Abstract
Aim
Soil microbial legacy is a potentially important regulator of the associations of plants and mycorrhizal fungi. However, our understanding of how plant performance and root-associated fungi react to distinct soil microbial legacies during subalpine forest succession remains unclear.
Methods
A pot experiment of two coniferous (Picea asperata Mast. and Abies fargesii var. faxoniana (Rehder & E. H. Wilson) Tang S. Liu) tree seedlings, using sterilized soil inoculated with the soil microbial legacy of herbs, shrubs, and trees, was conducted in a greenhouse. Plant biomass, root morphological traits (total root length, root surface area, and the number of root tips), the percentage of ectomycorrhizal (EcM) root colonization, root-associated fungal communities, and soil inorganic nitrogen content were measured.
Results
Both coniferous seedling performance and EcM colonization were facilitated when grown in the soil microbial legacies of shrubs and trees rather than herbs. Correspondingly, soil microbial legacy favored root-associated EcM Ascomycetes and EcM fungi with ‘short-distance’ exploration type. The soil microbial legacies of trees induced a greater relative abundance of Wilcoxina, while those of herbs and shrubs resulted in greater abundances of Trichophaea, Geopora, and Hebeloma (belonging to ‘short-distance’ exploration type). Notably, the relative abundances of ‘short-distance’ explorers were positively correlated with root biomass.
Conclusions
Soil microbial legacy may affect tree seedling establishment and modify plant performance across successional stages by regulating the colonization, composition, and exploration type of root-associated fungi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The natural regeneration of tree seedlings in degraded subalpine forests is recognized as a slow and difficult process because of the complex interactions between plants and soil microbial legacy (Reid and Holl 2013; Cahoon et al. 2018; Coban et al. 2022; Koyama et al. 2022). Conifer seeds that are entering the soil are exposed to a wide array of soil microbial legacy during forest secondary succession (Schmid et al. 2021). After germination, the roots are the first organ in contact with the soil microbial legacy of a pre-existing plant (Lozano et al. 2020). Changes in the soil microbial legacy induced by different plant species can in turn facilitate or inhibit the growth and survival of con- and hetero-specific plants (Teste et al. 2017; Wang et al. 2020; Jing et al. 2022). Given the variable effects of the soil microbial legacy on host plant fitness, it remains unclear how the changes in soil microbial legacy from various functional groups of pre-existing adult plants (i.e., herbs, shrubs, and trees) drive the establishment of coniferous tree seedlings.
Although tree seedlings are able to grow for a short period without mycorrhizal associations, conifer establishment is often limited by a lack of soil fungal communities, especially ectomycorrhizal (EcM) fungi (Collier and Bidartondo 2009; Nuñez et al. 2009; Hayward et al. 2015). Thus, the initial recovery of trees during forest secondary succession depends on whether appropriate EcM fungi in the soil microbial legacy persist and are able to serve as partners to regenerate tree seedlings (Glassman et al. 2016). The symbiosis between plants and EcM fungi is tightly coupled, and diverse EcM fungi acquire carbohydrates from the host plant in exchange for soil-derived nutrients (Corrales et al. 2016), resist soil-borne plant pathogens (Chakravarty and Unestam 2008), and improve tolerance to water stress (Parke et al. 1983). For obligate mycorrhizal species (e.g., Pinaceae), the infection status of EcM fungi has been demonstrated to enhance the nitrogen (N) supply for plants in N-limited forests (Vozzo and Hacskaylo 1971; Li et al. 2015; Zhang et al. 2019), which strengthens the positive effects of EcM fungi on vulnerable seedling growth (Bennett et al. 2017; Dong et al. 2021). As a result, an appropriate EcM fungi-host combination is critical for optimal tree seedling cultivation. However, it is still not clear to what extent the variations in soil microbial legacy (especially for EcM fungal communities) from different forest succession stages can regulate seedling establishment. Identifying the EcM fungal community compositions and the colonization status of roots can provide a basis for facilitating coniferous tree seedling establishment during the forest restoration process.
Exploration types, to some extent, could reflect the capability of EcM fungi to colonize new roots, or acquire and transport resources (Agerer 2001). The classification of those fungi based on the characteristics of extraradical hyphae and rhizomorphs. The ‘contact’ type has smooth, dense, hydrophilic mantles and only a few fungal taxa have emanating hyphae, while the ‘short-distance’ types (including coarse and delicate types) produce large, non-aggregating, short hyphae in close proximity of root tips. Those EcM ‘contact’ and ‘short-distance’ types with no or few cords have been found to maximize the hyphae area and thereby promote rapid uptake of mobile N in regions with high N availability (Fernandez et al. 2017). By contrast, the ‘medium-distance fringe’ forms extensive mycelia with many hydrophobic, aggregated cords. The ‘medium-distance smooth’ and ‘long-distance’ types form thickened mycelial cords and produce few extraradical mycelia near the root. ‘Medium-distance’ and ‘long-distance’ explorers focus on spatially concentrated and widely dispersed soil resources to enhance the nutrient acquisition of hosts (Hobbie and Agerer 2010), especially under N-limited conditions (Fernandez et al. 2017). It has also been suggested that exploration type is a pattern to reflect the community assembly through their different colonization abilities on roots (Peay et al. 2011). However, changes in the root-associated EcM exploration type have received little attention in the regeneration of coniferous tree seedlings. To improve our predictions of the establishment of tree seedlings in this region, it is necessary to understand how EcM function responds to soil microbial legacy.
Plants may sense the exploration types of specific EcM fungi, and their changes, to adjust and optimize the traits of their roots (Koide et al. 2014; Defrenne et al. 2019; Anthony et al. 2022; Chaudhary et al. 2022). These various root traits include total root length, root diameter, root mass fraction, surface area, and the number of tips (Stokes et al. 2009). A recent study highlighted that tree species with coarser roots are less capable of foraging and adsorbing soil nutrients; thus, they should benefit from ‘medium-’ and ‘long-distance’ explorers (Liu et al. 2019). Wasyliw and Karst (2020) found that increased areas of fine roots in maturing boreal forests may result in lower dependence of host trees on EcM fungi, evidenced by a decreased abundance of distance explorers. To our knowledge, few studies have linked root traits and EcM exploration types, especially in coniferous tree seedlings (Cheng et al. 2016; Chen et al. 2018). Importantly, changes in root abundance and morphology traits are likely to influence the belowground resources of hosts, and these shifts could be affected by the EcM exploration types. A better understanding of the responses of root biomass and morphological traits to EcM exploration types is important for predicting the nutrient uptake strategies of tree seedlings.
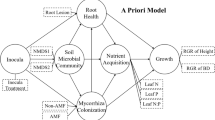
Here, a controlled soil inoculation experiment was conducted to examine the potential influence of the soil microbial legacy on coniferous tree seedling performance during forest succession. Picea asperata Mast. and Abies fargesii var. faxoniana (Rehder & E. H. Wilson) Tang S. Liu, which represent the most dominant coniferous tree species on the eastern Qinghai-Tibetan Plateau, were selected for this study. Differences in seedling performance (height, shoot and root biomass, root morphological traits (total root length, root surface area, and the number of root tips)) and soil nutrient availability (ammonium nitrogen (NH4+-N) and nitrate nitrogen (NO3−-N)) were determined. Additionally, we measured the colonization percentage of symbiotic EcM fungi and identified the fungal compositions using MiSeq sequencing. As seedling establishment is more strongly affected by the availability of mycorrhizal partners (Chen et al. 2018), we hypothesized that (i) relative to herb and shrub heterospecific soils, plants perform best in the soil microbial legacy of trees due to an accumulation of symbiotic EcM fungi on root tips that make them better acquire soil nutrients; (ii) divergence in root-associated fungal community compositions and exploration types would induce by different soil microbial legacies; (iii) host plants would benefit more when associated with ‘medium-’ and ‘long-distance’ explorers, because those fungi with rhizomorphs preferentially exploit soil resources for plant nutrient.
Materials and methods
Species selection and seed germination
The subalpine coniferous forests are located in the Miyaluo Provincial Nature Reserve on the eastern Tibetan Plateau in Sichuan, China (N 31°35′, E 102°35′; 2800 m a.s.l.). The annual temperature is 12.6 °C in July and − 8 °C in January. The annual precipitation at the sampling location ranges from 600 to 1100 mm. During the last century, part of the primary forests were deforested to meet the increasing demands for forest products. After deforestation, this area was fenced and afforested, forming a complete continuous secondary succession of early-successional herbs (dominated by Thalictrum uncatum, Rumex nepalensis, Kobresia pygmaea and multiple genera of Poaceae), mid-successional shrubs (dominated by Rubus sp., Berberis sichuanica, Rosa sweginzowii and Sibiraea angustata), and late-successional trees (dominated by Betula albosinensis, Betula platyphylla, Picea asperata and Abies faxoniana).
EcM tree species P. asperata and A. faxoniana were selected for the greenhouse experiment. Seeds were collected from the study site during the autumn of 2018, and then stored in a refrigerator at 4 °C. All seeds were surface sterilized in K2MnO4 (0.3%) for 15 min to remove microbial contamination, followed by rinsing with demineralized water four times. Seeds were germinated on sterilized soil in a growth chamber (25 °C constant, 12 h:12 h day:night, relative humidity 70%). Germinated seedlings were transplanted when they had reached an approximate height of 3 cm so that the seedlings were at a similar plant ontogenetic stage.
In May 2019, nine independent soil samples were collected as inoculum, including early-successional herbs (Poa annua (PAN), Koeleria macrantha (KM), Anemone rivularis (AR)), mid-successional shrubs (Berberis sichuanica (BS), Rhododendron fortunei (RF)) and late-successional trees (Betula platyphylla (BP), Betula albosinensis (BA), Picea asperata (PA) and Abies faxoniana (AF). Soils were collected from the rooting zone (5–20 cm) of several individuals of each target species present by excavating the plants. The soil was sieved through a 1 cm mesh to remove coarse stones and roots, transported to the laboratory in coolers and stored at 4 °C until the start of the experiment in June 2019.
Greenhouse sterilization experiment
To eliminate the effect of soil biota, we collected bulk soils from the study area (far away from the study species) and sterilized them with 25-kGy gamma irradiation. The bulk soil properties at the beginning of the experiment were as follows: pH = 6.4, soil organic matter = 7.43 g/kg dry soil, NH4+-N = 9.76 mg/kg dry soil, and NO3−-N = 10.78 mg/kg dry soil. Gamma radiation-treated bulk soils and rooting zone soils were mixed at a proportion of 93:7 volume/volume (Teste et al. 2017). The mixed soils were filled into plastic circular pots (9.8 × 11.6 × 11.6 cm), and one seedling was then transplanted into each pot. In total, the combined experiments (2 seedling species × 9 soil sources) were replicated eight times, resulting in 144 pots (Fig. S1). The pots were arranged in 16 blocks (i.e., eight replicated per seedling species) and the positions of the pots and the pots within a block were randomly shifted every month. Seedlings were grown in a climate-controlled greenhouse (25 °C constant, 12 h:12 h light:dark, relative humidity 70%) with water supply three times per week. After 13 months of growth, we measured the plant height from the substrate to the tip of the stem. Plants were harvested blockwise on the same day, and the roots were washed over a 53 μm sieve to capture any broken roots during the process. We extracted the soil soluble inorganic nitrogen (NH4+-N and NO3−-N) from fresh soil using 2 mol L−1 KCL. The concentrations of NH4+-N and NO3−-N in extract were determined by the indophenol-blue (Dorich and Nelson 1983) and phenol disulfonic acid colorimetry (Nicholas and Nelson 1957), respectively.
Root morphology and mycorrhizal colonization
Each root sample was scanned with a dedicated scanner (Epson Expression 11000XL, Seiko Epson Corporation, Nagano, Japan), and their total length, number of root tips, and total surface area were analysed with WinRhizo software (Regent Instruments, Inc., Quebec, Canada, 2012). Root tip-colonized EcM fungi were distinguished from uncolonized root tips by the presence of fungal mantle and Harting net (Smith and Read 2010). EcM colonization was calculated as the number of root tips colonized by EcM fungi divided by the total number of root tips (Brundrett et al. 1996). The corresponding root samples were then longitudinally divided into two equal parts that were weighed. The first set of subsamples was dried at 65 °C for 48 h, and the total dry weight of each sample was obtained based on the dry-to-wet ratio. The second set of subsamples was frozen (−20 °C) immediately for later examination of the fungal community composition.
Root tip fungal DNA extraction sequencing
Root tip samples were surface-sterilized in 99% ethanol for 1 min, washed with 2.63% NaOCl for 6 min, 99% ethanol for 30 s, and then rinsed with distilled water. We extracted total DNA from the root tip samples using the cetytrimethylammonium bromide (CTAB) method (Allen et al. 2006). The nuclear ribosomal internal transcribed spacer (ITS) region was amplified by polymerase chain reaction (PCR) using dual-indexed primers: ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5’-GCTGCGTTCTTCATCGATGC-3′) (Gardes and Bruns 1993). Each PCR (30 μl) contained 15 μl PhusionMasterMix 2× (Phusion® High-Fidelity PCR Master Mix with GC Buffer), 3 μl of 2 μM primers, 10 μl of DNA template (1 Ng μl−1), and 2 μl of H2O. The PCR amplification procedure was conducted with a temperature profile at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, primer annealing at 50 °C for 30 s and elongation at 72 °C for 30 s. The cycle was finished with an extension at 72 °C for 5 min to ensure complete amplification. After purification and quantification, high-throughput sequencing was done via Illumina HiSeq 2500 (San Diego, CA, USA; 2 × 300 bp).
The raw sequencing reads containing any nucleotide mismatches within the primers or the barcode and any ambiguous bases were removed. Usearch 10 was used to identify and remove chimeric sequences. The remaining sequences were clustered into operational taxonomic units (OTUs) by UPARSE analysis pipeline with the similarity of 97% sequence (Edgar 2013). The taxonomy of each OTU representative fungal sequence was analyzed by Ribosomal Database Project (RDP) Classifier, based on the ITS database (Unite v8.0) (Nilsson et al. 2019). Putative EcM fungi were further assigned using the FunGuild algorithm at the “high probable” and “probable” confidence rankings (Nguyen et al. 2016). On the other hand, OTUs assigned to similar guilds were retained, such as EcM fungi that were simultaneously identified as saprotrophs. Additionally, assignment of the exploration types (hyphal foraging distance) were based on previous studies (Agerer 2001; Tedersoo and Smith 2013; Moeller et al. 2014) and the DEEMY database (http://www.deemy.de/) (Rambold and Agerer 1997). Each ECM fungal species or genus was further classified into different exploration types.
Data analysis
All statistical analyses were conducted with R, v.4.1.2 (R Core Team 2020). To test how soil microbial legacy generated effects on plant biomass, root morphological traits and the percentage of EcM root colonization, models were created with soil inoculum origin as a fixed factor and block as a random factor. Whenever significant interactions were investigated, post hoc Tukey’s HSD tests were performed to test which treatments differed from each other. We used a linear regression to test whether the root biomass in the different soil microbial legacies explained the observed responses to root mycorrhizal colonization percentage and soil inorganic N content. The data were checked for normality using the Shapiro–Wilk test and, when needed, were log transformed before analysis to meet the assumption of normality.
To test the effects of the soil microbial legacy and plant species on the entire and EcM fungal community composition, a permutational multivariate analysis of variance (PERMANOVA) was performed using the function ‘adonis’ of the VEGAN package (Oksanen et al. 2019) based on the Bray-Curtis distance dissimilarity and 999 permutations. To visualize the influences of inoculated soil microbial legacies of various plant functional groups (i.e., herbs, shrubs, and trees) on the relative abundance of root-associated fungi, post hoc test was used to test which plant functional groups differed from each other. Because ‘medium-’ and ‘long-distance’ explorers were less abundant and present in a tiny minority of samples, linear regression was only used to analyse the correlations between root biomass and the log10-transformed relative abundances of ‘short-distance’ exploration types (including coarse and delicate types). Redundancy analysis (RDA) was used to assess the relationship between plant performance (root biomass and morphology) and EcM fungal community composition using the R package VEGAN.
Results
Effects of the soil microbial legacy on plant performance and EcM fungal colonization
The root biomass of both tree seedlings was strongly affected by soil microbial legacy (Fig. 1a, b, and S2; P < 0.001). Root dry biomass of P. asperata and A. faxoniana were mostly lower in the soil microbial legacy of herbs. P. asperata seedlings grew best in pots inoculated with the soil microbial legacy of AF (0.423 ± 0.023 g), but post hoc tests revealed no significant difference between AF, BS (0.405 ± 0.024 g) and PA (0.386 ± 0.041 g) treatment (Fig. 1a). A. faxoniana root biomass was significantly higher in BS treatment (0.303 ± 0.006 g) than in other treatments (Fig. 1b). Analyses of the shoot biomass, plant height, total root length, total root surface area, and root tip numbers showed similar patterns to the root biomass analysis (Fig. S2).
Root biomass (a, b) and EcM fungal colonization (c, d) of P. asperata and A. faxoniana seedlings grown in pots with inoculated soils originating from nine plant species. The inocula were the rooting zone soils from early-successional herbs (PAN, Poa annua; KM, Koeleria macrantha; AR, Anemone rivularis), mid-successional shrubs (BS, Berberis sichuanica; RF, Rhododendron fortunei), and late-successional trees (BP, Betula platyphylla; BA, Betula albosinensis; PA, Picea asperata; AF, Abies faxoniana). Data are mean values ± SEs (n = 8). Different lowercase letters indicate significant differences (P < 0.05) among the inoculated soil types using one-way ANOVA followed by Tukey’s HSD test
Similarity, the level of EcM root colonization differed based on soil microbial legacy (Fig. 1c, d). When averaged across all pots, EcM root colonization ranged from 10.05% to 45.05% in P. asperata and from 11.24% to 49.28% in A. faxoniana. For P. asperata, lower colonization was obtained when plants grew in soil microbial legacy of herbs, and post hoc tests revealed that colonization was significantly higher when plants grew in soil microbial legacies of shrubs and trees. On the other hand, P. asperata seedlings grown in inoculated soil with microbial legacy of AF exhibited the highest level of colonization (45.05 ± 1.55%), and post hoc tests revealed no differences between treatments of AF, BS (42.16 ± 1.73%), BA (40.72 ± 1.73%), and PA (41.97 ± 1.85%) (Fig. 1c). For A. faxoniana, in most cases, post hoc tests revealed that EcM colonization was lower when plants grew in soil microbial legacies of herbs than in shrubs and trees, and the highest colonization (49.28 ± 1.74%) was observed in BS treatment (Fig. 1d). Further analysis showed that for the seedlings in the pots, the percentage of root colonization by EcM fungi significantly positively correlated with their root biomass (Fig. 2a, b), but negatively correlated with NH4+ -N (Fig. 2c, d) and NO3−-N (Fig. 2e, f).
Root-associated fungal community composition and function
Sequencing of the ITS1 region yielded 5,490,779 reads, which were clustered into 2173 fungal OTUs. Of these, 1365 OTUs corresponded to unidentified fungi in UNITE. The composition of root fungal communities was dominated by Ascomycota (70.3% of total fungal sequences), followed by Basidiomycota (11.72%) and Chytridiomycota (0.19%) (Fig. S3). We were able to assign 349 OTUs to functional guilds, and 50 OTUs were classified as EcM (26.68% of total fungal sequences). After several months of seedling growth, the compositional differences in root-associated EcM fungal communities were significantly influenced by nine inoculated soil microbial legacies (P < 0.001), or two plant species (P = 0.027) (Table 1). These tree species colonized unique microbiomes in different soil treatments as indicated by the significant plant species × inoculated soil microbial legacies interactions (P = 0.009). Nearly identical results were obtained when using total fungal data in the PERMANOVA (Table 1).
Herein, we confirmed that a total of 17 genera of EcM fungi were encountered across both host seedlings, ten of which were shared between hosts (58.82%), with five (Amphinema, Boletellus, Entoloma, Sistotrema, and Tylospora) exclusive to P. asperata (29.41%) and two (Cortinarius and Sebacina) exclusive to A. faxoniana (11.6%). At the genus level, Trichophaea, Wilcoxina, Geopora, and Hebeloma were the four most abundant genera for both conifers (Fig. 3). Furthermore, the abundance proportions of several EcM fungal genera were greatly affected by the successional stages of the inoculated soils (Fig. 4). Colonization by Wilcoxina was higher in seedlings grown with the soil microbial legacy of trees than herbs and shrubs, especially for A. faxoniana. The abundance proportion of Hebeloma and Geopora was much more abundant when P. asperata received inoculants from soil microbial legacies of herbs and shrubs relative to trees (Fig. 4a). For A. faxoniana, the same decreasing trend was found for the abundance proportions of Trichophaea and Geopora (Fig. 4b).
Relative abundance of EcM fungi at the genus level in the P. asperata (a) and A. faxoniana (b) roots after growing in pots with inoculated soils originating from nine plant species. The inocula were the rooting zone soils from early-successional herbs (PAN, Poa annua; KM, Koeleria macrantha; AR, Anemone rivularis), mid-successional shrubs (BS, Berberis sichuanica; RF, Rhododendron fortunei), and late-successional trees (BP, Betula platyphylla; BA, Betula albosinensis; PA, Picea asperata; AF, Abies faxoniana). Blue (Ascomycota) and green (Basidiomycota) colours indicate different ECM fungi at the phylum level. Exploring type: S-D (short-distance delicate explorers), S-C (short-distance coarse explorers), M-S (medium-distance smooth explorers), M-F (medium-distance fringe explorers), L (long-distance explorers)
The relative abundances of top 8 EcM fungal genera on P. asperata (a) and A. faxoniana (b) roots. Error bars are ± SE of the mean with the different letters indicating significant differences (P < 0.05) of EcM fungal abundance among plant functional groups (e.g., three inoculated soil microbial legacies from three herb species) based on post-hoc analysis. Exploring type: S-D (short-distance delicate explorers), S-C (short-distance coarse explorers)
When classifying the EcM fungal OTUs into different exploration types, we found that the relative abundance of the ‘short-distance coarse’ type was dominant (24.56% fungal sequences), followed by the ‘short-distance delicate’ type (3.12% fungal sequences). The proportions of the different ‘short-distance’ exploration types were not significantly different between plant species. ‘Medium-’ and ‘long-distance’ exploration types were rare in the analysed samples (Fig. 4).
Variation in ectomycorrhizal (EcM) fungal community composition in the roots of two coniferous species in response to soil microbial legacy, as visualized by the fungal exploration type of Redundancy analysis (RDA) (a). The red straight arrow represents plant performances variables plotted in ordination space. The different colored points indicate different fungal exploration types of EcM fungi. The size of the points represents the relative abundance of EcM fungi. Exploration type abbreviations: S-D (short-distance delicate), S-C (short-distance coarse), M-S (medium-distance smooth), M-F (medium-distance fringe), L (long-distance). Liner relationship between the root biomass and the log10-transformed relative abundance of short-distance explorers is shown in Panel (b)
Relationships between root traits and functional traits of EcM
To better understand the differences in functional capacities between EcM fungal communities linked to root biomass and root morphological traits, we examined differences in exploration types among EcM communities using distance-based redundancy analysis (RDA). Axes 1 and 2 explained 55.17% and 21.05% of the variation, respectively. The EcM fungal community composition was significantly correlated with the biomass, surface area, number of tips, and total length of roots (P < 0.001, Fig. 5a), and the root biomass (48.26%) explained more variation than the root morphological traits. At the level of the exploration type, the root biomass was significantly and positively correlated with the abundance of genera belonging to the ‘short-distance’ (R2 = 0.640, P = 0.017) exploration type (i.e., Tricholophaea, Geopora and Tuber) (Fig. 5b).
Discussion
By measuring the seedling performance (e.g., height, biomass, total root length, root surface area, and number of root tips) and EcM fungal root colonization at each pot, we found that P. asperata and A. faxoniana seedlings performed better in the soil microbial legacies of shrubs and trees and worse in the soil microbial legacy of herbs (Fig. 1 and S2). Soil microbial legacy also led to marked differences in the abundance and composition of the root-associated fungi, which significantly influenced the biomass and morphological traits of roots. Importantly, the growth of seedlings was enhanced when the EcM genera belonging to ‘short-distance’ exploration types were increased. Overall, our results suggest that the soil microbial legacies of shrubs and trees facilitated the colonization of the specialist EcM species on root tips and greatly eased the establishment bottleneck. Using the soil microbial legacies from various functional groups of pre-existing adult plants, our work provides new insight into the significant role of root-associated fungi in tree seedling growth that may contribute to forest succession.
Our findings partly followed hypothesis (i) that soil microbial legacies of shrubs and trees facilitated tree growth through enhancing EcM root colonization, whereas those from herbs reduced plant performance. After 13 months of plant growth, seedlings mostly accumulated higher loads of EcM fungi on root tips in the soil microbial legacies of shrubs and trees (Fig. 1c, d). Interestingly, higher root biomass of conifers was significantly correlated with the increase of EcM fungal colonization percentage (Fig. 2a, b) as well as the decrease of soil inorganic N content (Fig. 2c-f). Hence, these results reinforced the view that increased mycorrhization may enhance the N capture of host plants in nutrient-poor soils, which provides a greater benefit for tree seedling establishment (Kuzyakov and Xu 2013; Wang et al. 2019; Huang et al. 2022). Meanwhile, higher levels of EcM fungi colonized on the roots are capable of hydrolyzing complex compounds (pectins, oil and cellulose) and increasing the resistance of seedlings to root-invading pathogens (Peterson 2012). This may explain previous field observations that EcM colonization was consistently greater in established shrubs and adult trees and could promote the subsequent establishment of neighbouring tree seedlings (Haskins and Gehring 2005; Dickie et al. 2012; Liang et al. 2020; Mekontchou et al. 2022).
The below interpretations and fungal community structure analysis are in line with hypothesis (ii) that the root-associated fungal communities were significantly different among inoculated soils. Although both plant species showed a similar response to EcM colonization and nutrient uptake (Fig. 2c-f), PERMANOVA analysis indicated that the soil microbial legacies (P < 0.001), plant species (P = 0.027) and their interactions (P = 0.009) as a significant factor explaining differences in fungal communities among samples (Table 1). The root-associated EcM fungal communities were dominated by Ascomycete genera under all treatments, indicating that members of these fungal taxa occupy a wide niche and strongly contribute to seedling growth (Patterson et al. 2019). In contrast, Basidiomycetes, such as the genera Hebeloma and Boletellus, were less abundant (Fig. 3a, b), which have been reported to display complex nutrient-acquiring enzymatic capabilities, and do not provide a strong net benefit to host species in low fertility soils (Finlay et al. 1992; Querejeta et al. 2021). Previous reports identified Ascomycota as possessing a higher number of genes involved in carbohydrate metabolism and nutrients than Basidiomycota, resulting in greater stress tolerance and competitive abilities (Egidi et al. 2019; Owen et al. 2019). Because of these effects, a higher proportion of Ascomycota may be also partly responsible for the greater biomass production. At the genus level, Trichophaea, Wilcoxina, and Geopora (Pyronemataceae, Pezizales, Ascomycota) were the most abundant on root tips. Taxa in the family Pyronemataceae have strong adaptability to stress (Mikryukov et al. 2021), and host plants can profit from mineral adsorption mediated by this fungal family (Hansen et al. 2013). Specifically, the abundance proportion of Wilcoxina was significantly higher in seedlings grown with the soil microbial legacy of trees than in seedlings inoculated with the soil microbial legacies of herbs and shrubs (Fig. 4a, b). Wilcoxina is known to be an excellent and rapid root colonizer on Pinaceae seedlings, as it has little host specificity and lower carbohydrate requirements than other EcM fungi (Jones et al. 2010; Wen et al. 2018; Milani et al. 2022). Wilcoxina have also been suggested to arise from a potential capacity for facultative saprotrophy and to increase the resistance of seedlings to pathogens (Yu et al. 2001; Rosenstock et al. 2019). Furthermore, Wilcoxina seem to be effective in degrading chitin, which is an abundant organic N reservoir in boreal forest soil (Velmala et al. 2014). A large amount of Wilcoxina may be a notable N contributor to plant N nutrition in forests. Studies comparing the benefits of different EcM fungal associations (Ascomycetes, Basidiomycetes, and non-mycorrhizal) have also observed that Wilcoxina can outcompete other EcM fungi in terms of the seedling biomass and nutrient uptake (Björkman 1949; Siemens and Zwiazek 2008). Hence, it is possible that the higher colonization of Wilcoxina in the soil microbial legacy of trees and its specific ecological functions may confer more beneficial repercussions for host tree species. As we continue to identify and understand the roles of these fungal taxa in seedling performance, the soil fungal legacy effects on forest regeneration will be better predicted.
We further suggest that the relative abundance of EcM exploration type was strongly related to the inoculation of soil microbial legacy. Specifically, fewer genera were considered to be the ‘medium-distance’ (e.g., Amphinema and Cortinarius) and ‘long-distance’ (e.g., Boletellus) exploration types, and the ‘short-distance’ (e.g., Trichophaea and Wilcoxina) exploration type harboured significantly higher numbers of sequences than the ‘medium-’ and ‘long-distance’ types in all treatment groups (P < 0.001). Additionally, ‘short-distance’ EcM explorers were significantly less abundant in the soil microbial legacies of herbs than those of trees (P < 0.001), indicating that the soil microbial legacy of trees promoted the establishment of a mycorrhizal species with ‘short-distance’ exploration type. Contrary to our third hypothesis, a positive relationship was observed between the abundance of the ‘short-distance’ exploration type and the root biomass (Fig. 5b). In addition to the root biomass, we propose that the ‘short-distance’ explorers induce higher total root length, root surface area and root tip number (Fig. 5a). Our greenhouse experiment expanded on similar previous conclusions drawn from field experiments of Pinus ponderosa Lawson & C. Lawson (Owen et al. 2019) and Pinus edulis seedlings (Patterson et al. 2019). The ‘short-distance’ exploration types have relatively low photosynthate costs to the host plants (Castaño et al. 2018) and thus may efficiently explore a larger soil volume without incurring higher carbon costs at the seedling stage (Hobbie and Agerer 2010; Näsholm et al. 2013). In addition, hydrophilic ‘short-distance’ explorers allow for the rapid intake of labile N, such as ammonium, nitrate, and amino acids (Morgado et al. 2016), via diffusion through the mantle to the plant host root (Nygren et al. 2008). In contrast, ‘medium-’ and ‘long-distance’ explorers favor N immobilization in mycelium, resulting in N-deficit in the medium (Agerer 2006; Näsholm et al. 2013), and have greater carbon requirements of their host plants than ‘short-distance’ explorers (Saikkonen et al. 1999). These processes may negatively affect N uptake by seedling roots (Chen et al. 2018; Mekontchou et al. 2022). Considering the dominance of EcM fungi in N-limited subalpine coniferous forests (Zhang et al. 2017; Guo et al. 2021), we propose that a higher abundance of ‘short-distance’ explorers can be less costly in terms of mobilizing labile N and have beneficial effects on host conifer seedlings (Rosinger et al. 2018).
Conclusion
With increasing levels of disturbances in forest ecosystems, understanding how soil microbial legacy affects coniferous tree seedling establishment during secondary forest succession is critically important. We provide clear evidence of strong associations between the root-associated EcM fungal community composition and function and root traits in coniferous tree seedlings via a greenhouse experiment. One important finding was that conifers apparently have greater compatibility with the soil microbial legacies of shrubs and trees, potentially due to the colonization percentage and functional shifts (based on exploration types) in EcM fungal communities over succession. EcM fungi provide nutrients to their host plants, and greater colonization by Ascomycetes or ‘short-distance’ explorers may further benefit seedlings grown in herb soils. Overall, this work implies that root-fungal symbiosis influence root performance, which may affect the prediction of vegetation dynamics over successional development in degraded forests.
References
Agerer R (2001) Exploration types of ectomycorrhizae. Mycorrhiza 11:107–114. https://doi.org/10.1007/s005720100108
Agerer R (2006) Fungal relationships and structural identity of their ectomycorrhizae. Mycol Prog 5:67–107. https://doi.org/10.1007/s11557-006-0505-x
Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1:2320. https://doi.org/10.1038/nprot.2006.384
Anthony MA, Crowther TW, Svd L, Suz LM, Bidartondo MI, Cox F, Schaub M, Rautio P, Ferretti M, Vesterdal L, Vos BD, Dettwiler M, Eickenscheidt N, Schmitz A, Meesenburg H, Andreae H, Jacob F, Dietrich H-P, Waldner P et al (2022) Forest tree growth is linked to mycorrhizal fungal composition and function across Europe. ISME J 16:1327–1336. https://doi.org/10.1038/s41396-021-01159-7
Bennett JA, Maherali H, Reinhart KO, Lekberg Y, Hart MM, Klironomos J (2017) Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355:181–184. https://doi.org/10.1126/science.aai8212
Björkman E (1949) The ecological significance of the ectotrophic mycorrhizal association in forest trees. Sven Bot Tidskr 43:223–286
Brundrett MC, Bougher N, Dell B, Grove T, Malajczuk N (1996) Working with Mycorrhizas in Forestry and Agriculture. ACIAR Monograph 32. Australian Centre for International Agricultural Research, Canberra
Cahoon SMP, Sullivan PF, Brownlee AH, Pattison RR, Andersen H-E, Legner K, Hollingsworth T (2018) Contrasting drivers and trends of coniferous and deciduous tree growth in interior Alaska. Ecology 99:1284–1295. https://doi.org/10.1002/ecy.2223
Castaño C, Lindahl BD, Alday JG, Hagenbo A, Aragón JM, Parladé J, Pera J, Bonet JA (2018) Soil microclimate changes affect soil fungal communities in a Mediterranean pine forest. New Phytol 220:1211–1221. https://doi.org/10.1111/nph.15205
Chakravarty P, Unestam T (2008) Differential influence of ectomycorrhizae on plant growth and disease resistance in Pinus sylvestris seedlings. J Phytopathol 120:104–120. https://doi.org/10.1111/j.1439-0434.1987.tb04423.x
Chaudhary VB, Holland EP, Charman-Anderson S, Guzman A, Bell-Dereske L, Cheeke TE, Corrales A, Duchicela J, Egan C, Gupta MM, Hannula SE, Hestrin R, Hoosein S, Kumar A, Mhretu G, Neuenkamp L, Soti P, Xie Y, Helgason T (2022) What are mycorrhizal traits? Trends Ecol Evol 37:573–581. https://doi.org/10.1016/j.tree.2022.04.003
Chen W, Koide RT, Eissenstat DM (2018) Nutrient foraging by mycorrhizas: from species functional traits to ecosystem processes. Funct Ecol 32:858–869. https://doi.org/10.1111/1365-2435.13041
Cheng L, Chen W, Adams TS, Wei X, Li L, McCormack ML, DeForest JL, Koide RT, Eissenstat DM (2016) Mycorrhizal fungi and roots are complementary in foraging within nutrient patches. Ecology 97:2815–2823. https://doi.org/10.1002/ecy.1514
Coban O, Deyn GBD, Mvd P (2022) Soil microbiota as game-changers in restoration of degraded lands. Science 375:abe0725. https://doi.org/10.1126/science.abe0725
Collier FA, Bidartondo MI (2009) Waiting for fungi: the ectomycorrhizal invasion of lowland heathlands. J Ecol 97:950–963. https://doi.org/10.1111/j.1365-2745.2009.01544.x
R Core Team (2020) R: A language and environment for statistical computing. Retrieved from https://www.R-project.org
Corrales A, Mangan SA, Turner BL, Dalling JW (2016) An ectomycorrhizal nitrogen economy facilitates monodominance in a neotropical forest. Ecol Lett 19:383–392. https://doi.org/10.1111/ele.12570
Defrenne CE, Philpott TJ, Guichon SHA, Roach WJ, Pickles BJ, Simard SW (2019) Shifts in ectomycorrhizal fungal communities and exploration types relate to the environment and fine-root traits across interior Douglas-fir forests of western Canada. Front Plant Sci 10:643. https://doi.org/10.3389/fpls.2019.00643
Dickie IA, Davis MR, Carswell FE (2012) Quantification of mycorrhizal limitation in beech spread. New Zeal J Ecol 36(2):210–215. http://www.jstor.org/stable/24060847
Dong H, Ge J, Sun K, Wang B, Xue J, Wakelin SA, Wu J, Sheng W, Liang C, Xu Q, Jiang P, Chen J, Qin H (2021) Change in root-associated fungal communities affects soil enzymatic activities during Pinus massoniana forest development in subtropical China. Forest Ecol Manag 482:118817. https://doi.org/10.1016/j.foreco.2020.118817
Dorich RA, Nelson DW (1983) Direct colorimetric measurement of ammonium in potassium chloride extracts of soils. Soil Sci Soc Am J 47:833–836. https://doi.org/10.2136/sssaj1983.03615995004700040042x
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996. https://doi.org/10.1038/nmeth.2604
Egidi E, Delgado-Baquerizo M, Plett JM, Wang J, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK (2019) A few Ascomycota taxa dominate soil fungal communities worldwide. Nat Commun 10:2369. https://doi.org/10.1038/s41467-019-10373-z
Fernandez CW, Nguyen NH, Stefanski A, Han Y, Hobbie SE, Montgomery RA, Reich PB, Kennedy PG (2017) Ectomycorrhizal fungal response to warming is linked to poor host performance at the boreal-temperate ecotone. Glob Chang Biol 23:1598–1609. https://doi.org/10.1111/gcb.13510
Finlay RD, Frostegard A, Sonnerfeldt A-M (1992) Utilization of organic and inorganic nitrogen sources by ectomycorrhizal fungi in symbiosis with Pinus contorta Dougl. Ex loud. New Phytol 120:105–115. https://doi.org/10.1111/j.1469-8137.1992.tb01063.x
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/j.1365-294x.1993.tb00005.x
Glassman SI, Levine CR, DiRocco AM, Battles JJ, Bruns TD (2016) Ectomycorrhizal fungal spore bank recovery after a severe forest fire: some like it hot. ISME J 10:1228–1239. https://doi.org/10.1038/ismej.2015.182
Guo W, Ding J, Wang Q, Yin M, Zhu X, Liu Q, Zhang Z, Yin H (2021) Soil fertility controls ectomycorrhizal mycelial traits in alpine forests receiving nitrogen deposition. Soil Biol Biochem 161:108386. https://doi.org/10.1016/j.soilbio.2021.108386
Hansen K, Perry BA, Dranginis AW, Pfister DH (2013) A phylogeny of the highly diverse cup-fungus family pyronemataceae (Pezizomycetes, Ascomycota) clarifies relationships and evolution of selected life history traitss. Mol Phylogenet Evol 67(2):311–335. https://doi.org/10.1016/j.ympev.2013.01.014
Haskins KE, Gehring CA (2005) Evidence for mutualist limitation: the impacts of conspecific density on the mycorrhizal inoculum potential of woodland soils. Oecologia 145:123–131. https://doi.org/10.1007/s00442-005-0115-3
Hayward J, Horton TR, Nñnez MA (2015) Ectomycorrhizal fungal communities coinvading with Pinaceae host plants in Argentina: Gringos bajo el bosque. New Phytol 208:497–506. https://doi.org/10.1111/nph.13453
Hobbie EA, Agerer R (2010) Nitrogen isotopes in ectomycorrhizal sporocarps correspond to belowground exploration types. Plant Soil 327:71–83. https://doi.org/10.1007/s11104-009-0032-z
Huang LL, Wang YL, Guerin-Laguette A, Wang R, Zhang P, Li YM, Yu FQ (2022) Ectomycorrhizal synthesis between two tuber species and six tree species: are different host-fungus combinations having dissimilar impacts on host plant growth? Mycorrhiza 32:341–351. https://doi.org/10.1007/s00572-022-01081-6
Jing J, Cong W-F, Bezemer TM (2022) Legacies at work: plant-soil-microbiome interactions underpinning agricultural sustainability. Trends Plant Sci 27:781–792. https://doi.org/10.1016/j.tplants.2022.05.007
Jones MD, Twieg BD, Ward V, Barker J, Durall DM, Simard SW (2010) Functional complementarity of Douglas-fir ectomycorrhizas for extracellular enzyme activity after wildfire or clearcut logging. Funct Ecol 24:1139–1151. https://doi.org/10.1111/j.1365-2435.2010.01699.x
Koide RT, Fernandez C, Malcolm G (2014) Determining place and process: functional traits of ectomycorrhizal fungi that affect both community structure and ecosystem function. New Phytol 201:433–439. https://doi.org/10.1111/nph.12538
Koyama A, Dias T, Antunes PM (2022) Application of plant-soil feedbacks in the selection of crop rotation sequences. Ecol Appl 32:e2501. https://doi.org/10.1002/eap.2501
Kuzyakov Y, Xu X (2013) Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytol 198:656–669. https://doi.org/10.1111/nph.12235
Li Y, Sun D, Li D, Xu Z, Zhao C, Lin H, Liu Q (2015) Effects of warming on ectomycorrhizal colonization and nitrogen nutrition of Picea asperata seedlings grown in two contrasting forest ecosystems. Sci rep-UK 5:17546. https://doi.org/10.1038/srep17546
Liang M, Johnson D, Burslem DFRP, Yu S, Fang M, Taylor JD, Taylor AFS, Helgason T, Liu X (2020) Soil fungal networks maintain local dominance of ectomycorrhizal trees. Nat Commun 11:2636. https://doi.org/10.1038/s41467-020-16507-y
Liu B, Li L, Rengel Z, Tian J, Li H, Lu M (2019) Roots and arbuscular mycorrhizal fungi are independent in nutrient foraging across subtropical tree species. Plant Soil 442:97–112. https://doi.org/10.1007/s11104-019-04161-3
Lozano YM, Aguilar-Trigueros CA, Flaig IC, Rillig MC (2020) Root trait responses to drought are more heterogeneous than leaf trait responses. Funct Ecol 34:2224–2235. https://doi.org/10.1111/1365-2435.13656
Mekontchou CG, Houle D, Bergeron Y, Roy M, Gardes M, Séguin A, Drobyshev I (2022) Contrasting structure of root mycorrhizal communities of black spruce and trembling aspen in different layers of the soil profile in the boreal mixedwoods of eastern Canada. Plant Soil. https://doi.org/10.1007/s11104-022-05410-8
Mikryukov VS, Dulya OV, Bergman IE, Lihodeevskiy GA, Loginova AD, Tedersoo L (2021) Sheltering role of well-decayed conifer logs for forest floor fungi in long-term polluted boreal forests. Front Microbiol 12:729244. https://doi.org/10.3389/fmicb.2021.729244
Milani T, Hoeksema JD, Jobbágy EG, Rojas JA, Vilgalys R, Teste FP (2022) Co-invading ectomycorrhizal fungal succession in pine-invaded mountain grasslands. Fungal Ecol 60:101176. https://doi.org/10.1016/j.funeco.2022.101176
Moeller HV, Peay KG, Fukami T (2014) Ectomycorrhizal fungal traits reflect environmental conditions along a coastal California edaphic gradient. FEMS Microbiol Ecol 87:797–806. https://doi.org/10.1111/1574-6941.12265
Morgado L, Semenova TA, Welker J, Walker M, Smets EF, Geml J (2016) Long-term increase in snow depth leads to compositional changes in arctic ectomycorrhizal fungal communities. Glob Chang Biol 22:3080–3096. https://doi.org/10.1111/gcb.13294
Näsholm T, Högberg P, Franklin O, Metcalfe D, Keel SG, Campbell C, Hurry V, Linder S, Högberg MN (2013) Are ectomycorrhizal fungi alleviating or aggravating nitrogen limitation of tree growth in boreal forests? New Phytol 198:214–221. https://doi.org/10.1111/nph.12139
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Nicholas DJD, Nelson A (1957) Determination of nitrate and nitrite. Method Enzymol 3:981–984. https://doi.org/10.1016/S0076-6879(57)03489-8
Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:259–264. https://doi.org/10.1016/10.1093/nar/gky1022
Nuñez MA, Horton TR, Simberloff D (2009) Lack of belowground mutualisms hinders Pinaceae invasions. Ecology 90:2352–2359. https://doi.org/10.2307/25592761
Nygren CMR, Eberhardt U, Karlsson M, Parrent JL, Lindahl BD, Taylor AFS (2008) Growth on nitrate and occurrence of nitrate reductase-encoding genes in a phylogenetically diverse range of ectomycorrhizal fungi. New Phytol 180:875–889. https://doi.org/10.1111/j.1469-8137.2008.02618.x
Oksanen AJ, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2019) vegan: Community ecology package. Retrieved from https://CRAN.R-project.org/package=vegan
Owen SM, Patterson AM, Gehring CA, Sieg CH, Baggett LS, Fulé PZ (2019) Large, high-severity burn patches limit fungal recovery 13 years after wildfire in a ponderosa pine forest. Soil Biol Biochem 139:107616. https://doi.org/10.1016/j.soilbio.2019.107616
Parke JL, Linderman RG, Black CH (1983) The role of ectomycorrhizas in drought tolerance of Douglas-fir seedlings. New Phytol 95:83–95. https://doi.org/10.1111/j.1469-81371983.tb03471.x
Patterson A, Flores-Rentería L, Whipple A, Whitham T, Gehring C (2019) Common garden experiments disentangle plant genetic and environmental contributions to ectomycorrhizal fungal community structure. New Phytol 221:493–502. https://doi.org/10.1111/nph.15352
Peay KG, Kennedy PG, Bruns TD (2011) Rethinking ectomycorrhizal succession: are root density and hyphal exploration types drivers of spatial and temporal zonation? Fungal Ecol 4:233–240. https://doi.org/10.1016/j.funeco.2010.09.010
Peterson RL (2012) 11 Ectendomycorrhizas: occurrence, structural characteristics, and possible roles. In: Fungal associations. Springer Berlin Heidelberg, pp 197–205
Querejeta JI, Schlaeppi K, López-García Á, Ondoño S, Prieto I, Heijden MGAD, Alguacil MM (2021) Lower relative abundance of ectomycorrhizal fungi under a warmer and drier climate is linked to enhanced soil organic matter decomposition. New Phytol 232:1399–1413. https://doi.org/10.1111/nph.17734
Rambold G, Agerer R (1997) DEEMY – the concept of a characterization and determination system for ectomycorrhizae. Mycorrhiza 7:113–116. https://doi.org/10.1007/s005720050171
Reid JL, Holl KD (2013) Arrival ≠ Survival. Restor Ecol 21:153–155. https://doi.org/10.1111/j.1526-100X.2012.00922.x
Rosenstock N, Ellström M, Oddsdottir E, Sigurdsson BD, Wallander H (2019) Carbon sequestration and community composition of ectomycorrhizal fungi across a geothermal warming gradient in an Icelandic spruce forest. Fungal Ecol 40:32–42. https://doi.org/10.1016/j.funeco.2018.05.010
Rosinger C, Sandén H, Matthews B, Mayer M, Godbold D (2018) Patterns in ectomycorrhizal diversity, community composition, and exploration types in European beech, pine, and spruce forests. Forests 9:445. https://doi.org/10.3390/f9080445
Saikkonen K, AhonenJonnarth U, Markkola AM, Helander M, Tuomi J, Roitto M, Ranta H (1999) Defoliation and mycorrhizal symbiosis: a functional balance between carbon sources and below-ground sinks. Ecol Lett 2:19–26. https://doi.org/10.1046/j.1461-0248.1999.21042.x
Schmid MW, Moorsel SJV, Hahl T, Luca ED, GBD D, Wagg C, Niklaus PA, Schmid B (2021) Effects of plant community history, soil legacy and plant diversity on soil microbial communities. J Ecol 109:3007–3023. https://doi.org/10.1111/1365-2745.13714
Siemens JA, Zwiazek JJ (2008) Root hydraulic properties and growth of balsam poplar (Populus balsamifera) mycorrhizal with Hebeloma crustuliniforme and Wilcoxina mikolae var. mikolae. Mycorrhiza 18:393–401. https://doi.org/10.1007/s00572-008-0193-2
Smith SE, Read DJ (2010) Mycorrhizal symbiosis. Third Edition. Academic press, London
Stokes A, Atger C, Bengough AG, Fourcaud T, Sidle RC (2009) Desirable plant root traits for protecting natural and engineered slopes against landslides. Plant Soil 324:1–30. https://doi.org/10.1007/s11104-009-0159-y
Tedersoo L, Smith ME (2013) Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol Rev 27:83–99. https://doi.org/10.1016/j.fbr.2013.09.001
Teste FP, Kardol P, Turner BL, Wardle DA, Zemunik G, Renton M, Laliberté E (2017) Plant-soil feedback and the maintenance of diversity in shrublands. Science 355:173–176. https://doi.org/10.1126/science.aai8291
Velmala SM, Rajala T, Heinonsalo J, Taylor AFS, Pennanen T (2014) Profiling functions of ectomycorrhizal diversity and root structuring in seedlings of Norway spruce (Picea abies) with fast- and slow-growing phenotypes. New Phytol 201:610–622. https://doi.org/10.1111/nph.12542
Vozzo JA, Hacskaylo E (1971) Inoculation of Pinus caribaea with ectomycorrhizal fungi in Puerto Rico. For Sci 17:239–245
Wang R, Guerin-Laguette A, Butler R, Huang LL, Yu FQ (2019) The European delicacy tuber melanosporum forms mycorrhizae with some indigenous Chinese Quercus species and promotes growth of the oak seedlings. Mycorrhiza 29:649–661. https://doi.org/10.1007/s00572-019-00925-y
Wang G, Bei S, Li J, Bao X, Zhang J, Schultz PA, Li H, Li L, Zhang F, Bever JD, Zhang J, Hannula E (2020) Soil microbial legacy drives crop diversity advantage: linking ecological plant–soil feedback with agricultural intercropping. J Appl Ecol 58:496–506. https://doi.org/10.1111/1365-2664.13802
Wasyliw J, Karst J (2020) Shift in ectomycorrhizal exploration types parallel leaf and fine root area with forest age. J Ecol 108:2270–2282. https://doi.org/10.1111/1365-2745.13484
Wen Z, Shi L, Tan Y, Hong L, Xue J, Xing J, Chen Y, Nara K (2018) Soil spore bank communities of ectomycorrhizal fungi in endangered Chinese Douglas-fir forests. Mycorrhiza 28:49–58. https://doi.org/10.1007/s00572-017-0800-1
Yu TEJC, Egger KN, Peterson RL (2001) Ectendomycorrhizal associations - characteristics and functions. Mycorrhiza 11:167–177. https://doi.org/10.1007/s005720100110
Zhang Z, Yuan Y, Zhao W, He H, Li D, He W, Liu Q, Yin H (2017) Seasonal variations in the soil amino acid pool and flux following the conversion of a natural forest to a pine plantation on the eastern Tibetan plateau, China. Soil Biol Biochem 105:1–11. https://doi.org/10.1016/j.soilbio.2016.11.002
Zhang Z, Yuan Y, Liu Q, Yin H (2019) Plant nitrogen acquisition from inorganic and organic sources via root and mycelia pathways in ectomycorrhizal alpine forests. Soil Biol Biochem 136:107517. https://doi.org/10.1016/j.soilbio.2019.06.013
Acknowledgements
This work was supported by National Natural Science Foundation of China (31870607, 41930645, 32171550), Youth Innovation Promotion Association of the Chinese Academy of Sciences (2019363), and Sichuan Science and Technology Program (2022YFS0485).
Author information
Authors and Affiliations
Contributions
Wenqiang Zhao and Qing Liu designed the study. Xiaohu Wang conducted the seedling growth experiment, data collection and analysis. The first draft of the manuscript was written by Xiaohu Wang and all authors commented on previous versions of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Responsible Editor: Janusz J. Zwiazek.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 482 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Kou, Y., Liu, J. et al. Soil microbial legacy determines mycorrhizal colonization and root traits of conifer seedlings during subalpine forest succession. Plant Soil 485, 361–375 (2023). https://doi.org/10.1007/s11104-022-05835-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-022-05835-1