COP9, COP10, COP11, HY5, CRY1, CRY2, PHYA, and PHYB in 10-day-old wild type and AHL22ox seedlings grown under long days, as detected by quantitative real-time RT-PCR. Error bars represent the standard deviation (n = 3). ZT Zeitgeber
The tissue specific expression patterns of AHL22
A ~2.3 kb fragment upstream of the AHL22 start codon was fused to a GUS reporter gene, and the resulting construct was transformed into the wild type plants to produce a set of 24 independent AHL22::GUS lines. GUS activity in these transgenic plants showed that expression of AHL22 was concentrated at the hypocotyl-root transition zone and the root hair zone. Some weak GUS activity was present in the vascular system, but no signal was observed in young leaves (Fig. 5A). A semi-quantitative RT-PCR and a quantitative real-time RT-PCR analysis confirmed the in situ localization result. The level of transcripts in the leaf was below the level of detection of standard RT-PCR, even after 40 amplification cycles (data not shown), but was finally detected by nested RT-PCR (Fig. 5B). However, strikingly, over 1,000-fold more transcript was detected in the root compared to that present in the leaf (Fig. 5C). After the root, the inflorescence showed the next highest level of AHL22 expression. So AHL22 is a ubiquitously expressed gene even though it has different transcriptional levels in different organs.
Tissue-specific expression of AHL22. A GUS activity in AHL22::GUS transgenic seedlings grown under long days. a 14-day-old whole seedlings. The black, blue and red arrows indicate the focus for the close-up images displayed in (b) the meristem, (c) and (d) the root. B Nested RT-PCR profiles detected AHL22 transcript in leaves of 10-day-old seedlings, ACT2 was acted as control gene. C The AHL22 mRNA level relative to ACT2 in various organs of the wild type WT, as detected by quantitative real-time RT-PCR, the relative expression was showed using Log value. R root, S stem, L leaf, F flower. Error bars represent the standard deviation (n = 3)
AHL22 is a nuclear localized AT-hook protein
AHL proteins may be transcriptional factors which function in nucleus. To detect the localization of the AHL22 protein, 35S::GFP:AHL22 construct was instructed into wild type plants mediated by Agrobacterium. The plants over-expressing GFP:AHL22 showed late flowering phenotype as the AHL22ox plants did (Fig. S5), suggesting that GFP:AHL22 is functional. The sub-cellular localization of AHL22 protein analyzed by confocal microscopy revealed clearly the nuclear localization of GFP:AHL22 protein in various organs (Fig. 6A), supporting that AHL22 may function in nucleus.
AHL22 as a nuclear-localized AT-hook-binding protein. A Nuclear localization of AHL22. Confocal images showed the presence of GFP in guard cells, hypocotyls, and roots of 3-day-old 35S::GFP:AHL22 seedlings (Leica). Left panels, fluorescence image of GFP–AHL22; middle panels, bright field image; right panel, merged fluorescence image of GFP–AHL22 and bright field image. Upper panels, guard cells; middle panels, hypocotyl cells; lower panels, root cells. B Binding of AHL22 to an AT-rich oligonucleotide. The oligonucleotide sequence corresponds to a part of the AT-rich sequence present in the promoter of the pea PRA2 gene. AHL27 protein was used as positive control (Lim et al. 2007). The 39-bp oligonucleotides of wild-type WT or mutant Mt were labeled with biotin for chemiluminescent detection. Competition experiment was carried out using the unlabeled wild-type oligonucleotide. The arrow indicates the location of the protein-bound, biotin labeled oligonucleotide
According to the classification of Aravind and Landsman (1998), AHL22 belongs to type I of AHL proteins. AHL22 protein has a single RGRP motif, and there is a glycine at the second position downstream of this motif and an extended module at the C-terminal of RGRP which includes basic residues forming a supporting polar network and additional contacts with DNA. These characters may confer to AHL22 protein high affinity of binding DNA. To detect this, an electrophoreticmobility shift assay (EMSA) was carried out with the A/T-rich DNA sequence from the promoter of the pea (Pisum sativum) PRA2 gene (Lim et al. 2007). As shown in Fig. 6B, the AHL22 protein was able to bind to biotin-labeled oligonucleotide (lane 3) as AHL27 did (lane 2, Lim et al. 2007), and this binding affinity could be greatly decreased by competitor DNA fragment (unlabled) with the same wild-type sequence (lane 6). However, different from AHL27 protein, AHL22 could also bind to the mutated sequence (lane 3 and 5), suggesting AHL22 may have different binding specificity from AHL27 because the A/T-rich DNA sequence employed in this study has three A/T-rich motif (see “Methods” section, bold letters). Thus, AHL22 is a nuclear-localized protein which may bind to A/T-rich DNA sequence.
Discussion
AHL22 is a highly conserved and nuclear localized AT-hook protein
Members of the AT-hook protein family are thought to connect the nuclear framework with the MAR sequences in interphase nuclei, and to coat the chromosomes during mitosis (Fujimoto et al. 2004). The At genome contains 29 AT-hook protein paralogs, one of which, AHL22, is the type I sequence (Aravind and Landsman 1998). The C-terminal flanking sequence of the AHL22 RGRP motif contains basic lysine residues, which are able to form a supporting polar network and to promote additional contacts with DNA. The PPC domain of AHL22 may be important for its nuclear localization (Fig. 6, S1). Several lines of evidence show that AHLs have transcriptional activity (Fujimoto et al. 2004; Matsushita et al. 2007; Street et al. 2008), and we have demonstrated here that the expression level of a number of genes is altered in AHL22ox plants and RNAi plants (Fig. 3, 4, S4). Further evidence confirmed that AHL22 protein is a nuclear localized protein and can bind to A/T rich DNA sequence (Fig. 6). Thus, AHL22 protein may regulate flowering and the growth of hypocotyls with its transcriptional activity.
AHL22 may have a role of flowering regulation and hypocotyl elongation
AHL22 appears to be functionally redundant with other AHL genes, because the single mutant reaches flowering at almost the same time as does the wild type. There are at least three candidates from the information of bioinformatics (Fujimoto et al. 2004) and literature (Street et al. 2008): AHL18, AHL27, and AHL29. However, both the ahl27ahl29 double mutant (Street et al. 2008) and RNAi plants silencing both AHL22 and AHL18 (Fig. 1C) did not show early-flowering phenotype. Interestingly, the plants of silencing AHL18, AHL22, AHL27, and AHL29 four genes obviously flowered earlier (Fig. 1D) and had higher transcripts of FT (Fig. S4) than the wild type did. Thus, their gene products may all represent proteins having functional redundancy with AHL22, and AHL22 regulates flowering in deed through FT pathway, even though the mRNA level of AHL22 in leaves is very low (Fig. 5) and single gene loss-of-function mutation (ahl22-1) does not produce marked alterations in flowering (Fig. 1B). We propose that in vivo, the AHLs are present at only a low level in leaves and function redundantly, and that ectopic over-expression disrupts the balance among the AHLs, with knock-on effects on plant development. Alternatively, AHL22 may regulate flowering from roots. In this case, the high level of AHL22 in roots may confer to the regulation of flowering.
In case of hypocotyl elongation, AHL22 may have much stronger effect on it than its effect on flowering regulation, because ahl22-1 mutant shows obviously longer hypocotyl phenotype. This phenotype is more obvious in AHL18-AHL22-AHL27-AHL29 quadruple RNAi silencing plants, suggesting that these four genes function redundantly on hypocotyl development either. This conclusion is also supported by previous results in which ahl27 and ahl29 mutants have longer hypocotyls (Street et al. 2008). From our results, AHL22 may control hypocotyl elongation mainly dependent on the activity of PIF4.
Overexpression of AHL22 has a pleiotropic effect on plant development
AHL27 is known to regulate flowering time, leaf longevity, and hypocotyl growth (Weigel et al. 2000; Lim et al. 2007; Street et al. 2008). The phenotype of AHL22ox plants differed in many ways including flowering and growth of hypocotyl from that of the wild type. AHL22ox plants also showed some different phenotypes in other developmental events including the growth of stem, roots and leaves, sterility, and greening (data not shown). Some of these differences were dependent on the light regime under which the plants were raised. Thus, like other AHL genes, it seems plausible that AHL22 regulates a number of disparate developmental events via a range of mechanisms. The delayed flowering of AHL22ox is dependent on the ambient photoperiod, and is more pronounced under long days than that under short days. This suggests that the biological clock in AHL22ox plants is still operational and that it controls the function of AHL22. CO/FT lies at the core of a flowering promotion pathway which responds to photoperiod (Turck et al. 2008). The significant changes in expression level of CO and FT in AHL22ox plants (Fig. 3) are evidence that AHL22 regulates flowering through these genes.
Rice PF1 is able to bind oat phyA and thereby enhance its activity, which has been taken to imply a regulatory role for AT-hook proteins in photomorphogenesis (Martinez-Garcia and Quail 1999). PIF3 and PIF4 proteins bind selectively to the biologically active form of phyB or phyA and function specifically in a branch of the phyB signalling network which regulates a subset of genes involved in cell expansion (Ni et al. 1999; Huq and Quail 2002). One of main regions of AHL22 activity is in the hypocotyl-root transition zone (Fig. 5), suggesting a function associated with hypocotyl elongation. The over-expression of AHL22 inhibits hypocotyl growth, irrespective of photoperiod or light quality, although short days and red light have a rather stronger effect than long days and blue light do (Fig. 2A, B). This behaviour mimics that of cop (Osterlund et al. 2000; Saijo et al. 2003) and pif (Ni et al. 1998; Huq and Quail 2002) mutants, but the expression of these genes (Fig. 4) implies that AHL22 regulates hypocotyl growth through neither COP nor photoreceptors, but via PIF3 and PIF4. It is also supported that pif3 and pif4 mutants were hypersensitive to red light (Huq and Quail 2002) and that AHL22ox showed much more sensitive to red light either (Fig. 2A, B). As photoreceptor gene expression is largely unaffected by the over-expression of AHL22, we conclude that these factors lie upstream of AHL22, or that AHL22 regulates hypocotyl growth via a mechanism independent of photoreceptor activity. Additional genetic and biochemical data are clearly needed to clarify the relationship between AHL22, PIF and COP.
The tissue-specific expression of AHL22 relates to its function
AHL22 expression is more tissue-specific than either AHL25 (AGF1) (seedlings, leaves, stems, floral tips, and flowers: Matsushita et al. 2007) or AHL27 and AHL29 (whole seedlings, Street et al. 2008). AHL22 promoter is mainly active in the root hair zone and the hypocotyl-root transition zone (Fig. 5). The AHL22ox phenotype is much clearer with respect to flowering time and hypocotyl length, even though transcript abundance is greater in the roots than in the leaves and flowers of the wild type plants (Fig. 5). However, the level of AHL22 transcript within a given tissue is in good correspondence with the strength of the phenotype—so for example, the higher the level of expression, the later the plant flowered (Fig. S1). Thus, AHL22 appears to enhance vegetative growth and inhibit reproductive growth in a dosage-dependent manner.
The ecc2 mutant, which is the initial material of this study, is an activation-tagging mutant in the background of cry1 cry2 double mutant, so we tried to explain the relationship between AHL22 and CRY1 and/or CRY2. Because cry1 cry2 mutant flowered too late, we firstly expressed AHL22 in wild type plants and got three types of transgenic lines as shown in Fig. S1. Then we crossed type II into cry2 mutant. Interestingly, F1 plants showed type I phenotype (late flowering and no seeds, data not shown), suggesting that cry2 may enhance the phenotype of AHL22ox, even though cry1 was heterozygous in F1 plants. However, it is needed further extensive evidence to elucidate the relationship between AHL22 and CRY1 and/or CRY2.
References
Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166. doi:10.1038/366162a0
Aravind L, Landsman D (1998) AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res 26:4413–4421. doi:10.1093/nar/26.19.4413
Bastow R, Dean C (2003) Plant sciences. Deciding when to flower. Science 302:1695–1696. doi:10.1126/science.1092862
Cattaruzzi G, Altamura S, Tessari MA, Rustighi A, Giancotti V, Pucillo C, Manfioletti G (2007) The second AT-hook of the architectural transcription factor HMGA2 is determinant for nuclear localization and function. Nucleic Acids Res 35:1751–1760. doi:10.1093/nar/gkl1106
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi:10.1046/j.1365-313x.1998.00343.x
Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97:3718–3723. doi:10.1073/pnas.97.7.3718
El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Wagemaker C, Weller JL, Koornneef M (2003) The role of cryptochrome 2 in flowering in Arabidopsis. Plant Physiol 133:1504–1516. doi:10.1104/pp.103.029819
Ferrario S, Immink RG, Shchennikova A, Busscher-Lange J, Angenent GC (2003) The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15:914–925. doi:10.1105/tpc.010280
Fujimoto S, Matsunaga S, Yonemura M, Uchiyama S, Azuma T, Fukui K (2004) Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol Biol 56:225–239. doi:10.1007/s11103-004-3249-5
Grasser KD, Launholt D, Grasser M (2007a) High mobility group proteins of the plant HMGB family: dynamic chromatin modulators. Biochim Biophys Acta 1769:346–357
Grasser M, Christensen JM, Peterhansel C, Grasser KD (2007b) Basic and acidic regions flanking the HMG-box domain of maize HMGB1 and HMGB5 modulate the stimulatory effect on the DNA binding of transcription factor Dof2. Biochemistry 46:6375–6382. doi:10.1021/bi6024947
Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–1363. doi:10.1126/science.279.5355.1360
Harrer M, Luhrs H, Bustin M, Scheer U, Hock R (2004) Dynamic interaction of HMGA1a proteins with chromatin. J Cell Sci 117:3459–3471. doi:10.1242/jcs.01160
Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21:2441–2450. doi:10.1093/emboj/21.10.2441
Launholt D, Merkle T, Houben A, Schulz A, Grasser KD (2006) Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell 18:2904–2918. doi:10.1105/tpc.106.047274
LeClere S, Bartel B (2001) A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol 46:695–703. doi:10.1023/A:1011699722052
Lim PO, Kim Y, Breeze E, Koo JC, Woo HR, Ryu JS, Park DH, Beynon J, Tabrett A, Buchanan-Wollaston V, Nam HG (2007) Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J 52:1140–1153
Lin C, Ahmad M, Cashmore AR (1996) Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J 10:893–902. doi:10.1046/j.1365-313X.1996.10050893.x
Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95:2686–2690. doi:10.1073/pnas.95.5.2686
Lu Q (2005) Seamless cloning and gene fusion. Trends Biotechnol 23:199–207. doi:10.1016/j.tibtech.2005.02.008
Martinez-Garcia JF, Quail PH (1999) The HMG-I/Y protein PF1 stimulates binding of the transcriptional activator GT-2 to the PHYA gene promoter. Plant J 18:173–183. doi:10.1046/j.1365-313X.1999.00440.x
Matsushita A, Furumoto T, Ishida S, Takahashi Y (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 143:1152–1162. doi:10.1104/pp.106.093542
Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126:2073–2082
Nagy F, Schafer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53:329–355. doi:10.1146/annurev.arplant.53.100301.135302
Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95:657–667. doi:10.1016/S0092-8674(00)81636-0
Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400:781–784. doi:10.1038/23500
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466. doi:10.1038/35013076
Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26:385–394. doi:10.1046/j.1365-313X.2001.2641042.x
Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17:2642–2647. doi:10.1101/gad.1122903
Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V, Bedford MT, Manfioletti G (2006) The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem 281:3764–3772. doi:10.1074/jbc.M510231200
Street IH, Shah PK, Smith AM, Avery N, Neff MM (2008) The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J 54:1–14. doi:10.1111/j.1365-313X.2007.03393.x
Su Y, Kwon CS, Bezhani S, Huvermann B, Chen C, Peragine A, Kennedy JF, Wagner D (2006) The N-terminal ATPase AT-hook-containing region of the Arabidopsis chromatin-remodeling protein SPLAYED is sufficient for biological activity. Plant J 46:685–699. doi:10.1111/j.1365-313X.2006.02734.x
Sung S, Amasino RM (2004) Vernalization and epigenetics: how plants remember winter. Curr Opin Plant Biol 7:4–10. doi:10.1016/j.pbi.2003.11.010
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: flowering locus T moves center stage. Annu Rev Plant Biol 59:573–594. doi:10.1146/annurev.arplant.59.032607.092755
Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101:1397–1402. doi:10.1073/pnas.0308044100
Vom Endt D, Soares e Silva M, Kijne JW, Pasquali G, Memelink J (2007) Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol 144:1680–1689. doi:10.1104/pp.107.096115
Weigel D, Glazebrook J (2002) TAIL-PCR. In: Weigel D, Glazebrook J (eds) Arabidopsis, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 144–153
Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, Nguyen JT, Sato S, Wang ZY, Xia Y, Dixon RA, Harrison MJ, Lamb CJ, Yanofsky MF, Chory J (2000) Activation tagging in Arabidopsis. Plant Physiol 122:1003–1013. doi:10.1104/pp.122.4.1003
Zhao X, Yu X, Foo E, Symons GM, Lopez J, Bendehakkalu KT, Xiang J, Weller JL, Liu X, Reid JB, Lin C (2007) A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol 145:106–118. doi:10.1104/pp.107.099838
Acknowledgments
Thanks to Drs. Bekir Ülker and Jane Parker for kindly providing pJawohl8-RNAi and pLeela vectors, and to Dr. Robert Koebner for his critical reading of the manuscript. This work was supported in part by the National ‘‘863’’ Program of China (2006AA10A111, 2006AA10Z107, and 2007AA10Z119), the National Key Basic Research ‘973’ Program of China (2004CB117206), the Key Technology R & D Program (2007BAD59B02), National Science Foundation of China (30671245), National Institute of Health (GM56265), and UCLA faculty research and Sol Leshin BGU-UCLA Academic Cooperation programs.
Author information
Authors and Affiliations
Corresponding authors
Additional information
C. Xiao, F. Chen and X. Yu contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, C., Chen, F., Yu, X. et al. Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana . Plant Mol Biol 71, 39–50 (2009). https://doi.org/10.1007/s11103-009-9507-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9507-9