Abstract
Background and aims
The production of maize (Zea mays L.) is restricted by various edaphic stresses, including drought and low-fertility soil. Searching for genotypes with optimal root traits is a promising practice when breeding for improved adaptation to abiotic stress and resource-use efficiency.
Methods
Using an established semi-hydroponic phenotyping technique, we assessed root trait variability across 174 maize genotypes including 113 cultivars and 11 breeding lines from northern China and 50 Serbian hybrids.
Results
Large variation in root architecture traits was observed among the tested genotypes 28 days after transplanting. Sixteen of the characterized traits had coefficients of variation greater than 0.25, especially local root traits. Root traits including total root length, root length at various depths, total shoot mass and nodal root angle, should be considered in maize breeding programs. Genotype ranking data based on a composite score was used to assist in the selection of genotypes with contrasting root architecture traits for future studies.
Conclusion
This study identified genotypic variation in root architecture traits in a diverse genotypes of maize. The outcomes of this study could form a basis for maize breeding programs aimed at producing maize cultivars for improved adaptation to target environments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Plant roots play an essential role in water and nutrient acquisition and thus influence plant health, environmental adaptation, and productivity (Hinsinger et al. 2011; Lynch and Wojciechowski 2015; Paez-Garcia et al. 2015). Crop growth and grain yield rely on the capacity of the root system to forage and capture soil resources (Dunbabin et al. 2003; Osmont et al. 2007; Sadras et al. 2016). Progress in root research is well-behind that of aboveground plant parts since accessing roots for research is inherently difficult (Smith and De Smet 2012). Alterations to root growth and root system architecture (RSA) are a critical adaptive strategy of crops to cope with drought, soil infertility and other edaphic stresses (Mi et al. 2016). Therefore, it is important to breed cultivars that have root systems with improved adaptation to edaphic stresses and water and nutrient efficiencies (Siddique et al. 2001). It is estimated that, on average, grain yield increases result from an equal contribution of farm management and plant breeding (Duvick 2005). Developing future crop genotypes with efficient root systems for enhanced abiotic stress tolerance is vital for improved crop adaptation.
Maize (Zea mays L.), the largest crop in plant shoot size, is widely planted around the world with annual productivity of ~ 700 million metric tons (Ranum et al. 2014). Maize has an embryonic root system (Hochholdinger 2009). A series of root traits have been identified that are associated with plant performance in specific environments (Ogawa et al. 2014; Ruta et al. 2010; Valliyodan et al. 2017). Variations in total root number, root angles, seminal root length, and root dry weights observed in some maize genotypes showed correlations to nitrogen acquisition, drought resistance and/or grain yield in several studies (Abdel-Ghani et al. 2013; Chun et al. 2005; Li et al. 2015; Wang et al. 2016).
The development of a semi-hydroponic phenotyping system (Chen et al. 2011a) serves as an efficient tool for characterizing root traits in large numbers of germplasm in various crop species, including narrow-leafed lupin (Lupinus angustifolius L.) (Chen et al. 2011a, 2011b, 2012, 2016), chickpea (Cicer arietinum L.) (Chen et al. 2017), wheat (Triticum aestivum L.), and barley (Hordeum vulgare L.) (unpublished). This study used the semi-hydroponic phenotyping system to characterize root trait variability in 174 maize genotypes. We hypothesized that root architecture traits in a large set of maize germplasm have significant variation, and that genotypes differ in the traits at both global and local levels. The variation in different traits may influence root function in exploiting water and nutrients in the soil.
This study aimed to characterize several root-related traits at both global and local levels across genotypes, identify variation among genotypes, and discuss on how the differences in some traits affect their function. The outcomes of this study will help in the selection of representative genotypes with contrasting root properties for further study in soil under both controlled and field environments, and enhance our understanding of the relative contribution of individual root traits to root architecture and plant performance.
Materials and methods
Maize genotypes
A collection of 174 maize (Zea mays L.) genotypes was used in this study, which included 124 genotypes from northern China (113 local cultivars and 11 breeding lines) and 50 hybrid lines from Serbia (Table S1). The Chinese cultivars were released from 2000 to 2015. These genotypes were selected based on their expansive growth area, grain yield, drought tolerance and disease resistance (Table S1). Seeds of the Chinese genotypes were gathered from various sources, including maize breeding research groups at the Northwest A&F University and the Crop Research Institute of Gaoling, China, and the Serbian hybrid lines were imported from the Institute of Field and Vegetable Crops, Serbia.
Experimental design and growth conditions
The experiment was conducted in a glasshouse at Northwest A&F University, Yangling (34° 16′ N, 108° 4″ E) using a semi-hydroponic phenotyping system (Fig. 1a) (Chen et al. 2011a). Four plants of each genotype planted in four replicate bins were arranged in a randomized block design in 24 wheelie bins (240-L capacity) with six bins as one replicate. Each bin system accommodated 36 plants with two plants in each growth unit and buffer plants were used when required to ensure an equal number of plants allocated to each bin.
The details of the setting of the semi-hydroponic system are described in Chen et al. (2011a). Each bin was filled with 30 L of low-concentration nutrition solution containing (μM): K (1220), P (20), S (1802), Ca (600), Mg (200), Cu (0.2), Zn (0.75), Mn (0.75), B (5), Co (0.2), Na (0.06), Mo (0.03), Fe (40), and N (2000). The calico black cloth in the plant growth units maintained moisture via automatic pumping systems. The solution was renewed weekly, and the pH was monitored frequently. The average daily temperature during the experimental period was approximately 25/15 °C (day/night).
Planting and harvesting
Surface-sterilized seeds were sown in washed river sand. Four days after sowing, the germinated seeds were carefully washed with DI water to remove the sand and transplanted into the growth pouches of the phenotyping system. The 24 bins were arranged in four rows in a glasshouse and randomized weekly. Plants were harvested 28 days after transplanting (when the maximal roots of approximately half the plants had reached the bottom of the 60-cm long black cloth).
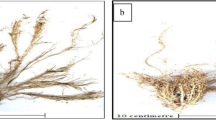
Maize has an embryonic root system that includes the primary root and seminal roots, as well as a post-embryonic root system of shoot-borne nodal roots that emerge over time (Fig. 1b) (Mi et al. 2016). At harvest, the maximum physical height of plant shoots (SH) and leaf number were measured manually. Root systems were photographed using a portable photo box. Maximal root depth (the vertical depth of the primary root), total seminal and nodal root depth (the sum of vertical depths of all individual seminal and nodal root), total root number (nodal roots less than 3 cm were not counted), and maximal root width (the maximal horizontal width of a root system) were measured manually at harvest. Nodal root angle (the maximal growth angle between two outer nodal roots) was measured manually from the root images in the lab. After photographing, shoots were cut from the roots and shoot dry mass was determined after drying in an air-forced oven at 75 °C for 72 h. Subsamples of roots were collected by cutting the root system (every 10 cm along the glass sheet) into 10-cm sections starting from the base which were then stored in a cool room until scanning.
Root section samples were scanned in greyscale at 300 dpi using a desktop scanner (Epson Perfection V800, Long Beach, CA, USA), and root images for each 10-cm sections were analyzed using WinRHIZO Pro (v2009, Regent Instruments, Montreal, QC, Canada). Imaging analyses used the debris removal filter of discounting objects less than 1 cm2 with a length-width ratio less than 8 in the image. After scanning, root subsamples from the same plant were combined into one root sample and dried in an oven to obtain root dry mass (RM). Root morphology data, such as root length, root surface area, root volume, average root diameter for each root section, and root length for different diameter classes were generated in the WinRHIZO program. The following root traits were calculated from the measured data:
Specific root length (SRL) = total root length divided by root dry mass; root tissue density (RTD) = root dry mass divided by total root volume; root to shoot mass ratio (RSM) =: root dry mass divided by shoot dry mass; root length ratio (RLR-upper/lower) = root length in top 20-cm section divided by root length in the lower section below 20 cm; root surface area ratio (RAR-upper/lower) = topsoil root surface area divided by subsoil root surface area; root volume ratio (RVR-upper/lower) = topsoil root volume divided by subsoil root volume.
The 30 root traits were divided into two general categories: 18 global traits (whole plant level) and 12 local traits (Tables 1 and 2). Global traits refer to the whole root system and whole shoots, and local root traits are referred to roots in different depths and diameter classes.
Data statistical analysis
One-way ANOVA was conducted using SPSS Statistics 20 (IBM, USA) for significant differences among the tested genotypes for each trait. Figures were plotted using SigmaPlot 12.5 (Systat, USA). Correlations were considered statistically significant at P ≤ 0.05. Traits with coefficient of variation (CV) values ≥ 0.25 were selected for principal component analysis to identify the determinants of root architecture variability across genotypes (Jolliffe and Cadima 2016). Hierarchical cluster analysis was used to determine the variance among selected root traits and homogeneous groups among genotypes using the average linkage method. The composite score based on principal component analysis was used to obtain a general ranking order for each genotype. The ranking value was based on the actual composite score from the least to the most. The composite score was calculated as the sum of variability × component score, where component score was the sum of weight coefficient × standardization of the mean value for each trait. Variability, weight coefficient and standardization of the mean value for each trait were acquired from principal component analysis.
Results
Global traits
Global root traits reflected the pattern of root development and distribution, and most differed among genotypes. Variation in root traits including total root length (CV = 0.26), root mass (CV = 0.24), total root depth (CV = 0.29), and total root number (CV = 0.20) were relatively high (all p < 0.05) (Table 2), while others were relatively low. The three shoot traits measured at harvest e.g., leaf number (CV = 0.06), shoot height (CV = 0.15), and shoot mass (CV = 0.26), differed significantly among genotypes (all P ≤ 0.05) (Table 2).
The maize plants grew vigorously with an average root growth rate (based on daily elongation of the maximal root, cm day−1) of 2.02 (range 1.24–2.56) (Table 2). Total root length (cm plant−1) ranged from 336 (genotype #063) to 1478 (genotype #054) (mean 824) (Table 2). Total root length and root mass differed fourfold between the largest and the smallest genotypes. The 174 genotypes had an average root length of 824 cm plant−1. The Chinese cultivars had relatively larger root systems with average root length of 889 cm plant−1 followed by the 11 Chinese breeding lines (718) and the Serbian hybrids (702). Genotypic variation in root mass exhibited a similar trend as total root length (Fig. 2). There was no apparent correlation between root length (or root mass) and the year of release (Fig. S1).
Genotypic variation in root length and root mass (RM) of 174 maize genotypes grown in a semi-hydroponic phenotyping platform. Mean values were plotted by three groups of genotypes: 50 Serbian hybrids, 11 Chinese breeding lines, and 113 Chinese cultivars released from 2000 to 2015. Mean values of total root length for each genotype group (colored bars) and that of the all genotypes (dashed line) are presented. Mean data of root length in each genotype group with different letters differ significantly between genotype groups (P ≤ 0.05). Both RL-upper (root length in 0–20 cm depth) and RL-lower (root length below 20-cm depth) are presented. Trend lines of RM for each genotype groups with correlation value (R2) are also given
The spatial distribution of root systems varied among genotypes for total root depth (TRD), root width (RW), and the root angle between the top two outer nodal roots (NA) (Table 2). TRD ranged from 34.3 (genotype #035) to 225 cm (#153) (mean 124 cm). RW ranged from 8.37 (#096) to 19.2 cm (#161) (mean 13.2), and NA (degrees) ranged from 54 (#008) to 104 (#126) (mean 78.7). The 65–90° NA classes contained 76% of the genotypes, ≥ 90° accounted for 13.8%, while those with steep (< 65°) root systems (e.g., #118: 58°, #168: 61.7°, #009: 62°, #016: 64.7°) represented 9.8% of all the genotypes in this study (Fig. 3).
Local traits
Root traits at different depths (root sections) and root lengths in different diameter classes (Tables 1 and 2) had relatively high variations with CVs > 0.25 except for the root length of medium diameter roots where CV = 0.22. The root length ratio (RLR-upper/lower) (CV = 0.42) ranged from 0.61 (genotype #168) to 4.08 (genotype #149) (mean 1.49) (Table 2). Root length, root area, and root volume varied significantly among genotypes at respective depths. For example, root length in the upper layer (RL-upper) ranged from 201 to 818 cm (mean 443 cm; CV = 0.28) and root length in the lower layer (RL-lower) ranged from 106 to 772 cm (mean 381 cm; CV = 0.31) (Table 2). For the combined data of all genotypes in each section, the 10–20 cm section had the maximum root length accounting for 31% of the total root length, with more than half the root length in the top 20 cm and 93% in the top 40 cm (Table 2, Fig. 4). The mean root length of Chinese cultivars in each 10 cm section (except for 50–60 cm) was higher than those of Serbian hybrids and Chinese breeding lines (Fig. 4).
Root length distribution in 10-cm increments for three groups of genotypes (50 Serbian hybrids, 11 Chinese breeding lines, and 113 Chinese cultivars released from 2000 to 2015) grown in a semi-hydroponic phenotyping platform. Data are means ± standard error. Significant differences are shown for 0–10-cm and 10–20-cm depths among the three genotype groups, respectively (P ≤ 0.05)
Root diameter data showed that maize plants had relatively thin roots with an average root diameter of 0.5 mm for all genotypes (Table 2). Most of the root lengths was in the 0- to 0.5-mm diameter class of thin roots (RL-thin), which accounted for 75% of the total root length. The root length of thick roots (RL-thick) contributed only 0.7% to total root length, and was primarily proximal (at the top, near the shoot) (Table 2).
Correlation among traits
A subset of 16 traits including 15 root traits and one shoot trait (i.e., shoot mass, SM) with larger coefficients of variation (CVs ≥ 0.25, Table 2) were selected for Pearson correlation analysis to identify relationships among the traits. Strong correlations were observed for most of the selected traits (Table 3). Total root length (RL) was strongly associated with all other traits (all P < 0.01) except root volume ratio (P < 0.05). For example, RL was strongly correlated with total root depth and SM, respectively. Root length, root surface area, and root volume in upper and lower layers were strongly associated with all other traits (all P < 0.01) except for the mentioned traits in the upper layer with root length ratio (P < 0.05). In addition, nodal root angle showed good correlation with root width (P < 0.01).
The same set of traits with CV ≥ 0.25 (Table 2) was included in the principal component analysis (PCA). Two principal components (PCs) were identified with eigenvalues > 1, capturing 84.2% of the total variation in these traits across the 174 maize genotypes (Table 4). The first component (PC1) captured 62.0% of the variability, accounting for most of the root-related traits, including total root length (RL), root surface area (RA), root volume (RV), total root depth (TRD), root traits in the upper and lower layers, and shoot mass (SM) (Table 4). PC2 represented 22.2% of the total variation and accounted for root length ratio (RLR-upper/lower), root area ratio (RAR-upper/lower), and root volume ratio (RVR-upper/lower) (Table 4; Fig. 5a).
The loading plot for PC1 vs. PC2 representing 84.2% of the variability identified four general clades of the tested 16 traits. Clade I includes RL, RA, RV, RL-thin, RL-thick, TRD and SM (Fig. 5a). Root length, area, and volume in the upper and lower layers were grouped into clades II and III, respectively (Fig. 5a), and root trait ratios were classified as a separate clade (clade IV). Genotype distribution based on PCA regression scores of the 16 traits is shown in Fig. 5b. The relative distance among the 174 genotypes is displayed for each combination of root traits. The outer genotypes with particular traits are shown in red, and should be selected for further study.
Genotype homogeneous grouping based on root trait variation
An agglomerative hierarchical clustering (AHC) dendrogram was constructed using the average linkage method of the 16 selected traits, which revealed large diversity among the 174 maize genotypes (Fig. S2). Three general clades (clades I to III) were determined using a distance of 18 and further separated into six groups (G1 to G6) at a middle distance of 12. These six groups were then divided into 14 sub-groups at a shorter distance of 6, with 1 to 74 genotypes per sub-group. Group 1 had five sub-groups with 129 genotypes, group 2 had three sub-groups with 33 genotypes, group 3 had one sub-group with seven genotypes, group 4 had two sub-groups with two genotypes (genotype #038 and #049), group 5 had two sub-groups with two genotypes (genotype #066 and #118), and group 6 had one sub-group with one genotype (genotype #100).
Composite score and genotype distribution
The composite score (CS) based on PCA was used to rank the 174 maize genotypes from the least to the most (Fig. 6). The Chinese cultivars had a significantly higher average CS (0.11) than the Serbian hybrids (− 0.19) and Chinese breeding lines (− 0.23) (Fig. 6).
The ranking of composite score based on principal component analysis of 174 maize genotypes grown in a semi-hydroponic phenotyping platform. Data were plotted by five groups of genotypes: 50 Serbian hybrids, 11 Chinese breeding lines, 35 Chinese cultivars released from 2000 to 2005, 40 Chinese cultivars released from 2006 to 2010, and 38 Chinese cultivars released from 2011 to 2015. Group means (red bars) with different letters differ significantly between genotype groups (P ≤ 0.05)
The experimental data were further analyzed based on origin and breeding status. The 174 maize genotypes were assigned to six groups at a middle distance of 12 (Fig. S2): Serbian hybrids in groups 1 to 4, Chinese breeding lines in groups 1 to 3, and the Chinese cultivars released from 2000 to 2005 in groups 1 and 2, from 2006 to 2010 in groups 1, 2, and 5, and from 2011 to 2015 in all groups except group 4 (Fig. 7). Seventeen genotypes, with the highest CS values in each category for each group of genotypes, were selected for further study (Fig. 7). The widely planted cultivar #091 (Zhendan 958) in north China was considered the reference genotype.
The distribution of 174 maize genotypes in six groups at a middle distance of 12 based on a combination of principal component analysis, hierarchical clustering analysis and composite score. Data were plotted by five groups of genotypes: 50 Serbian hybrids, 11 Chinese breeding lines, 35 Chinese cultivars released from 2000 to 2005, 40 Chinese cultivars released from 2006 to 2010, and 38 Chinese cultivars released from 2011 to 2015. The labeled genotypes from each genotype groups can be selected for future studies
Genotypic variation in root traits for selected genotypes
Large differences in root-related traits exist among the 17 selected genotypes and the reference cultivar (genotype #091). Their ranking scores for the 16 selected traits (CV ≥ 0.25) are in Table S2. The 17 genotypes varied in their rooting patterns; for example, genotype #100 (Chinese cultivar released in 2015) and #049 (Serbian hybrid) had similar root lengths (ranked 40th and 39th longest roots, respectively, based on actual mean values) but different root length ratios (RL-upper/lower, ranked 44th and 168th), and had similar total root depths (ranked 166th and 149th) but different maximal root depths (ranked 128th and 13th) (Table S2). Variations in root length, root length-upper, root length-lower, root mass, shoot mass, and nodal root angle in the pre-selected 18 genotype representatives are in Fig. S3.
Discussion
Variation among root architecture traits
Collecting genetic data from large genotypes is increasingly becoming a strategy in plant breeding. The development of automated phenotyping platforms improves the efficiency and accuracy of collecting genetic data (Kuijken et al. 2015). This study used a semi-hydroponic phenotyping platform (Chen et al. 2011a) to characterize a range of root architecture traits in 174 maize genotypes. Plant genetic variation needs to be considered in phenotyping experiments. Since the root system is plastic and sensitive to G × E effects (Avramova et al. 2016), the ability of roots to adapt to varying environmental conditions may differ among genotypes and growth environments. One study identified a basic pattern of maize root distribution in the soil that was modified, but not fundamentally changed when plants were grown under varying conditions (Liedgens and Richner 2001). The efficiency and reliability of the semi-hydroponic phenotyping platform used in this study to reveal root system architecture (RSA) and increase genetic gain have been tested on a large number of germplasm from various crop species (Chen et al. 2011a, 2011b, 2011, 2012, 2016, 2017).
We measured 30 root-related traits, including 18 global and 12 local traits, and identified large variation in these traits among the 174 maize genotypes; 16 traits had CVs ≥ 0.25, 11 of which were local traits (Table 2). Studies of RSA are concerned with the entire root system of an individual plant as well as fine details of the root structure (Lynch 1995). However, variation in local root traits is of particular interest as it indicates differences in some underlying genes among genotypes that control root functions and contribution (Oikeh et al. 1999). Significant genetic variation in local root traits in the tested maize genotypes, especially root distribution and density at various depths may provide useful data for breeding programs by utilizing potential root traits.
Contribution of individual root traits
The 30 root-related traits measured in this study reflect root growth (such as total root length, total root number, and root growth rate), root distribution (such as maximum root depth, nodal root angle, and root width), and shoot growth (such as shoot mass, shoot height, and leaf number) (Table 2). Root size (total root length and total root dry mass) is often associated with plant foraging for soil resources (Wijesinghe et al. 2001). In barley, genotypes with contrasting root system size influenced grain yield in barley (Svacina et al. 2014). Total root length at the seedling stage has been associated with drought tolerance under field conditions (Avramova et al. 2016; He et al. 2017; Palta et al. 2011). In this study, the largest root systems had more than four times the total root length (RL) and root mass (RM) of the smallest root systems (Table 2). Root growth rates varied up to twofold among the genotypes (Table 2). In chickpea, root length tends to increase with increasing root growth rate (Chen et al. 2017). In our study on maize, there was a positive trend between RL and total root number. However, genotypes with more roots did not always have larger root systems in terms of RL, and vice versa. Our study observed up to three-fold difference in total root numbers among genotypes (Table 2). Decreases in nodal root number may relate to enhanced nitrogen-use efficiency when maize plants are grown in low-nitrogen soils (Saengwilai et al. 2014). Total root number has been positively correlated with grain yield in maize (Wang et al. 2015). The contribution of root genetic variation to plant drought tolerance has been verified in maize at the seedling stage (Wang et al. 2016).
It is well-known that crop production is restricted by drought, soil compaction, and low-fertility stresses (Chen et al. 2013a, 2014; Comas et al. 2013; Fang et al. 2017). Root growth into deep horizons is often affected by one or more constraints, which influence plant acquisition of water and nutrients from the soil (Adcock et al. 2007). In our study, root distribution in the soil profile was divided into vertical and horizontal directions. Other studies have shown that, in the vertical direction, root depth and root angle are the most important traits in terms of their influence on crop yield; while in the horizontal direction, root angle and root width are considered the most important traits (Colombi and Walter 2017; Lynch 2013; Lynch and Wojciechowski 2015). In this study, deep-rooting genotypes had almost twice the maximal root depth of shallow-rooting genotypes, and the total root vertical depth differed up to sevenfold among the genotypes (Table 2). Deeper root systems are preferred for improved water and nitrogen uptake in leaching environments (Ytting et al. 2014), and maximum rooting depth is commonly used to characterize the susceptibility of plant species to drought (Nippert and Holdo 2015). Root angles play an important role in determining rooting depth and drought tolerance in crops (Ali et al. 2015). Our data showed that most of the genotypes were in the 65–90° angle classes and only 9.8% had steep angles (≤ 65°) (Fig. 3). Root width differed about twofold among the genotypes (Table 2). There was a general trend between nodal root angle and root width. Under high plant density and sufficient fertilization, maize root systems tend to distribute narrower and deeper in semi-arid areas (Wang et al. 2015). Nodal root angle in young plants is an important trait for drought tolerance (Singh et al. 2012). The maize genotypes with steep angles in this study could be used for further study of drought tolerance. Genotypic variation in shoot traits was strongly correlated some important root traits. Studies have shown that root traits in seedlings grown under controlled conditions mirrored plant performance in the field (Landi et al. 1998; Tuberosa et al. 2002). A significant reduction in total root length, root depth, and root width was reflected in the corresponding reduction in root dry mass and shoot production in maize under drought stress (Avramova et al. 2016).
Genotype selection based on root trait properties for breeding programs
Some root traits such as total root length, shoot mass, nodal root angle, and local root traits at various depths can potentially serve as parameters for future screening programs. One study showed that root mass deep in the soil profile, total root mass, and root length are associated with maize grain yield under water-deficit field conditions (Ali et al. 2016). Phenotypic differences among genotypes under drought conditions in the field can be identified from total root length and total shoot dry mass at the seedling stage (Avramova et al. 2016). A strong positive correlation was observed between nodal root angle and grain yield under drought, and high-yielding hybrids had steeper root growth angles than low-yielding hybrids (Ali et al. 2015). These results indicate that such root traits can be selected to breed cultivars for target environments in future studies.
Crop breeders and researchers are increasingly interested in phenotyping for root architecture traits in their breeding programs (Kuijken et al. 2015). Identifying genotypes with suitable root traits for improved adaptation to environmental stresses and improved grain yield remains challenging. Based on the hierarchical cluster and composite score analyses, representative genotypes with contrasting root traits were selected, although the total number of genotypes to be selected relies on the requirements and feasibility of future studies. There may be a possible association in some root-related traits with year of release among genotypes under edaphic stresses (Aziz et al. 2017) although no apparent trend was observed in this study. In this study, cultivars generally had higher values for total root length, root length at various depths, and composite scores than the breeding lines (Figs. 2, 4, and 6), which suggests that maize cultivars may capture more suitable root properties through the breeding programs. One study showed that indirect selection of the maize root system in the past 100 years had evolved phenotypes suited to more intense competition for nitrogen (York et al. 2015).
Phenotyping for root trait variability in a relatively large number of genotypes in maize provides opportunities to select genotypes with suitable root properties for improved adaptation to abiotic stress and enhanced crop yield. Root trait data collected in this phenotyping study forms a basis for breeding new cultivars through marker-assisted selection programs. Root trait-marker association and linkage mapping analyses have already identified the genetic basis of a few root traits in maize (Manavalan et al. 2011) and other crops (Arifuzzaman et al. 2014; Henry 2013; Sanguineti et al. 2007; Uga et al. 2013; Varshney 2016).
The selected 16 genotypes with contrasting root architecture traits, together with the widely planted cultivar #091 (Zhengdan 958) in north China (Fu et al. 2011; Qi et al. 2012; Wu et al. 2011; Zhu et al. 2011) as a reference cultivar are being used in the follow-up studies. These 17 genotypes are being further examined for plant adaptation and root responses to polyethylene glycol induced drought, low phosphorus and salinity stresses, using the same semi-hydroponic phenotyping system. Preliminary analyses of root trait data in non-stressed plants showed consistent rankings of genotypes in these separate studies (unpublished data). Previous studies in selected genotypes of narrow-leafed lupin also reported a relatively consistent ranking of genotypes in some important root traits between the semi-hydroponic system and two soil media in glasshouse environment and field conditions, which confirms the advances of this phenotyping technique for screening large numbers of genotype for characterizing root trait variability, and its capacity to represent growth conditions in simple, uniform soil environments (Chen et al. 2011b, 2012, 2014). Our recent study showed that genotypic variation in root system size among five selected genotypes from a phenotyping experiment using the same semi-hydroponic system was reproducible at the booting stage (Z4.9; 63 days after sowing) grown in rhizoboxes filled with soil (Figueroa-Bustos et al. 2018). Despite some studies confirming the correlation between root traits at the seedling stage under controlled conditions and those obtained at various growth stages in the field, future studies are needed to validate root trait variability in soil and under field conditions. The selected genotypes can be further assessed for their adaptation to drought stress, low soil fertility, and other edaphic stresses in soils in both controlled and field environments as we did recently in narrow-leafed lupin (Chen et al. 2011b, 2013a, b, 2014, 2016).
Conclusion
Our study identified substantial variation in a wide range of root-related traits across the tested 174 maize genotypes. Findings in genotypic variation in root architecture traits, particularly, local root traits at various depths, form a basis for future study. Genotypic variation in root traits among diverse maize lines could be used to identify potential quantitative trait loci controlling root architecture. Some typical traits such as total root length, root length at various depths, nodal root angle, and shoot mass can potentially serve as parameters for future breeding programs aimed at producing cultivars with improved adaptation to adverse environments and better resource-use efficiency and grain yield.
References
Abdel-Ghani AH, Kumar B, Reyes-Matamoros J, Gonzalez-Portilla PJ, Jansen C, San Martin JP, Lee M, Lubberstedt T (2013) Genotypic variation and relationships between seedling and adult plant traits in maize (Zea mays L.) inbred lines grown under contrasting nitrogen levels. Euphytica 189:123–133
Adcock D, McNeill AM, McDonald GK, Armstrong RD (2007) Subsoil constraints to crop production on neutral and alkaline soils in south-astern Australia: a review of current knowledge and management strategies. Aust J Exp Agric 47: 1245–1261
Ali ML, Luetchens J, Nascimento J, Shaver TM, Kruger GR, Lorenz AJ (2015) Genetic variation in seminal and nodal root angle and their association with grain yield of maize under water-stressed field conditions. Plant Soil 397:213–225
Ali ML, Luetchens J, Singh A, Shaver TM, Kruger GR, Lorenz AJ (2016) Greenhouse screening of maize genotypes for deep root mass and related root traits and their association with grain yield under water-deficit conditions in the field. Euphytica 207:79–94
Arifuzzaman M, Sayed MA, Muzammil S, Pillen K, Schumann H, Naz AA, Leon J (2014) Detection and validation of novel QTL for shoot and root traits in barley (Hordeum vulgare L.). Mol Breed 34:1373–1387
Avramova V, Nagel KA, AbdElgawad H, Bustos D, DuPlessis M, Fiorani F, Beemster GTS (2016) Screening for drought tolerance of maize hybrids by multi-scale analysis of root and shoot traits at the seedling stage. J Exp Bot 67:2453–2466
Aziz MM, Palta JA, Siddique KHM, Sadras VO (2017) Five decades of selection for yield reduced root length density and increased nitrogen uptake per unit root length in Australian wheat varieties. Plant Soil 413:181–192
Chen YL, Dunbabin VM, Diggle AJ, Siddique KHM, Rengel Z (2011a) Development of a novel semi-hydroponic phenotyping system for studying root architecture. Funct Plant Biol 38: 355–363
Chen YL, Dunbabin VM, Postma JA, Diggle AJ, Palta JA, Lynch JP, Siddique KHM, Rengel Z (2011b) Phenotypic variability and modelling of root structure of wild Lupinus angustifolius genotypes. Plant Soil 348:345–364
Chen YL, Dunbabin VM, Diggle AJ, Siddique KHM, Rengel Z (2012) Assessing variability in root traits of wild Lupinus angustifolius germplasm: basis for modelling root system structure. Plant Soil 354:141–155
Chen YL, Dunbabin VM, Diggle AJ, Siddique KHM, Rengel Z (2013a) Phosphorus starvation boosts carboxylate secretion in P-deficient genotypes of Lupinus angustifolius with contrasting root structure. Crop & Pasture Science 64: 588–599
Chen YL, Dunbabin VM, Postma JA, Diggle AJ, Siddique KH, Rengel Z (2013b) Modelling root plasticity and response of narrow-leafed lupin to heterogeneous phosphorus supply. Plant Soil 372:319–337
Chen YL, Palta J, Clements J, Buirchell B, Siddique KHM, Rengel Z (2014) Root architecture alteration of narrow-leafed lupin and wheat in response to soil compaction. Field Crop Res 165:61–70
Chen YL, Shan F, Nelson MN, Siddique KHM, Rengel Z (2016) Root trait diversity, molecular marker diversity, and trait-marker associations in a core collection of Lupinus angustifolius. J Exp Bot 67:3683–3697
Chen YL, Ghanem ME, Siddique KHM (2017) Characterising root trait variability in chickpea (Cicer arietinum L.) germplasm. J Exp Bot 68:1987–1999
Chun L, Mi GH, Li JS, Chen FJ, Zhang FS (2005) Genetic analysis of maize root characteristics in response to low nitrogen stress. Plant Soil 276:369–382
Colombi T, Walter A (2017) Genetic diversity under soil compaction in wheat: root number as a promising trait for early plant vigor. Front Plant Sci 8:420
Comas LH, Becker SR, Cruz VV, Byrne PF, Dierig DA (2013) Root traits contributing to plant productivity under drought. Front Plant Sci 4:442
Dunbabin V, Diggle A, Rengel Z (2003) Is there an optimal root architecture for nitrate capture in leaching environments? Plant Cell and Environment 26:835–844
Duvick DN (2005) The contribution of breeding to yield advances in maize (Zea mays L.). In: Sparks DL, ed. Advances in agronomy, volume 86, Vol. 86. San Diego: Elsevier Academic Press Inc: 83–145
Fang Y, Du YL, Wang J, Wu AJ, Qiao S, Xu BC, Zhang SQ, Siddique KHM, Chen YL (2017) Moderate drought stress affected root growth and grain yield in old, modern and newly released cultivars of winter wheat. Front Plant Sci 8:872
Figueroa-Bustos V, Palta JA, Chen Y, Siddique KH (2018) Characterization of root and shoot traits in wheat cultivars with putative differences in root system size. Agronomy 8:14. https://doi.org/10.3390/agronomy8070109
Fu ZY, Jin XN, Ding D, Li YL, Fu ZJ, Tang JH (2011) Proteomic analysis of heterosis during maize seed germination. Proteomics 11:1462–1472
He YB, Lin LR, Chen JZ (2017) Maize root morphology responses to soil penetration resistance related to tillage and drought in a clayey soil. J Agric Sci 155:1137–1149
Henry A (2013) IRRI’s drought stress research in rice with emphasis on roots: accomplishments over the last 50 years. Plant Root 7:92–106
Hinsinger P, Brauman A, Devau N, Gerard F, Jourdan C, Laclau JP, Le Cadre E, Jaillard B, Plassard C (2011) Acquisition of phosphorus and other poorly mobile nutrients by roots. Where do plant nutrition models fail? Plant Soil 348:29–61
Hochholdinger F (2009) The maize root system: morphology, anatomy, and genetics: Springer, 233 Spring Street, New York, NY 10013, United States
Jolliffe IT, Cadima J (2016) Principal component analysis: a review and recent developments. Philosophical Transactions of the Royal Society A−Mathematical Physical and Engineering Sciences 374:20150202
Kuijken RCP, van Eeuwijk FA, Marcelis LFM, Bouwmeester HJ (2015) Root phenotyping: from component trait in the lab to breeding. J Exp Bot 66: 5389–5401
Landi P, Albrecht B, Giuliani MM, Sanguineti MC (1998) Seedling characteristics in hydroponic culture and field performance of maize genotypes with different resistance to root lodging. Maydica 43:111–116
Li RY, Zeng YJ, Xu J, Wang Q, Wu FK, Cao MJ, Lan H, Liu YX, Lu YL (2015) Genetic variation for maize root architecture in response to drought stress at the seedling stage. Breed Sci 65:298–307
Liedgens M, Richner W (2001) Minirhizotron observations of the spatial distribution of the maize root system. Agron J 93: 1097–1104
Lynch J (1995) Root architecture and plant productivity. Plant Physiol 109:7–13
Lynch JP (2013) Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann Bot 112:347–357
Lynch JP, Wojciechowski T (2015) Opportunities and challenges in the subsoil: pathways to deeper rooted crops. J Exp Bot 66:2199–2210
Manavalan LP, Musket T, Nguyen HT (2011) Natural genetic variation for root traits among diversity lines of maize (Zea mays L). Maydica 56:59–68
Mi G, Chen F, Yuan L, Zhang F (2016) Ideotype root system architecture for maize to achieve high yield and resource use efficiency in intensive cropping systems. In: Advances in Agronomy 139:73–97. Academic Press
Nippert JB, Holdo RM (2015) Challenging the maximum rooting depth paradigm in grasslands and savannas. Funct Ecol 29:739–745
Ogawa S, Valencia MO, Ishitani M, Selvaraj MG (2014) Root system architecture variation in response to different NH4+ concentrations and its association with nitrogen−deficient tolerance traits in rice. Acta Physiol Plant 36:2361–2372
Oikeh SO, Kling JG, Horst WJ, Chude VO, Carsky RJ (1999) Growth and distribution of maize roots under nitrogen fertilization in plinthite soil. Field Crop Res 62(1):1–13
Osmont KS, Sibout R, Hardtke CS (20070 Hidden branches: developments in root system architecture. Annu Rev Plant Biol, Vol. 58. Palo Alto: annual reviews: 93–113, 2007
Paez-Garcia A, Motes CM, Scheible WR, Chen R, Blancaflor EB, Monteros MJ (2015) Root traits and phenotyping strategies for plant improvement. Plants−Basel 4:334–−355
Palta JA, Chen X, Milroy SP, Rebetzke GJ, Dreccer MF, Watt M (2011) Large root systems: are they useful in adapting wheat to dry environments? Funct Plant Biol 38: 347–354
Qi WZ, Liu HH, Liu P, Dong ST, Zhao BQ, So HB, Li G, Liu HD, Zhang JW, Zhao B (2012) Morphological and physiological characteristics of corn (Zea mays L.) roots from cultivars with different yield potentials. Eur J Agron 38:54–63
Ranum P, Pena-Rosas JP, Garcia-Casal MN (2014) Global maize production, utilization, and consumption. In: PenaRosas JP, GarciaCasal MN, Pachon H (eds) Technical considerations for maize flour and corn meal fortification in public health, vol 1312. Blackwell Science Publ, Oxford, pp 105–112
Ruta N, Stamp P, Liedgens M, Fracheboud Y, Hund A (2010) Collocations of QTLs for seedling traits and yield components of tropical maize under water stress conditions. Crop Sci 50: 1385–1392
Sadras VO, Villalobos FJ, Fereres E (2016) Crop Development and. Growth pp:141–158
Saengwilai P, Tian X, Lynch JP (2014) Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiol 166:581–589
Sanguineti MC, Li S, Maccaferri M, Corneti S, Rotondo F, Chiari T, Tuberosa R (2007) Genetic dissection of seminal root architecture in elite durum wheat germplasm. Ann Appl Biol 151:291–305
Siddique KHM, Regan KL, Tennant D, Thomson BD (2001) Water use and water use efficiency of cool season grain legumes in low rainfall Mediterranean-type environments. Eur J Agron 15:267–280
Singh V, van Oosterom EJ, Jordan DR, Hammer GL (2012) Genetic control of nodal root angle in sorghum and its implications on water extraction. Eur J Agron 42:3–10
Smith S, De Smet I (2012) Root system architecture: insights from Arabidopsis and cereal crops Introduction. Philosophical Transactions of the Royal Society B−Biological Sciences 367: 1441–1452
Svacina P, Streda T, Chloupek O (2014) Uncommon selection by root system size increases barley yield. Agron Sustain Dev 34:545–551
Tuberosa R, Sanguineti MC, Landi P, Michela Giuliani M, Salvi S, Conti S (2002) Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol Biol 48:697–712
Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Yano M (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45:1097–1102
Valliyodan B, Ye H, Song L, Murphy M, Shannon JG, Nguyen HT (2017) Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J Exp Bot 68:1835–1849
Varshney RK (2016) Exciting journey of 10 years from genomes to fields and markets: some success stories of genomics-assisted breeding in chickpea, pigeonpea and groundnut. Plant Sci 242:98–107
Wang ZG, Ma BL, Gao JL, Sun JY (2015) Effects of different management systems on root distribution of maize. Can J Plant Sci 95:21–28
Wang XL, Wang HW, Liu SX, Ferjani A, Li JS, Yan JB, Yang XH, Qin F (2016) Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet 48:1233–1241
Wijesinghe DK, John EA, Beurskens S, Hutchings MJ (2001) Root system size and precision in nutrient foraging: responses to spatial pattern of nutrient supply in six herbaceous species. J Ecol 89:972–983
Wu XL, Liu HY, Wang W, Chen SN, Hu XL, Li CH (2011) Proteomic analysis of seed viability in maize. Acta Physiol Plant 33:181–191
York LM, Galindo-Castaneda T, Schussler JR, Lynch JP (2015) Evolution of US maize (Zea mays L.) root architectural and anatomical phenes over the past 100 years corresponds to increased tolerance of nitrogen stress. J Exp Bot 66:2347–2358
Ytting NK, Andersen SB, Thorup-Kristensen K (2014) Using tube rhizotrons to measure variation in depth penetration rate among modern North−European winter wheat (Triticum aestivum L.) cultivars. Euphytica 199:233–245
Zhu XC, Song FB, Liu SQ, Liu TD (2011) Effects of arbuscular mycorrhizal fungus on photosynthesis and water status of maize under high temperature stress. Plant Soil 346:189–199
Acknowledgments
We acknowledge Ivica Djalovic of the Institute of Field and Vegetable Crops, Serbia, and Dongjing Xi of the Crop Research Institute of Gaoling, China, for supplying maize seeds. Jianwei Chen, Mengxuan Yuan, Min Li, Hui Li, Jiyue Kang, Peifeng Xiong, Yadong Xu, and Zhifei Chen assisted in this study.
Funding
This work was supported by the National Natural Science Foundation of China (31471946) and Chinese Academy of Sciences (“100 Talent” Program, A315021449).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Martin Weih
Electronic supplementary material
ESM 1
(DOCX 78 kb)
Fig. S1.
Genotypic variation in total root length (RL) and root mass (RM) among 113 Chinese cultivars for year of release grown in a semi-hydroponic phenotyping platform. (JPG 889 kb)
Fig. S2.
Dendrogram of agglomerative hierarchical clustering (AHC) using the average linkage (between groups) method. The 174 maize genotypes were assigned to three general clades (Clade I, II, or III) at a distance of 18 (right dashed line) containing six groups (G1 to G6) at a middle distance of 12 (middle dashed line), which can be further separated into 14 sub-groups at a shorter distance of 6 (left dashed line). The 16 traits are the same as those used for PCA in Table 4. (JPG 595 kb)
Fig. S3.
Root length (a), root length in upper 20 cm layer (b) and lower section below 20 cm (c), root mass (d), shoot mass (e) and nodal root angle (f) in 18 selected genotypes grown in a semi-hydroponic phenotyping platform. Data for the reference cultivar (genotype #091) are indicated as closed red dot. (JPG 638 kb)
Rights and permissions
About this article
Cite this article
Qiao, S., Fang, Y., Wu, A. et al. Dissecting root trait variability in maize genotypes using the semi-hydroponic phenotyping platform. Plant Soil 439, 75–90 (2019). https://doi.org/10.1007/s11104-018-3803-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-018-3803-6