Abstract
Background and aims
Root-rot disease, a catastrophic disease of Panax quinquefolium L. causes yield reduction and serious economic losses. However, knowledge of the relationship between rhizosphere microbial community and root-rot disease is limited. This study is aim to test whether the bacteria and fungi community differed between the soil attached to healthy and rotten roots of American ginseng. Moreover, the effects of American ginseng cultivation for 4 years on changes of soil physiochemical properties and microbial community were also investigated.
Methods
High-throughput sequencing (Illumina MiSeq) was used to investigate the difference of microbial communities in the soils of new farmland (C) and the rhizosphere soils around healthy (H) and root rot diseased ginseng (R).
Results
Cultivation of American ginseng for 4 years not only changed the soil physicochemical properties, but also significantly increased the richness of the soil bacteria and decreased the fungal richness and diversity. Compared with other genera, the bacterial genera Nitrospira and the fungal genera Gibberella and Podospora were strongly enriched in the soil of new farmland. However, the relative abundance of Janthinobacterium, Nitrospira and Pedomicrobium in bacterial community, and Mrakia, Paradendryphiella, Sporopachydermia, Myrothecium and Racocetra in fungal community were significantly decreased after culture of American ginseng. The results also showed that the bacteria and fungi community differs between the soil attached to healthy and rotten roots of American ginseng. The richness indices of fungal community showed a significant decrease in rhizosphere soils of R comparing with H. The bacteria Rhodoplanes and Kaistobacter were the dominant genera in the H sample, whereas Sphingobium was dominant in the R sample. Notably, Monographella was significantly higher in the R sample (23.13%) than that of H sample (2.90%). In addition, the fungi Melanophyllum and Staphylotrichum were the most differently abundant in the H sample, whereas Mortierella and Cistella were the differently abundant genera in the R sample.
Conclusions
Our results indicate that cultivation of American ginseng changed the edaphic factors and the soil microbial community, and there are significant differences in the microbial community between the soil attached to healthy and rotten roots of American ginseng.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
American ginseng, or xiyangshen (Panax quinquefolius L.), is a highly valuable herb in the Araliaceae family, as its dry root and root extracts have been popular health supplements in Asia for hundreds of years. Undoubtedly the biggest challenge to American ginseng production is the management of diseases which render the ginseng root unmarketable due to infection primarily by fungal pathogens (Punja 2011). High planting density, over fertilization, and growth condition that are damp and warm with reduced sunlight and air flow all provide a favorable environment for plant pathogens, especially root-rot disease, which would consequently lead to crop yield and quality reduction (Wu et al. 2016). These diseases are primarily caused by fungal pathogens, including several species of Fusarium (causing root rot or rusty root) (Bi et al. 2011; Punja et al. 2007; Rahman and Punja 2005), Rhexocercosporidium panacis (causing rusty root) (Reeleder 2007), Cylindrocarpon destructans (causing root rot) (Punja 1997; Reeleder and Brammall 1994), and Phytophthora cactorum (causing Phytophthora root rot) (Punja 1997; Darmono et al. 1991). Among these pathogens, Fusarium solani and F. oxysporum are highly aggressive fungi causing American ginseng root rot in the Beijing ginseng producing region of China (Bi et al. 2011). Our survey found that root rot of American ginseng has become an important factor limiting the production of American ginseng in the Liuba area, Hanzhong, a major producing area in the Northwest of China. Typical early symptoms of root rot diseases in American ginseng are reddish-brown to orange-brown discolored areas on the root surface (Jiao et al. 2015). Rotten symptoms include dry rot in both exterior and interior root tissues and loss of fibrous roots with the development of disease (Jiao et al. 2015).
Although these pathogens are frequently isolated from the roots of decaying American ginseng, whether these pathogens are dominant microbial communities in the rhizosphere soil has rarely been reported. In addition, the bacterial community in the rhizosphere soil of root rot is often overlooked due to the isolation of fungi from American ginseng. The initiation of root rot has been associated with deterioration of the physicochemical properties of the soil especially in a soil with long-term inorganic fertilization and continuous cropping, autotoxicity, and changes in the soil microbial community (Ogweno and Yu 2006; Huang et al. 2013). Conventional farming practices with time have led to decline in soil structure, fertility and microbial diversity and simultaneously given rise to many soil and root borne diseases (Ahanger et al. 2014). Root diseases are more damaging when soil conditions are poor as a result of inadequate drainage, poor soil structure, low organic matter and low soil fertility (Ahanger et al. 2014). Autotoxicity is a type of intraspecific allelopathy where a plant species inhibits the growth of its own or relatives through the release of toxic chemicals into the environment (Singh et al. 1999; Yu et al. 2000). Some reports illuminated that autotoxins in root tissue of asparagus show synergism with Fusarium oxysporum spp. asparagi causing root rot and increasing the incidence of the disease (Hartung and Stephens 1983; Peirce and Colby 1987; Nigh 1990). Soil microorganisms have profound effects on the growth, nutrition and health of plants in natural and agricultural ecosystems (Garbeva et al. 2004; Almario et al. 2013). Among these, imbalances (the increase of potential pathogens and the decrease of beneficial soil organisms) in soil microbial communities were shown to be the pivotal event for infection of peanuts by root rot-causing microbes (Chen et al. 2012). Tan et al. (2017) investigated the rhizospheric and root endophytic fungi during continuous cropping of sanchi (Panax notoginseng), and the result demonstrated that root-rot disease affected the community structure and diversity of rhizospheric and root endophytic fungi. Xiong et al. (2017) researched synthetically the soil microbial communities and analyzed the relative importance of bacterial and fungal communities for the suppression of Fusarium wilt through Illumina MiSeq sequencing, and indicated that fungal communities may be important in the development of soil suppressiveness against vanilla Fusarium wilt disease. Dong et al. (2017) revealed that the diversity and composition of the soil bacterial and fungal communities changed during the continuous cropping of American ginseng compared to the communities present during cultivation of traditional crops. However, these reports mainly focused on the microbial community of rhizospheric soil during repeated cropping or crop rotation. At this time, it is unknown if there are any differences in the microbial communities in soils that have been planted with American ginseng for 4 years and in soils that have never been used to cultivate American ginseng. Since we also observed that healthy and rotted American ginseng roots are both present in the same land, we speculated that there may be differences in the microbial community of the rhizospheric soil of healthy and diseased ginseng from the same plot.
In this study, we compared the soil physicochemical propertiessoil and microbial communities of barren and cultivated fields. In addition, we also tested whether the rhizospheric soil of healthy and rotten roots differed in bacterial and fungal abundance, diversity and taxonomic composition.
Materials and Methods
Soil collection
The sampling site located in Liuba County (a American ginseng research farm) (33° 38' N, 106° 43' E), Hanzhong, Shaanxi province, which is one of the main areas of American ginseng production in China. The mean annual temperature and altitude in this area are 22.1°C and 1, 722 m. In order to investagate the effects of American ginseng cultivation for 4 years on changes of soil physiochemical properties and microbial community, a plot of land that has never been planted in any crop was chosen as the blank control plot (C). This barren land had been treated to prepare for a ginseng crop in accordance with the similar cultivation methods including edaphic treatment, agronomic management and fertilization regimes. Farmland that has been planted in American ginseng (CAG) for 4 years and is 50 m from the control soil was selected for the experimental samples (Fig. 1a). In this farmland, it was found that there were some sporadic lands (about 15%) in which American ginseng did not grow new seedlings on April 30, 2017, and the roots of these places dug out were found to root rot. Soil samples were collected from the four corners and the diagonal center of each plot (five-point sampling method) (C and CAG) (Fig. 1a) to analyze the physicochemical properties. Control soil samples (C) were collected by digging 10-20 cm into the fallow plot for a total of 8-10 g of soil, and soil from the 5 sampling points were random chosen to 3 samples (C1, C2, C3). Rhizosphere soil samples around 5 healthy ginseng (H) and 5 root rot-diseased ginseng (R) were collected by digging them up (Fig. 1c, d) and shaking off the soil into a plastic bag accordling to five-point sampling method. After identification, rhizosphere soil samples of 3 healthy (H1, H2, H3) and 3 diseased ginsengs (R1, R2, R3) were collected for experimental analysis. After excavating the healthy or diseased American ginseng roots, 8 to 10 g of soil were taken from where the ginseng had been growing for determination of the physicochemical properties of the soils in the CAG fields. Samples for microbial testing were placed on ice before transport to the laboratory of Plant Biotechnology Research Center of Shaanxi University of Technology, Shaanxi, China. One portion of each sample was air-dried and refrigerated before chemical and soil property analysis. The other portions were kept in aseptic valve bags, transported in an ice box, and immediately processed upon return to the laboratory.
Collection of the rhizosphere soil of American ginseng from two sampling sites. Five samples were collected according to the diagonal method (a) from a fallow field and a field growing American Ginseng for 4 years (b). Rhizosphere soil samples from healthy ginseng (H) and root-rot ginseng (R) were collected by digging up ginseng roots, identifying them as healthy (c) or root-rot diseased (d), and shaking the soil from the root
Soil physicochemical properties
Three soil parallel samples were collected from the C plot and the CAG plot to measure the physicochemical properties. The samples were homogenized by being passed through a 2-mm sieve. The soil pH was determined using a glass electrode pH meter (Mettler-Toledo FE20-Five Easy PlusTM, Schwerzenbach, Switzerland) in a 1: 2.5 soil:water (w/v) suspension (Qiu et al. 2012). The amount of organic matter was determined by quantifying the amount of oxidized soil carbon based on its reaction to acidic dichromate (Cr2O72−). Total nitrogen was measured based on direct combustion using an elemental analyzer (Vario EL III, Germany). The hydrolytic nitrogen in the soil was determined based on the transformation of hydrolyzed nitrogen into ammonia nitrogen through sodium hydroxide (Tan et al. 2017). Total phosphorus was measured by acid digestion (Grimshaw 1987). Total potassium was measured after digesting the soils in a mixture of concentrated nitric acid (HNO3) and perchloric acid (HClO4) using a soil-acid ratio of 1:10 (Tan et al. 2017). Available phosphorus (P) was determined using the sodium hydrogen carbonate solution-Mo-Sb anti-spectrophotometric method (Shen et al. 2011). Soil available potassium (K) was measured using a standard protocol according to McLean and Watson (1985).
Genomic DNA extraction, PCR amplification and High throughput amplicon sequencing
About 0.25 g of the chilled soil samples were used to extract total genomic DNA using PowerSoil™ DNA Isolation Kits (MoBio Laboratories, Solana Beach, CA, USA) according to the manufacturer’s protocol. Total DNA was used to analyze the changes in the microbial communities among the control soil (C), healthy ginseng rhizosphere soil (H), and root rot ginseng rhizosphere soil (R) samples. Genomic DNA concentration and purity were measured using a NanoDrop ND-2000 (NanoDrop Technologies, Wilmington, DE) spectrophotometer. The V3-V4 region of the 16S rRNA gene was amplified using the primer set 338F (5′- ACTCCTACGGGAGGCAGCA -3′) and 806R (5′- GGACTACHVGGGTWTCTAAT-3′) for bacterial community analysis. For fungal community analysis, the Internal Transcribed Spacer (ITS1) sequence, including the partial 18S rRNA gene, was amplified with primer set ITS 5F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS 1R (5′-GCTGCGTTCTTCATCGATGC-3′). Sample-specific 7-bp barcodes were incorporated into the primers for multiplex sequencing. The PCR amplification was carried out in a total volume of 25 μL containing 5 μL 5 × reaction buffer, 5 μL 5 × GC buffer, 2 μL dNTPs (2.5 mM), 1 μL forward primer (10 μM), 1 μL reverse primer (10 μM), 2 μL DNA template, 8.75 μL ddH2O and 0.25 μL Q5 High-Fidelity DNA Polymerase (NEB, USA). The PCR protocol consisted of an initial denaturation step at 98 °C for 2 min, 25⁓30 cycles of denaturation at 98 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, a final extension at 72 °C for 5 min, and a hold at 10 °C. Three replicates of the amplifications were pooled together to minimize the PCR bias. PCR products were detected by electrophoresis through 2.0% agarose gels and appropriately sized fragments (about 450 bp for bacteria and 350 bp for fungi) were purified with DNA Gel Extraction Kit (Axygen, Union City, CA, USA). PCR amplicons were quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Finally, paired-end 2×300 bp sequencing of fungal and bacterial amplicons were carried out on the Illumina MiSeq sequencer at Personal Biotechnology Co., Ltd (Shanghai, China). The detailed experimental procedure is shown in Fig. S1.
Sequence analysis
After removing the adaptors and primer sequences, the raw sequnences were assembled for each sample according to the unique barcode using the QIIME pipeline (Quantitative Insights Into Microbial Ecology, v 1.8.0, qiime.org) (Caporaso et al. 2010). The low-quality sequences were filtered through following criteria (Gill et al. 2006; Chen and Jiang 2014): sequences that had a length of <150 bp, sequences that had average Phred scores of <20, sequences that contained ambiguous bases, and sequences that contained mononucleotide repeats of >8 bp. Paired-end reads were assembled using FLASH (v1.2.7) (Magoč and Salzberg 2011). After chimera detection, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity by UCLUST (Edgar 2010 ). A representative sequence was selected from each OTU using default parameters. OTU taxonomic classification was conducted by BLAST searching the representative sequences set against the Greengenes Database (DeSantis et al. 2006) using the best hit (Altschul et al. 1997) and the fungal sequences were matched against the UNITE database (Release 5.0, https://unite.ut.ee/) (Kõljalg et al. 2013). An OTU table was further generated to record the abundance of each OTU in each sample and the taxonomy of these OTUs. OTUs containing less than 0.001% of total sequences across all samples were discarded. To minimize the difference of sequencing depth across samples, an averaged, rounded rarefied OTU table was generated by averaging 100 evenly resampled OTU subsets under the 90% of the minimum sequencing depth for further analysis.
Bioinformatics analysis
Sequence data analyses were mainly performed using QIIME (v1.8.0) and R packages (v3.2.0). OTU-level alpha diversity indices, such as Chao1 richness estimator, ACE metric (Abundance-based Coverage Estimator), Shannon diversity index, and Simpson index, were calculated using the OTU table in QIIME (v1.8.0). OTU-level ranked abundance curves were generated to compare the richness and evenness of OTUs among samples (Caporaso et al. 2010). To explore variation in fungal and bacterial community structures across the soil samples analyzed, weighted UniFrac distance was also performed in Mothur (v.1.25.1). PCoA (Principal Coordinate Analysis) was performed on distance matrices, and coordinates were used to draw 2D graphical outputs (Lozupone et al. 2007). The linear discriminant analysis (LDA) effect size (LEfSe) method was used to detect differentially abundant taxa across groups using the default parameters through the Galaxy online analytics platform (http://huttenhower.sph.harvard.edu/galaxy/) (Segata et al. 2011). An alpha value of 0.05 was used for the factorial Kruskal-Wallis test, and the threshold was set at 3.0 for the logarithmic LDA score for a discriminative feature. Hierarchical clusters were generated in Mothur (v.1.25.1) using cluster and heat map was drawn for each each sample in terms of abundance using R packages (v3.2.0). Co-occurrence analysis was performed by calculating Spearman’s rank correlations between dominant genera of the top 50 abundances.
Correlations with |RHO| > 0.6 and P < 0.01 were visualized as co-occurrence network using Cytoscape software (www.cytoscape.org) (Shannon et al. 2003).
Statistical analysis
Soil physicochemical characteristics were compared using Student's t-test (P < 0.05). The number of OTUs, alpha diversity indices, and the taxa (phyla and genus) bacterial and fungal relative abundances were calculated for all replicates and subjected to an analysis of variance by one-way ANOVA (P < 0.05). These analyses were performed in SPSS version 16.0 (SPSS Inc., Chicago, IL, USA).
Data accession numbers
All raw sequences were deposited in the NCBI Sequence Read Archive (SRA) database under the accession number SRP163101.
Results
The effect of cultivation of American ginseng on soil physiochemical properties
Soil characteristics for the two types of fields are summarized in Table 1. Compared with the uncultivated soil (Control), the soil from farmland used to cultivate American ginseng for 4 years (CAG) had a significantly (P < 0.05, Student's t-test) higher pH, electrical conductivity (EC), organic matter (OM) content, and available P. The available N and K values showed no significant (P < 0.05, Student's t-test) difference between the control and CAG soils.
Analysis of amplicon-data and microbia community diversity
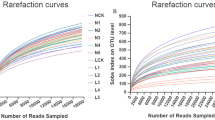
High quality sequences of 381, 992 reads for 16S rRNA genes and 303, 581 reads for ITS genes were obtained for an average of 42, 444 sequences of bacterial 16S rRNA genes (most common length ranged from 420⁓460 bp) and 33, 731 fungal ITS reads (most common length range 200⁓320 bp) (Table S1 and Fig. S2 ). A total of 25, 918 OTUs of bacterial phyla and 3, 907 OTUs of fungal phyla from the 9 samples were obtained, and 17, 405 OTUs of bacterial phyla and 3, 175 OTUs of fungal phyla were obtained after discarding OTUs less than 0.001% of total sequences across all samples (Table S1). Although there were nearly ten thousand sequences per soil sample representing the bacterial community, the slope of the rarefaction curve did not reach linearity at different similarity cutoff values, indicating that there were bacteria that were not detected (Fig. S3 a). Conversely, the slope of the rarefaction curve for the fungal species was flat at different similarity cutoff values, indicating that the identified fungal diversity was close to saturation and that an increase in the sequencing depth would not help to observe more fungal species (Fig. S3 b).
Cultivation of American ginseng significantly increased the number of bacterial OTUs in the soil, but decreased the number of fungal OTUs (Table 2). For the bacterial community, the Simpson and Shannon indices showed no significant changes (ANOVA, P < 0.05), but when the number of OTUs were calculated by Chao1 and ACE richness estimators, the numbers increased significantly (ANOVA, P < 0.05) in the soil culturing American ginseng compared to the control soil samples. This indicated that culture of American ginseng resulted in an increase in the richness of the bacterial community. The Simpson, Chao1, ACE and Shannon values for the fungal community all significantly decreased (ANOVA, P < 0.05) in the soil used for cultivating American ginseng for 4 years compared with the control soil sample, indicating that culture of American ginseng caused a decline in diversity and richness of the fungal community. Additionally, the diversity and richness indices showed no significant changes (ANOVA, P < 0.05) in the bacterial communities between the rhizosphere soils of healthy and diseased roots, whereas the richness indices (Chao1 and ACE) in the fungal community showed a significant decrease in rhizosphere soils of root rot diseased ginseng than that of healthy ginseng.
Microbial community similarities in different samples
The similarity of the microbial communities in the soil samples of C, H and R was determined by weighted-UniFrac principal coordinates analysis (PCoA; Fig. 2). As shown in Fig. 2, the weighted-UniFrac PCoA showed that both bacterial and fungal communities from the soil of C, H and R were clearly separated from each other (Fig. 2), and the difference was statistically significant (ANOSIM, R = 0.967, P = 0.006 for bacteria; R = 0.893, P = 0.001 for fungi). The principal coordinates PC1 and PC2 represented 65.69% and 21.33%, respectively, of the variance, and the contribution of the cumulative variance of the two principal coordinates (PC1 and PC2) accounted for 87.02%. As for the fungal communities, PC1 and PC2 represented 74.47% and 14.70%, respectively, of the variance, and the contribution of the cumulative variance of the two principal coordinates (PC1 and PC2) accounted for 89.17%. This was also demonstrated by hierarchical trees constructed through weighted pair-group method using arithmetic averages (WPGMA) calculations. This clustering showed that the bacterial and fungal communities in the rhizosphere soil of both healthy and diseased ginseng appeared more similar to each other than to those in the control, indicating that culture of American ginseng remarkably changes the microbial community in the soil (Fig. S4 ).
UniFrac-weighted principle coordinate analysis (PCoA) of bacterial and fungal community structures in the control soil (C) and the rhizosphere soils of healthy (H) and root rot-diseased (R) ginseng. Analysis of similarities (ANOSIM) was performed to evaluate the significant differences in microbial communities using QIIME (v1.8.0) (P < 0.05)
Microbial community composition and structure
The microbial populations in the different samples were analyzed by submitting the obtained 16S rRNA and ITS sequences to a local blast search against the Greengenes Database and UNITE databases (Fig. 3). The dominant bacterial phyla were Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, Gemmatimonadetes and Nitrospirae across the three sample groups (Fig. 3a). Proteobacteria was predominant in each of the samples, at 31.41, 36.39 and 36.44% of the total bacterial phyla detected in the C, H and R samples, respectively (Fig. 3a). The most populous bacterial genera were also the same across the 3 samples, namely Kaistobacter, Rhodoplanes, Sphingobium, Arthrobacter, Blastococcus, Candidatus solibacter, Nitrospira, Mycobacterium, Phenylobacterium and Bacillus (Fig. 3b). The proportions of Nitrospira (1.78%) and Kaistobacter (1.55%) were highest in the control sample (C). In the healthy ginseng soil samples (H), the proportions of Rhodoplanes (5.51%) and Kaistobacter (3.78%) were more abundant than other bacteria. And the proportions of Sphingobium (4.01%) and Rhodoplanes (1.46%) in the rhizosphere soil of rotten plants (R) were predominant.
The composition and structure of the bacterial and fungal community from the control soil sample (C) and the rhizosphere soil samples from healthy ginseng (H) and root rot-diseased ginseng (R). Bacterial community at the phylum level (a) or the genus level (b); Fungal community at the phylum level (c) or the genus level (d). The phyla with relative abundances in the top 20 were chosen to exhibit. Others represents phyla of low relative abundance that ranks lower than 20. *, # and ※ represent significant differences (P < 0.05) according to one-way ANOVA (n=3). *, # and ※ stands for C:H, C:R and H:R, respectively
For the fungal community, we detected three main phyla in the samples: Ascomycota, Basidiomycota and Zygomycota (Fig. 3c). Ascomycota was the most dominant group in all three soil samples at the phylum level, representing 68.06%, 40.77% and 55.90% in the C, H and R groups, respectively (Fig. 3c). The limited resolution of ITS sequencing meant that there was a notable fraction of the ITS sequences in each sample that could not be exactly classified, especially in the rhizosphere samples, with 46.07 (H) and 33.25% (R) of the sequences undefined in these two samples, respectively (Fig. 3c). Similarly, undefined sequences accounted for a very high proportion in the soil samples at the genus level (Fig. 3d). The most abundant genera detected in the control soil were Podospora, Gibberella and Schizothecium, at 2.52% of the total sequences, while other genera, such as Devriesia (2.27%), Monographella (1.91%) and Bullera (1.88%) were also detected. The main genera detected in the rhizosphere soil of healthy ginseng were Cadophora (5.32%) and Monographella (2.90%) except for unidentified genera. In the root rot ginseng sample, Monographella (23.13%) and Mortierella (6.00%) were the main genera detected.
In order to the further analysis a significant difference of relative abundances, we performed a significance analysis for the relative abundance of the microflora using one-way ANOVA, and the results had been marked in the Fig. 3 with different symbols. In addition, a heat map was also used to intuitively show the differences of the relative abundances of bacterial and fungal genera observed, and to highlight the particularly high or low genus in each sample using a colored frame (Fig. S5 ). The relative abundances with the top 50 of the highest contribution to the ordination were selected.
The LEfSe analysis of the differentially abundant feature from bacteria and fungal communities
LEfSe uses LDA scores to estimate the effect size of each differentially abundant feature, and to rank the relative difference of microbial taxa that are discriminative with biological consistency and statistical significance. The LEfSe analysis of the bacterial communities from C, H and R soil samples shows that there are 47 differentially abundant taxonomic clades with a LDA score higher than 3.0 (Fig. 4). Out of the 47 different bacterial genera, 21 genera were differentially abundant in the control soil sample (C) (LDA score > 3.0), including Nitrospira, Janthinobacterium, Arthrobacter, Candidatus_Solibacter, Pedomicrobium, Flavobacterium and so on. The most differentially abundant bacterial taxa in the H soil sample belong to genera: Rhodoplanes and Kastobacter (LDA score > 4.0). The differential genera overrepresented in the rhizosphere soils of R samples include Sphingobium, Pedobacter, Pseudomonas and Sphingopyxis (LDA score > 4.0).
The LEfSe analysis of the fungal communities from C, H and R samples shows that there are 28 differentially abundant taxonomic clades with a LDA score higher than 3.0 (Fig. 4). Out of the 28 different fungal genera, 16 genera were differentially abundant in the control soil samples (C) (LDA score > 3.0), including Mrakia, Gibberella, Lasiosphaeriaceae, Devriesia, Racocetra, Acicuseptoria, Pyrenochaetopsis, Myrothecium and so on. The genera overrepresented in the rhizosphere soils of H samples include Melanophyllum and Staphylotrichum (LDA score > 3.5). The most differentially abundant fungal taxa in the rhizosphere soils of R sample belong to genera: Mortierella (LDA score > 4.0).
In order to further exhibit the possible "collaborative" or "competing" relationships among different communities, Spearman’s rank correlation coefficients between the most abundant genera were calculated using the Mothur software. The correlations among the 49 dominant bacterial genera and the 22 fungal genera were analyzed (Fig. S6 ).
Discussion
Soil pH was a major factor in defining microbial diversity through impacting pH homeostasis of microbial cell or regulating availability of soil nutrients (Fierer and Jackson 2006; Rousk et al. 2010). In this study, cultivation of American ginseng for 4 years significantly improved (P < 0.05; Student's t-test) the soil pH compared with the control (Table 1). This is also in line with a report of Tan et al. (2017), and their results showed that the soil pH significantly increased after 3 years of cultivation of Panax notoginseng (Tan et al. 2017). Zhalnina et al. (2015) reported that pH is the main driver of microbial community in the park grass soil and pH changes in soils are the result of nutrient management, and these pH changes affect nutrient availability to both plant and microbial communities. Agricultural soils are commonly fertilized with nitrogen, and this could be another possible factor regulating the diversity of microorganisms (Zhalnina et al. 2015). However, there are no significant difference (P < 0.05; Student's t-test) between the control and CAG sample soil. Additionally, the CAG soil had a significantly (P < 0.05; Student's t-test) higher EC, OM and available P content compared to the control soil. Whether these changes (including EC, OM and available P) can affect the soil microbiota community still need to further study.
Soil microorganisms have profound effects on the growth, nutrition and health of plants in natural and agricultural ecosystems (Garbeva et al. 2004; Almario et al. 2013). Characterizing the diversity and richness of the microbial communities in the rhizosphere soils of healthy ginseng and root rot ginseng is an important first step toward understanding whether these communities influence the occurrence of root rot diseases. In this study, soil growing American ginseng for 4 years harbored a significantly (ANOVA, P < 0.05) higher bacterial richness and lower fungal richness and diversity than the control soil (Table 2). In contrast, Dong et al. (2017) showed that less diverse bacterial communities and more diverse fungal communities were found in the rhizosphere soil of American ginseng compared to the rhizosphere soil of maize. Compared with the rhizosphere soil of a healthy ginseng root (H), the soil of diseased ginseng (R) showed a significant decrease (ANOVA, P < 0.05) in the fungal richness (Table 2). This result is in line with a study in vanilla showing that soil conducive to Fusarium wilt disease harbored a significantly lower fungal abundance and diversity than the suppressive soil (Xiong et al. 2017). Tan et al. (2017) also reported that fungal diversity in the root-rot diseased notoginseng rhizospheric soil decreased, in contrast, fungal diversity in healthy notoginseng rhizospheric soil increased with increasing years of continuous cropping years. On the other hand, soil microbial communities are also influenced by multiple factors such as plant type, climate, soil properties and agricultural practice (Burton et al. 2010; Hao et al. 2009).
Changes in the composition of the bacterial community may lead to variations in metabolic capacity, biodegradation, and disease-suppression abilities (Garbeva et al. 2004; Bell et al. 2013). Analysis of the bacterial proportions showed that Proteobacteria and Actinobacteria were the most high at phyla level in the three group samples (Fig. 3a). Actinobacteria, producing a large and diverse array of bioactive compounds that inhibit the development of pathogens in the soil, are some of the microorganisms with a role of biological control that have been extensively studied (Sharma et al. 2005). And at the genus level, Nitrospira (1.78%), Rhodoplanes (5.51%) and Sphingobium (4.01%) was predominant in the C, H and R sample, respectively (Fig. 3b). As for fungi, Ascomycota, Basidiomycota and Zygomycota were the most abundant fungal phyla identified in the different samples (Fig. 3c). Li et al. (2014) reported that Ascomycota and Basidiomycota were the top two prevalent fungal phyla in a continuous cropping peanut system. Tan et al. (2017) also reported that Ascomycota, Basidiomycota and Zygomycota were dominant phyla during the continuous cropping of P. notoginseng. This result is also in agreement with a previous study in which Ascomycota and Basidiomycota were the most abundant fungal phyla identified in soil conducive to Fusarium wilt disease (Xiong et al. 2017). Ascomycetes occur in terrestrial, marine, and freshwater habitats, and many species play a major ecological role as decomposers (Miadlikowska et al. 2006). Notably, Monographella was significantly higher in the rhizosphere soil of diseased ginseng than healthy ginseng (Fig. 3d). It is reported that Monographella is a fungal plant pathogen and contains many plant pathogenic species (Aveskamp et al. 2010).
The composition of the bacterial community analysis showed that Nitrospira, Janthinobacterium and Pedomicrobium were significantly (ANOVA, P < 0.05) decreased after culture of American ginseng (Fig. 4). Nitrospira and Pedomicrobium are ubiquitous bacteria that function in the nitrogen cycle and take advantage of the nutrients, respectively. Nitrospira has been shown to perform nitrite oxidation in the second step of nitrification (Koch et al. 2015). Larsen et al. (1999) showed that Pedomicrobium is able to greatly enhance the rate of Mn oxidation. It is reported that Janthinobacterium, a genus of gram-negative soil bacteria, can produce a purple-violet pigment, manifest diverse energy metabolism abilities, and tolerate cold, ultraviolet radiation, and other environmental stressors (Koo et al. 2016). With respect to fungi, Paradendryphiella, Psilocybe, Sporopachydermia, Myrothecium, Racocetra, Aureobasidium and Coprinellus were remarkable decreased after culture of American ginseng. Psilocybe and Coprinellus are saprotrophic species, deriving nutrients from dead and decomposing organic matter, and grow in and around stumps or logs of broad-leaved trees or attached to buried wood (Borovička et al. 2014; Redhead et al. 2001). Other newly discovered genera such as Sporopachydermia, Paradendryphiella, Myrothecium, Aureobasidium and Racocetra were also significantly (ANOVA, P < 0.05) decreased after culture of American ginseng, but their potential functions are not yet clear.
The LEfSe analyses revealed that there were indeed differences in the relative abundances of both bacterial and fungal genera between the soils accompanying healthy and root rot-diseased ginseng roots. Rhodoplanes, Kaistobacter, Dokdonella and Actinoplanes bacteria were more differently abundant in the rhizosphere soils of healthy ginseng, whereas the Pedobacter, Pseudomonas, Sphingopyxis and Dactylosporangium bacteria were more differently abundant in the rhizosphere soils of root rot ginseng (Fig. 4). Rhodoplanes and Kaistobacter were identified as indigenous degraders for atrazine and useful for atrazine bioremediation (Lodha et al. 2015; Wei et al. 2018). Dokdonella is related to pyridine biodegradation and could be a potential alternative for the enhancement of pyridine removal from waste water in anaerobic systems (Liu et al. 2013). Actinoplanes, a genus in the family Micromonosporaceae, is able to produce the pharmaceutically important compounds valienamine (a precursor to the anti-diabetic drug acarbose and the antibiotic validamycin), teicoplanin and ramoplanin (Laube 2002). In spite of these bacteria might exhibit the feature of degrading harmful compounds in the soil, such as atrazine and pyridine (Liu et al. 2013; Lodha et al. 2015; Wei et al. 2018), whether the species associated with root-rot disease of American ginseng still needs further investigations. Surprisingly, the Pseudomonas bacteria is rich in the rhizosphere soils of diseased ginseng roots. Because many Pseudomonas produce the antifungal compounds 2,4-diacetylphloroglucinol (Phl) and/or hydrogen cyanide (HCN) and were antagonistic to T. basicola in vitro and to protect tobacco from black root rot (Stutz et al. 1986; Ramette et al. 2003).
Mortierella genus was the most dominant fungal genus in the rhizosphere soils of root rot-diseased American ginseng (Fig. 4). A culture-dependent study showed that Phoma was the main fungal genera observed as obstacles in continuous cropping (Miao et al. 2006). Some studies have shown that some species of Mortierella can produce antibiotics, and several isolates have been investigated as potential antagonistic agents against various plant pathogens (Tagawa et al. 2010; Wills and Lambe 1980). Additionally, Melanophyllum, a genus of fungi in the family Agaricaceae, is a hyper-dominant in the rhizosphere soils of healthy ginseng. Although Fusarium spp., Phytophthora spp. and Cylindrocarpon spp. were often reported to cause severe root rot and could be frequently isolated from cultivated American ginseng root (Reeleder and Brammall 1994; Punja 1997; Punja et al. 2007; Bi et al. 2011), these fungi were not detected from the rhizosphere soils of the diseased ginseng in this study. The research results of Agler et al. (2016) dementated that not all pathogens share the hub microbe or even keystone status, so pathogenicity cannot be taken as a rule to detect “hub” or “keystone” species. The microbia diversity manipulation might be a key battleground where hosts and various hubs cooperate or compete with one another (Agler et al. 2016). Phoma is a genus of common coelomycetous soil fungi and contains many plant pathogenic species (Aveskamp et al. 2010). For example, Phoma beta is the cause of the heart rot and blight of beets and Phoma batata can cause a dry rot of sweet potato. Conversely, the fungi that secrete antibiotics, such as Trichoderma and Penicillium, were relatively higher in the rhizosphere soil of healthy American ginseng compared with that of diseased American ginseng.
In addition, microbe-microbe interactions generally increase host effects due to the community correlation network topology (Agler et al. 2016). In this study, a map of the microbial networks was also provided to exhibit a possible "collaborative" or "competing" correction within the microbial communities in the rhizosphere soils of American ginseng (Fig. S6 ). For example, the bacterial genera Pedomicrobium, Janthinobacterium, Nitrospira, Lysobacter, Candidatus. Xiphinematobacter and Flavobacterium might kept a "collaborative" relationship with each other in bacterial community. These relationships between bacteria and fungi might provide some clues for the further screening of bacteria antagonistic to root rot disease in American ginseng.
Conclusions
In this study, our results indicated that the microbial community differs between the soil attached to healthy and rotted roots of American ginseng, and some hyper-dominant bacteria and fungi were also founded in the rhizosphere soils of health and rotten root. However, soil microbial communities are influenced by multiple factors such as plant type, climate, soil properties and agricultural practice (Burton et al. 2010; Hao et al. 2009). In addition, complex interactions take place between microorganisms and roots, and the effective plant-protecting populations may be influenced by accompanying microbiota (Duijff et al. 1999; Raaijmakers et al. 2008). Therefore, further studies identifying the hyper-dominant taxon isolates with effective rotten root disease suppression ability and revealing their interactions may open new avenues for the development of informed bio-control strategies.
Abbreviations
- ACE :
-
Abundance based coverage estimator
- ANOSIM:
-
Analysis of similarities
- ANOVA :
-
Analysis of variance
- CAG :
-
Cultivate American ginseng
- EC :
-
Electrical conductivity
- HCN:
-
Hydrogen cyanide
- ITS :
-
Internal transcribed spacer
- LDA :
-
Linear discriminant analysis
- LEfSe :
-
Linear discriminant analysis effect size
- OM :
-
Organic material
- OTUs :
-
Operational taxonomic units
- PCoA :
-
Principal co-ordinates analysis
- PCR :
-
Polymerase chain reaction
- Phl :
-
2,4-diacetylphloroglucinol
- QIIME :
-
Quantitative insights into microbial ecology
- SRA :
-
Sequence Read Archive
- SPSS :
-
Statistical product and service solutions
- UNITE :
-
unite.ut.ee
- WPGMA :
-
Weighted pair group method with arithmetic averages
References
Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM (2016) Microbial hub taxa link host and abiotic factors to plant microbiome variation. Plos Biol 14:e1002352
Ahanger RA, Bhat HA, Ganie SA, Shah AA (2014) Division of plant pathology, shalimar, kashmir integrated disease management in organic agriculture. Glob J Agr Food Sci Res 1:28–42
Almario J, Moenne-Loccoz Y, Muller D (2013) Monitoring of the relation between 2,4-diacetylphloroglucinol-producing Pseudomonas and Thielaviopsis basicola populations by real-time PCR in tobacco black root-rot suppressive and conducive soils. Soil Biol Biochem 57:144–155
Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aveskamp MM, De GJ, Woudenberg JH, Verkley GJ, Crous PW (2010) Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related Pleosporalean genera. Stud Mycol 65:1–60
Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Gree CW (2013) Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7:1200–1210
Bi W, Chen J, Jiao XL, Gao WW (2011) Identification of the pathogens causing the root rot and their pathogenicity on American ginseng in Beijing. Plant Prot 37:135–138
Borovička J, Oborník M, Stříbrný J, Noordeloos ME, Sánchez LP, Gryndlger M (2014) Phylogenetic and chemical studies in the potential psychotropic species complex of Psilocybe atrobrunnea with taxonomic and nomenclatural notes. Persoonia 34:1–9
Burton J, Chen C, Xu Z, Ghadiri H (2010) Soil microbial biomass, activity and community composition in adjacent native and plantation forests of subtropical Australia. J Soils Sediments 10:1267–1277
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chen H, Jiang W (2014) Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol 5:6
Chen M, Li X, Yang Q, Chi X, Pan L, Chen N, Yang Z, Wang T, Wang M, Yu S (2012) Soil eukaryotic microorganism succession as affected by continuous cropping of peanut-pathogenic and beneficial fungi were selected. PLoS One 7:e40659
Darmono TW, Owen ML, Parke JL (1991) Isolation and pathogenicity of Phytophthora cactorum from forest and ginseng garden soils in Wisconsin. Plant Dis 75:610–612
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Dong LL, Xu J, Zhang LJ, Yang J, Liao BS, Li XW, Chen SL (2017) High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chin Med 12:18
Duijff BJ, Recorbet G, Bakker PA, Loper JE, Lemanceau P (1999) Microbial antagonism at the root level is involved in the suppression of Fusarium wilt by the combination of nonpathogenic Fusarium oxysporum Fo47 and Pseudomonas putida WCS358. Phytopathology 89:1073–1079
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631
Garbeva P, Van Veen JA, Elsas JDV (2004) Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42:243–270
Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359
Grimshaw HM (1987) The determination of total phosphorus in soils by acid digestion. In: Rowland, A.P. (ed.), Chem. Environ Res:92–95
Hao ZP, Christie P, Zheng F, Li JL, Chen Q, Wang JG, Li XL (2009) Excessive nitrogen inputs in intensive greenhouse cultivation may influence soil microbial biomass and community composition. Commun Soil Sci Plant Anal 40:2323–2337
Hartung AC, Stephens CT (1983) Effects of allelopathic substances produced by asparagus on incidence and severity of asparagus decline due to Fusarium, crown rot. J Chem Ecol 9:1163–1174
Huang LF, Song LX, Xia XJ, Mao WH, Shi K, Zhou YH, Yu JQ (2013) Plant-Soil feed-backs and soil sickness: from mechanisms to application in agriculture. J Chem Ecol 39:232–242
Jiao XL, Lu XH, Chen AJ, Luo Y, Hao JJ, Gao WW (2015) Effects of Fusarium solani and F. oxysporum infection on the metabolism of Ginsenosides in American Ginseng roots. Molecules 20:10535–10552
Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, Nielsen P, Wagner M, Daims H (2015) Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus. Proc Natl Acad Sci USA 112:11371–11376
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Grif fith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277
Koo H, Strope BM, Kim EH, Shabani AM, Kumar R, Crowley MR, Andersen DT, Bej AK (2016) Draft genome sequence of Janthinobacterium sp. Ant5-2-1, isolated from proglacial lake podprudnoye in the schirmacher oasis of east Antarctica. Genome Announc 4:e01600–e01615
Larsen EI, Sly LI, McEwan AG (1999) Manganese (II) adsorption and oxidation by whole cells and a membrane fraction of Pedomicrobium sp. ACM 3067. Arch Microbiol 171:257–264
Laube H (2002) Acarbose: an update of its therapeutic use in diabetes treatment. Clin Drug Investig 22:141–156
Li X, Ding C, Zhang T, Wang X (2014) Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol Biochem 72:11–18
Liu Y, Jin JH, Liu HC, Liu ZP (2013) Dokdonella immobilis sp. nov. isolated from a batch reactor for the treatment of triphenylmethane dye effluent. Int J Syst Evol Microbiol 63:1557–1561
Lodha TD, Srinivas A, Sasikala C, Ramana CV (2015) Hopanoid inventory of Rhodoplanes spp. Arch Microbiol 197:861–867
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585
Magoč T, Salzberg SL (2011) Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
McLean EO, Watson ME (1985) Soil measurements of plant available potassium. Potassium Agric:277–308
Miadlikowska J, Kauff F, Hofstetter V, Fraker E, Grube M, Hafellner J, Reeb V, Hodkinson BP, Kukwa M, Lücking R, Hestmark G, Otalora MG, Rauhut A, Büdel B, Scheidegger C, Timdal E, Stenroos S, Brodo I, Perlmutter GB, Ertz D, Diederich P, Lendemer JC, May P, Schoch CL, Arnold AE, Gueidan C, Tripp E, Yahr R, Robertson C, Lutzoni F (2006) New insights into classification and evolution of the Lecanoromycetes (Pezizomycotina, Ascomycota) from phylogenetic analyses of three ribosomal RNA and two protein-coding genes. Mycologia 98:1088–1103
Miao ZQ, Li SD, Liu XZ, Chen YJ, Li YH, Wang Y, Guo RJ, Xia ZY, Zhang KQ (2006) The causal microorganisms of Panax notoginseng root rot disease. Sci Agric Sin 39:1371–1378
Nigh ELJ (1990) Stress factors influencing Fusarium infection in asparagus. Acta Hortic (271):315–322
Ogweno JQ, Yu JQ (2006) Autotoxic potential in soil sickness: a re-examination. Allelopathy J 18:93–101
Peirce LC, Colby LW (1987) Interaction of asparagus root filtrate with fusarium oxysporum f. sp. asparagi. J Am Soc Hortic Sci 112:35–40
Punja ZK (1997) Fungal pathogens of American ginseng (panax quinquefolium) in british columbia. Can J Plant Pathol 19:301–306
Punja ZK (2011) American ginseng: research developments, opportunities, and challenges. J Gins Res 35:368–374
Punja ZK, Wan A, Goswami RS, Verma N, Rahman M, Barasubiye T, Seifert KA, Lévesque CA (2007) Diversity of Fusarium species associated with discolored ginseng roots in British Columbia. Can J Plant Pathol 29:340–353
Qiu MH, Zhang RF, Xue C, Zhang SS, Li SQ, Zhang N, Shen QR (2012) Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol Fert Soils 48:807–816
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2008) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361
Rahman M, Punja ZK (2005) Biochemistry of ginseng root tissues affected by rusty root symptoms. Plant Physiol Bioch 43:1103–1114
Ramette A, Moënneloccoz Y, Défago G (2003) Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. Fems Microbiol Ecol 44:35–43
Redhead SA, Vilgalys R, Moncalvo JM, Johnson J, Hopple JS (2001) Coprinus pers. and the disposition of coprinus species sensu lato. Taxon 50:203–241
Reeleder RD (2007) Rhexocercosporidium panacis sp. nov. a new anamorphic species causing rusted root of ginseng (panax quinquefolius). Mycologia 99:91–98
Reeleder RD, Brammall RA (1994) Pathogenicity of Pythium species, Cylindrocarpon destructans and Rhizoctonia solani to ginseng seedling in Ontario. Can J Plant Pathol 14:311–316
Rousk J, Baath E, Brooks P, Lauber CL, Lozupone C, Caporaso GJ, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in arable soil. ISME J 4:1340–1351
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res 13:2498–2504
Sharma S, Aneja MK, Mayer J, Much JC, Schloter M (2005) Characterization of bacterial community structure in rhizosphere soil of grain legumes. Microb Ecol 49:407–415
Shen ZQ, Zhang Q, Liu LJ, Qiu Y (2011) Determination of available phosphorus in soil by sodium bicarbonate extraction Mo-Sb anti-spectrophotometry method. Environ Monit Forew 3:12–15
Singh HP, Batish DR, Kohli RK (1999) Autotoxicity: Concept, Organisms, and Ecological Significance. Crit Rev Plant Sci 18(6):757–772
Stutz EW, Défago G, Kern H (1986) Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76:181–185
Tagawa M, Tamaki H, Manome A, Koyama O, Kamagata Y (2010) Isolation and characterization of antagonistic fungi against potato scab pathogens from potato field soils. FEMS Microbiol Lett 305:136–142
Tan Y, Cui YS, Li HY, Kuang AX, Li XR, Wei YL (2017) Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol Res 194:10–19
Wei Y, Wu Y, Yan Y, Zou W, Xue J, Ma W, Wang W, Tian G, Wang L (2018) High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS One 13:e0193097
Wills WH, Lambe RC (1980) Mortierella antagonism to oomycetes. Phytopathology 70:694–694
Wu ZX, Hao ZP, Sun YQ, Guo LP, Huang LQ, Zeng Y, Wang Y, Yang L, Chen BD (2016) Comparison on the structure and function of the rhizosphere microbial community between healthy and root-rot Panax notoginseng. Appl Soil Ecol 107:99–107
Xiong W, Li R, Ren Y, Liu C, Zhao QY, Wu HS, Jousset A, Shen QR (2017) Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol Biochem 107:198–207
Yu JQ, Shou SY, Qian YR, Zhu ZJ, Hu WH (2000) Autotoxic potential of cucurbit crops. Plant Soil 223(1/2):147–151
Zhalnina K, Dias R, Quadros PDD, Davis-Richardson A, Camargo FAO, Clark IM, McGrath SP, Hirsch PR, Triplett E (2015) Soil pH determines microbial diversity and composition in the park grass experiment. Microb Ecol 69:1–12
Acknowledgements
We thank Anita K. Snyder, M.Sc. for the helpful suggestions and language polishing on this manuscript. This research was supported by the National Natural Science Foundation of China (31660153), Shaanxi Province Key Research and Development Project (2018NY-042), China, Shaanxi Key Projects of International Cooperation in Science and Technology Innovation (2015KTTSSF01-02), China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Responsible Editor: Stéphane Compant.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 30 kb)
Fig. S1
The flow chart of the experimental procedure (PNG 412 kb)
Fig. S2
Length distribution of bacterial and fungal sequences. (PNG 123 kb)
Fig. S3
Rarefaction curves measuring the observed species in bacterial and fungal communities. (PNG 583 kb)
Fig. S4
Weighted Pair-Group Method with Arithmetic Averages (WPGMA) dendrogram constructed from ThetaYC distances of bacterial and fungal communities in each sample. (PNG 133 kb)
Fig. S5
Hierarchical clustering of the bacterial and fungal communities based on the top 50 genera in the different soil samples. (PNG 825 kb)
Fig. S6
Spearman’s correlation network analysis of dominant bacterial (top) and fungal (bottom) species interactions. Nodes represent dominant genera. A line between two nodes indicates that there is a correlation between the two genera. A negative correlation is indicated by a green line, while a positive correlation is indicated by orange. The more connections to a node, the more associations this genus has with other members of the flora. (PNG 19899 kb)
Rights and permissions
About this article
Cite this article
Jiang, J., Yu, M., Hou, R. et al. Changes in the soil microbial community are associated with the occurrence of Panax quinquefolius L. root rot diseases. Plant Soil 438, 143–156 (2019). https://doi.org/10.1007/s11104-018-03928-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-018-03928-4