Abstract
Background and aims
Alyssum section Odontarrhena is the largest single clade of Ni-hyperaccumulator plants, most of which are endemic to ultramafic (serpentine) soils. Alyssum serpyllifolium is a facultative hyperaccumulator able to grow both on limestone-derived and ultramafic soils. Analysis of different populations of this species with contrasting phenotypes could allow the identification of genes involved in Ni-hyperaccumulation and serpentine tolerance.
Methods
A glasshouse pot experiment on compost-amended ultramafic soil was carried out with three ultramafic (U) and two non-ultramafic (NU) populations of A. serpyllifolium. The leaf ionome was determined by elemental analysis and used as a proxy for serpentine adaptation. A Ni-hyperaccumulating phenotype was estimated from leaf Ni concentrations. Cultured plants were genotyped using Amplified Fragment Length Polymorphism (AFLP) markers. Outlier analysis and regressions of leaf ionome over band distribution were applied to detect markers potentially involved in Ni-hyperaccumulation and serpentine tolerance.
Results
As well as U populations, some plants from NU populations were found to be able to hyperaccumulate Ni in leaves to concentrations exceeding 0.1% (w/w). U populations had a higher Ca/Mg leaf ratio than NU populations, mainly due to Mg exclusion. 374 AFLP markers were amplified and a potential adaptive value was identified in 34 of those markers.
Conclusions
Phenotype regression analyses were found to be more powerful than outlier analyses and indicated that regulation of foliar concentrations of Ni, Ca, Mg and P are the main factors involved in serpentine adaptation. More research is needed in order to resolve the ancestral or recently -evolved nature of Ni-hyperaccumulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trace metal(loid)s are known to be toxic to plants. However, some plant species have developed tolerance to these elements and are able to grow on metalliferous (i.e. metal-rich) soils. Among these plants, some species, known as hyperaccumulators, have evolved effective mechanisms of uptake of particular metals from the soil and their translocation to aerial parts, thereby accumulating very high concentrations of metals in their shoots, typically one to two orders of magnitude higher than in normal (i.e. non-accumulator) plants (Baker 1981; Van der Ent et al. 2013). More than 500 species of vascular plants have been described as hyperaccumulators, distributed across phylogenetically distant families and growing over metalliferous and sometimes non-metalliferous (e.g. zinc hyperaccumulation) substrates in different areas of the world (Krämer 2010; Van der Ent et al. 2013). Approximately 75% of these are Ni hyperaccumulators (Van der Ent et al. 2013). The Brassicaceae contains 25% of reported Ni hyperaccumulators so far (Krämer 2010), although new techniques (i.e. X-ray fluorescence applied to large-scale screening of herbarium specimens) show that the hyperaccumulation phenomenon has been strongly underestimated, especially in tropical regions, thus decreasing the proportion of Brassicaceae in the total number of hyperaccumulators globally (Guillaume Echevarria, personal communication).
By far the largest single clade of Ni hyperaccumulators in Brassicaceae is found in the section Odontarrhena of the genus Alyssum, which contains 48 Ni-hyperaccumulating taxa whose distribution is strongly correlated with the occurrence of ultramafic (= serpentine) outcrops around the Mediterranean basin (Brooks 1987). Apart from high Ni contents (often available in temperate conditions), ultramafic soils are characterized by high contents of other trace metals (Co, Cr), a low Ca/Mg ratio, a low bioavailability of other essential plant nutrients (K, P, Fe), and physical properties that make them prone to erosion and drought (Kruckeberg 2002; Brady et al. 2005; Mengoni et al. 2012). As a result, the plants inhabiting these sites must often tolerate drought as well as chemical imbalances (Brady et al. 2005; Kazakou et al. 2008). Indeed, the richness of hyperaccumulators in section Odontarrhena has been explained as a result of the occurrence of several preconditioning traits in this group (high Ca uptake efficiency, tolerance to ultrabasic pH, dry environments, and, especially, high soil Mg) that first allowed the colonization of ultramafic areas in the Mediterranean basin, and then the evolution of serpentinophile (and finally hyperaccumulator) plants (Mengoni et al. 2012).
Not all the Ni-hyperaccumulators are strict serpentine endemics. Some species grow on different kind of substrates and hyperaccumulate Ni when growing on ultramafic soils. These facultative hyperaccumulators constitute from 10 to 25% of known hyperaccumulators in areas such as Cuba, Turkey, or Brazil (Pollard et al. 2014).
Knowledge of the molecular mechanisms and genetics of metal hyperaccumulation by plants has advanced considerably in the last decade (Verbruggen et al. 2009; Krämer 2010; Centofanti et al. 2013; Merlot et al. 2014; Deng et al. 2016), but the phenomenon is still incompletely understood. One cause may be that hyperaccumulation is a complex multigenic trait, involving many component processes. Another reason may be that some model hyperaccumulators usually lack intraspecific variation in this trait (Verbruggen et al. 2009). The intraspecific variation in the hyperaccumulation phenotype found in facultative hyperaccumulators make them potentially very valuable model organisms for studying the genetic basis and physiology of metal (Ni) hyperaccumulation and serpentine adaptation. In addition, interest in metal hyperaccumulators has increased recently due to their potential application in soil phytoremediation (phytoextraction) techniques (Kidd et al. 2015) and the development of phytomining/agromining using hyperaccumulators as ‘metal crops’ (Van der Ent et al. 2013; Nkrumah et al. 2016).
The complexity of these adaptations, involving many genes in different plant organs, has been recently assessed using Next Generation Sequencing (NGS) approaches, which have been applied to studying serpentine adaptation in the (non-hyperaccumulator) Arabidopsis lyrata (Turner et al. 2010), while transcriptomic analyses of the Zn/Cd hyperaccumulator Sedum alfredii (Gao et al. 2013) and Zn/Cd/Ni Noccaea caerulescens (Halimaa et al. 2014) have been recently reported. Moreover, transcriptomic analyses have identified a gene potentially involved in Ni tolerance and hyperaccumulation by comparing hyperaccumulating and non-hyperaccumulating species of Psychotria (Rubiaceae) (Merlot et al. 2014).
Although less powerful than NGS technologies, the Amplified Fragment Length Polymorphism (AFLP) technique (Vos et al. 1995) allows generation of hundreds of molecular markers from the genomes of non-model organisms. AFLPs are generated from anonymous regions of the genome, so different approaches have been developed to identify AFLP markers potentially linked to adaptive loci. One approach is a ‘population genomics framework’, in which AFLPs (applied to several populations and many individuals) are used to infer a baseline neutral genetic variation across the genome and identify deviant loci potentially involved in adaptation to different factors. Another ‘population free’ approach directly correlates band presence/absence with an environmental variable or a phenotypic trait using different regression models (references in Bonin et al. 2007; Holderegger et al. 2008). AFLPs have been applied to the study of plant adaptation to different factors (see Strasburg et al. 2012 for a review). More specifically, they have been applied to the study of metal tolerance in the facultative hyperaccumulator Arabidopsis halleri (Meyer et al. 2009) and in the pseudometallophyte Cistus ladanifer (Quintela-Sabarís et al. 2012).
Alyssum serpyllifolium Desf. is a Ni-hyperaccumulator species from section Odontarrhena of the genus distributed in the western Mediterranean region from southern France across the Iberian Peninsula to Morocco. It is able to grow both on limestone-derived and ultramafic soils. According to current taxonomic treatments, the Ni-tolerant and -hyperaccumulating ultramafic populations are treated as separate subspecies, namely ssp. lusitanicum T.R.Dudley & P. Silva (NW Spain and NE Portugal) and ssp. malacitanum Rivas Goday (S Spain), whereas limestone populations are referred to the type subspecies serpyllifolium (Ball and Dudley 1993; Küpfer and Nieto-Feliner 1993). Recent molecular studies confirm the conspecific nature of the different subspecies (Cecchi et al. 2013; Sobczyk et al. 2017).
In addition to its broad ecological range, A. serpyllifolium has been found to show a significant degree of intraspecific variation in its responses to Ni (Pollard et al. 2014 and references therein), even among ultramafic populations and subspecies (Cabello-Conejo 2015).
Such intraspecific variability could make this species a valuable model to study genes involved in Ni-hyperaccumulation and serpentine adaptation in section Odontharrena. We therefore sought to identify molecular markers linked to Ni hyperaccumulation and serpentine adaptation of A. serpyllifolium using an exploratory genome scan with AFLP markers. Plants from ultramafic and non-ultramafic origin were grown in pots with compost-amended ultramafic soil under glasshouse conditions. By growing the different population types using a common-garden approach, we hope to eliminate the putative effect of the soil of origin of each population (ultramafic or limestone-derived) on the population phenotype. Moreover, we have favoured the use of ultramafic soil to ensure a medium with a sufficiently high exchangeable Ni pool and capacity for Ni re-supply to the soil solution in order to allow the expression of the hyperaccumulator phenotype. The leaf ionome was investigated by elemental analysis and the resulting data were used as a proxy for Ni hyperaccumulation and for serpentine adaptation (which we define as the ability to maintain a balanced ionome under nutrient limiting conditions -especially with unfavourable Ca/Mg ratio- and in the presence of elevated concentrations of Ni). A genome scan was then applied using AFLP markers to search for markers potentially linked to Ni hyperaccumulation and serpentine adaptation using either a ‘population genomics’ approach (outlier detection by population-pairwise comparisons) or a ‘population free’ approach (regression of leaf ionome data on marker distribution).
Material and Methods
Seed sampling and germination
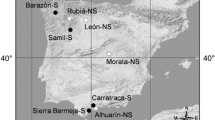
Ripe fruits (silicles) were sampled in summer 2013 from more than 30 mother plants (separated by at least 5 m) in each of five populations of Alyssum serpyllifolium. These populations were growing either on ultramafic (U) or non-ultramafic (NU) limestone-derived soils. U populations were sampled from the three main ultramafic areas of the Iberian Peninsula, where the Ni-hyperaccumulating subspecies lusitanicum and malacitanum grow. NU populations belong to the type subspecies serpyllifolium, which is considered a non-hyperaccumulator (Fig. 1). Previous reports confirmed the Ni-hyperaccumulating phenotype of the U populations (Kidd et al. 2011).
In the laboratory, seeds were separated from fruits and, for each of the five populations, seeds from different mothers were pooled. From each population, 150 seeds were randomly selected and surface-sterilised with a 10% (w/v) sodium hypochlorite solution, and rinsed with deionised water. Seeds were then germinated in plastic containers filled with a 1:1 (v/v) mixture of perlite and vermiculite and covered with a 0.5 cm-thick layer of acid-washed sea sand (SiO2). The five containers (one per population) were kept in a growth chamber at a constant temperature of 25 °C and a day: night cycle of 16 h: 8 h. Seeds were watered periodically with deionised water. Germination percentage was around 90% for all populations except SE, whose germination percentage was around 50%. When most plantlets had reached the 4-leaf stage, deionised water was substituted by irrigation with a 0.5-strength Hoagland solution, with a slight modification depending on the nature of bedrock material from which each population was originally sampled: Ca/Mg ratio in the nutrient solution was 0.4 for the populations from ultramafic soils, and 2.5 for the populations from limestone soils. After 8 weeks, plantlets with similar size (height approx. 3–4 cm and with more than 10 leaves) were selected (15 per population) and planted in plastic pots containing 1 kg of ultramafic soil sampled from Barazón, Melide ultramafic area (L in Fig. 1), where A. serpyllifolium ssp. lusitanicum grows naturally, amended with 5% (w/w) compost and 10% (v/v) of perlite. Three aliquots of unplanted soils were collected before planting for physicochemical characterisation (Table 1). The addition of 5% (w/w) compost to ultramafic soil greatly increased the Ca/Mg ratio (up to 1.7 in our amended soils compared to usual ultramafic values which are lower than 1). However, other characteristics of ultramafic soils, such as high Ni, Co and Cr concentrations remained mostly unchanged by compost addition. Based on previous experiments (Kidd et al. unpublished), the addition of compost was necessary to aid the growth of NU populations.
Pots were randomized and maintained in a glasshouse with natural light until the end of the experiment (July 2014) and were watered regularly with tap water. The experiment lasted for 16 weeks.
Plants that did not survive the first 5 days of the experiment were replaced by new plants; moreover a high percentage of plants from the two non-ultramafic populations died during the first few weeks of growth on the compost-amended ultramafic soil (almost 50% mortality in population SE and around two-thirds in population SN). These plants were not replaced. In addition, the surviving NU plants had a shoot biomass around 25% of U plants. Thus, a large number of NU plants were only collected for molecular analyses at the end of the experiment, which resulted in a different number of plants being analysed for leaf ionome per population (15 in each of the three U populations, 10 in population SE, and 5 in population SN).
Collection and elemental analysis of plants
At the end of the experiment, plant shoots were harvested, washed with tap water and rinsed with distilled water. A subsample of fresh leaves was collected from each plant and kept in plastic bags at −20 °C for use in the AFLP analysis. The remainder was kept in paper envelopes and dried at 40 °C for 72 h. After drying, leaves were separated from stems and 0.1 g of dry leaf biomass from each plant were digested in a 2: 1 (v/v) concentrated HNO3: HCl mixture on a hot plate at 120 °C. Foliar concentrations of Ca, Co, Cu, Fe, K, Mg, Mn, Ni, P and Zn were determined by inductively coupled plasma optical emission spectrometry (ICP-OES, model Vista-PRO, Varian Inc., Australia).
DNA extraction, AFLP protocol and data scoring
DNA was extracted from 200 mg of frozen leaves using the DNeasy® Plant Mini Kit (QIAGEN), following the manufacturer’s instructions.
AFLP markers were obtained using the AFLP Plant Mapping Kit (LifeTechnologies), following the manufacturer’s protocol with the some modifications: (1) dilution ratios of the products of the restriction-ligation step and pre-selective amplification step were 1: 2 and 1: 5, respectively; and (2) selective amplifications were performed in a final volume of 11.34 μl instead of 15 μl.
For the selective amplifications, six primer combinations were used (Table 2), four of which had been previously applied with success in other Alyssum species (Gerth et al. 2011; Spaniel et al. 2011a, b, 2012).
Fluorescently labelled selective PCR products were separated using an ABI 3130 xl (Applied Biosystems) DNA analyser. The ABI files with electropherograms were visualized and subsequently scored using the open source program Genographer v 2.1 (authored by Travis Banks and James Benham; available at http://sourceforge.net/projects/genographer/). Genographer converts the electropherograms into a gel-like image for scoring by visual screening.
The AFLP markers have two alleles, scored as presence (1) or absence (0) of bands. Plants showing weak or problematic profiles were removed from the analysis. The quality of PCR amplifications and bin scoring was assessed as following: 12 plants (17% of plants analysed) were re-extracted and amplified independently. An error rate was computed as the sum of errors/total number of comparisons (Bonin et al. 2004). Bins with an error rate equal to or higher than 0.2 were removed from the analysis. All fragments from 50 to 500 bp were considered.
Data analysis: elemental concentrations in leaves
Multivariate leaf ionome data were assessed by Principal Component Analysis (PCA). Differences in the concentrations of Ca, K, Mg, Ni and P and in the Ca/Mg ratio were assessed by one-way ANOVA analyses, using ‘population’ as factor. Significant differences were further explored by post-hoc analyses using the Bonferroni correction for multiple comparisons. When needed, data were transformed to meet ANOVA assumptions. All analyses were performed with SPSS 15.0.1 (SPSS Inc.).
Data analysis: AFLP markers
Population genetic diversity was estimated using the allele frequency-based metric % of polymorphic loci (PPL), and Nei’s gene diversity per locus (HE), modified after Lynch and Milligan (1994). These indices were computed using AFLP-SURV 1.0 software (Vekemans 2002). To estimate allele frequencies, a Bayesian method was used with non-uniform prior distribution of allele frequencies (Zhivotovsky 1999), assuming Hardy–Weinberg genotypic proportions (FIS = 0). Band richness (an analogue of allelic richness) was also computed for a population size of 10, using the rarefaction method of Coart et al. (2005) implemented in the software AFLPdiv.
Genetic differentiation among populations was estimated as the parameter ΦPT, an analogue of FST for binary markers. Paired ΦPT values were computed and were a measurement of the genetic differentiation among populations. Analysis of Molecular Variance (AMOVA, Excoffier et al. 1992) was performed to test the partition of genetic variance at different levels: (i) within and among populations; (ii) between geographic areas (NW and S of Iberian Peninsula); and (iii) between population types (U vs. NU populations). The significance of the model and of each estimate were validated by 9999 permutations over the whole dataset. The multivariate AFLP genotypes dataset was further explored by Principal Coordinate Analysis (PCoA) using a covariance matrix. This technique identifies the major axes of variation in the dataset and allows the assessment of the main trends in the data by graphical representation in a reduced-dimension space (e.g. a two dimension scatterplot). ΦPT computation, AMOVAs and PCoA were performed using GenAlEx 6.5 (Peakall and Smouse 2006, 2012).
In order to detect markers potentially involved in serpentine adaptation and Ni-hyperaccumulation, two different approaches were followed. Firstly, a population-based FST outlier loci approach was applied using the DFDIST method. This method is an extension of FDIST (Beaumont and Nichols 1996) for dominant markers (such as AFLPs). It consists of the computation of FST and heterozygosity of each AFLP marker (in population-pairwise comparisons) to compare afterwards those values with a neutral distribution of FST estimated from the whole dataset. Those markers with a deviant FST value represent outliers that can be considered to be affected by selection. DFDIST was implemented in the MCHEZA selection workbench (Antao and Beaumont 2011), which includes some improvements that ameliorate previous versions of DFDIST: (i) it performs a first run to remove candidate selected loci for computing the mean neutral FST value; (ii) it has an algorithm to force simulated FST to have similar values to the FST computed from the empirical data (adaptation to experimental conditions diverging from theoretical conditions); and (iii) it includes a False Discovery Rate (FDR) correction for multiple comparisons to avoid false positives.
Given the high geographic distances and possible genetic isolation between populations from the Northwest (L, S, SE) and South (SB, SN) Iberian Peninsula, separate runs were performed for each area, and in each run a pairwise comparison of a U and a NU population was carried out. An additional run, comparing limestone populations (SE, SN) was performed and compared to previous runs in order to eliminate possible outliers due to population-specific differences and not to soil type. The default parameters in MCHEZA were used for the computations, with the exceptions that 100,000 simulations were chosen to obtain the FST distribution and that an FDR of 0.05 was used.
Secondly, an individual-based phenotype to genotype association analysis was performed following the approach of Herrera and Bazaga (2009). For each locus, a series of logistic regressions with binomial distribution of residuals were computed, using band presence/absence as the dependent variable and leaf concentrations of each of the measured elements as fixed parameters. For this analysis, AFLP loci present in <5% or >95% of the individuals were discarded, since parameter estimates might be excessively influenced by outlying data, producing spurious results without ecological meaning. Thus, the final number of AFLP loci considered for the genetic–phenotypic association was 99 out of the 374 initially investigated. The significance of the GLM regressions was obtained after accounting for the possibility of obtaining false significant regressions (i.e. committing type I errors) due to the large number of computed p-values, using the False Discovery Rate (Benjamini and Hochberg 1995) at α = 0.05. The statistical analyses for this second approach were performed using the functions glm and p.adjust included in the package stats, R software (version 3.0.3, Foundation for Statistical computing, Vienna, Austria). Results of regression analysis were graphically summarised by an UpSet plot (Lex et al. 2014) elaborated using the online available UpSetR Shiny App (https://gehlenborglab.shinyapps.io/upsetr/).
Results
Leaf ionome
Principal Component Analysis (PCA) on leaf ionome data inferred two principal components (PC) that together explained 46.0% of the total variance (Fig. 2). The first PC was mainly influenced by foliar concentrations of macronutrients, with positive loadings for Ca, P and the Ca/Mg ratio. The second PC was influenced by trace element foliar concentrations (mainly Co, Mn, Ni and Zn, all with positive loadings). Mg and K concentrations had intermediate contributions to each PC, whereas foliar concentrations of Cu and Fe had little or no contribution to PCs. Accordingly, plants are positioned in different quadrants depending on the population type (Fig. 2): plants from ultramafic populations had positive values for one or both axes (i.e. high foliar concentrations of macronutrients – except Mg – and/or trace elements), whereas almost all plants from non-ultramafic populations showed negative values of both PCs (i.e. high Mg concentrations, low concentrations of other macronutrients and trace elements). Some plants of population SE were positioned close to the ultramafic populations in the PCA: these plants had high leaf Ni concentrations, above those considered to represent the hyperaccumulation threshold (see below).
Principal Component Analysis of leaf ionome data. Ultramafic populations are indicated by triangles, whereas non-ultramafic ones are indicated by circles. All individual points are connected with coloured lines to each population’s centroid. Loadings of leaf variables in each of the principal components are indicated in the inset at the bottom-right corner of the figure. Population codes and colours (POP) are presented in a small inset at the top-right corner of the figure
In accordance with the patterns observed in the PCA, ANOVA analyses revealed significant differences in leaf concentrations of macronutrients and Ni (Fig. 3 and Supplementary Material, Table S1), with U and NU populations showing contrasting strategies for Ca and Mg accumulation. Ca tended to be higher in U populations, but the main difference appeared in Mg concentrations: the median Mg concentration in the three U populations was around 6 g kg−1, while in NU populations the values were two-fold higher (median > 16.5 g kg−1) (Fig. 3). This also resulted in a marked difference in leaf Ca/Mg molar ratios (median values around 1 in NU populations and >4 in U populations, Fig. 3). Moreover, NU populations showed low intra-population variation in Ca/Mg ratio, whereas in U populations the Ca/Mg ratio among individuals varied from 1 to >7 (Fig. 3). In the case of K and P, the differences between types of populations were less clear, although there was a tendency for U populations to show higher leaf concentrations of both elements.
Leaf concentrations of different elements. The results of the ANOVA analyses for each element, with F ratios and p-values, are presented in each boxplot. All concentrations (except Ca/Mg) are expressed as g kg−1 dry mass. Ca/Mg is presented as the molar ratio. Ultramafic populations are indicated by dark-grey boxes, whereas non-ultramafic ones are marked in pale-grey boxes. The bottom and top of each box indicate the first and third quartiles, and the band inside the box marks the median for each population. Dashed lines in the Ca/Mg boxplot mark values of 1.0 (molar equality between the two elements) and 1.7 (molar ratio in the compost-amended ultramafic soil). The Ni-hyperaccumulation threshold of 1000 mg kg−1 is shown in the Ni boxplot as a horizontal dashed line. In each plot, different letters indicate significant differences between populations as indicated by post-hoc analyses with Bonferroni correction
Regarding leaf Ni concentrations, the hyperaccumulator phenotype (defined as values >1 g kg−1) was confirmed for all U populations, with median values ranging from 5.7 to around 9.9 g kg−1, and the highest leaf Ni concentrations being found for population SB. Population SN, from limestone soils, was below the Ni-hyperaccumulating threshold, although it showed notable Ni foliar concentrations (from 0.5 to 0.8 g kg−1). In contrast, the SE population, which also originated from limestone soils, showed an unexpectedly high foliar Ni concentration: seven of the 10 plants had values above the Ni-hyperaccumulator threshold of 1.0 g kg−1; three of them showing concentrations higher than 1.8 g kg−1, with a maximum of 2.12 g kg−1 (Fig. 3).
Genetic diversity and differentiation
The six primer combinations tested provided a total of 374 AFLP markers, with 50 to >70 markers per combination. The average genotyping error rate for each combination was between 2.8 and 5.3% (Table 2), well below the 10% maximum acceptable error rate proposed by Bonin et al. (2004).
Although there were no significant differences in genetic diversity between U and NU populations (Student’s t-test, data not shown), the non-ultramafic population SN had a genetic diversity (measured as PLP or Hj) twice that of the ultramafic population L. The other three populations (two U and one NU) had intermediate diversity values (Table 3). Differences in band richness, computed for population sizes of 10 plants, were less apparent.
Genetic differentiation between populations was quite high, with an overall ΦPT value of 0.27, with individual population-pairwise values ranging from 0.18 to 0.40 (Table 4). Population L showed the highest degree of differentiation (average ΦPT = 0.326). All estimates were significantly different from 0 (p < 0.001) (Table 4).
AMOVA analysis without considering groups of populations indicated that most of the variance (73.1%) occurred within populations (data not shown). Inclusion in the model of higher hierarchical levels (soil type or region) explained only a small amount of additional variance (Fig. 4). Differences between regions accounted for 7.57% of the total variance, whereas differences between soil types only accounted for 3.18%. Differences between population within groups accounted for 22 to 25% of the variance (Fig. 4).
Genetic variation and relative position of five A. serpyllifolium populations. (a) Principal Coordinate Analysis of AFLP data from five A. serpyllifolium populations, using the complete dataset of 374 loci. Ultramafic populations are indicated by triangles, and non-ultramafic ones by circles. All individual points are connected with coloured lines to each population’s centroid. Population codes and colours (POP) are presented in a small inset at the top-right corner of the fig. (b) Partition of genetic variance obtained by hierarchical AMOVAs between soil types (top) and between regions (bottom)
The first three axes identified by Principal Coordinate Analysis (PCoA) explained 26.7% of total variation. Projection of the plants onto the first two axes (Fig. 4) shows that the five populations were well separated, although two plants (one from population S and the other from SN) overlapped with the other populations. In accordance with the AMOVA results, the relative positions of the populations in the PCoA plot reflected mainly their geographic distribution rather than soil type. For example, populations SB and SN, which both originate from the South of the Iberian Peninsula, were positioned close together in the PCoA plot despite coming from different soil types.
AFLP analysis: outliers and genotype to phenotype association
The significant effect of regional origin on the partitioning of molecular variance led us to undertake separate outlier analyses for populations from the NW (L, S, SE) and S of the Iberian Peninsula (SB and SN). A marker was considered a potential positive outlier if its FST value was higher than expected at a 99.5% confidence interval, and applying a FDR of 0.05. Ten loci were identified as outliers, one in the comparison L vs. SE and nine in the comparison of SB vs. SN populations. Comparison of the S vs. SE populations revealed no outlier loci (Fig. 5). However, one of these loci was also identified in the comparison between non-ultramafic populations, so it was not considered to be related to soil adaptation. None of the identified outlier loci was selected in more than one comparison. Two of these outlier loci were present only in non-ultramafic populations, whereas none of the outliers was exclusive to ultramafic populations.
Results of DFDIST analyses comparing pairs of populations in Mcheza. The first three plots show comparisons of U vs. NU populations, whereas the fourth shows the comparison of two NU populations. FST is plotted against heterozygosity (He). The area above the continuous line in each graph represents the 0.995th quantile of the conditional distribution obtained from DFDIST simulations. Each marker is represented by a dot. Empty dots represent presumably neutral loci, whereas black-filled dots indicate outlier loci with potential adaptive value
Regarding phenotype to band distribution regression analysis, 26 loci showed significant regressions (either positive or negative) over 5 of the 11 tested variables: leaf concentrations of Ca, Mg, P and Ni, and molar Ca/Mg ratio (Fig. 6, Supplementary material Table S2 and Fig. S1). Ca and P were the elements with more significant loci (14 and 11, respectively). The four loci that had significant regressions with Ni were exclusive to this element. In contrast, loci related to the other four variables were shared amongst them in some cases (e.g. three markers were shared by Ca/Mg, P and Ca and even one locus –AB 146– had significant regressions with Ca, Mg, Ca/Mg and P). One locus (BA 326), with a significant positive regression with Ca, was previously detected as an outlier in the comparison between populations L and SE.
Summary of phenotype to band distribution regression analysis. Bottom: Combination matrix. Matrix rows indicate elements of leaf ionome, whereas matrix columns indicate (black dots) variables (or combinations of variables) with significant regressions over AFLP markers. Top: Barplot indicating the number of bands significantly related to each combination of leaf ionome variables
Overall, the two methods yielded a total of 34 potentially adaptive loci. Principal Coordinate Analyses were conducted to summarize genetic distance between plants on the basis of these potentially adaptive 34 loci (adaptPCoA, Fig. 7a) and on the basis of the 340 presumably neutral loci (neutPCoA, Fig. 7b). The first three axes of neutPCoA explained 24.1% of total variation, slightly less than the complete dataset. In contrast, the first three axes of adaptPCoA explained 47.6% of total variation, almost double the variation explained by the PCoA based on all markers (Fig. 4). Whereas neutPCoA yielded a plant grouping similar to that obtained with the complete dataset, the projection of plants on the two axes of adaptPCoA showed a plant grouping as a function of soil type: NU populations are placed in a dense swarm with negative values for both axes, whereas U populations are more dispersed with positive values for at least one of the two axes. One plant from population SN is placed in the middle of the area occupied by population S. This plant showed high concentrations of Ca, K and P in its leaves, and also a high Ca/Mg molar ratio (3.19, an extreme value for a plant from non-ultramafic population). Hierarchical AMOVA developed with presumably neutral bands inferred that 0% of variance occurred between soil types. Conversely, soil types explained 14% of molecular variance when considering potentially adaptive loci.
Genetic variation and relative position of five A. serpyllifolium populations. a Analysis based on 34 potentially adaptive loci (detected by outlier analysis or genotype to phenotype association). b Analysis based on 340 presumably neutral loci. Left, partitioning of genetic variance obtained by hierarchical AMOVAs. Right, plants plotted against the two first axes identified by PCoA
Discussion
We have performed research on Ni hyperaccumulation and serpentine adaptation in various populations of the facultative hyperaccumulator Alyssum serpyllifolium Desf., based on a glasshouse pot experiment using compost-amended ultramafic soil, followed by the analysis of leaf ionome and AFLP genotyping of cultured plants.
Based on previous studies (Morrison et al. 1980; Brooks et al. 1981; Kidd et al. 2011; Cabello-Conejo 2015), we assumed that Ni-hyperaccumulation was restricted to ultramafic populations (i.e. from the subspecies lusitanicum and malacitanum), whereas non-ultramafic populations were non-hyperaccumulators. Consistent with this expectation, foliar Ni concentrations in plants from three ultramafic populations ranged from 2.3 to 14 g kg−1 in the present study (Fig. 3). Similar values were also observed in field-sampled A. serpyllifolium plants in their native serpentine habitats (Cabello-Conejo 2015), as well as in other Ni-hyperaccumulator species of Alyssum (Galardi et al. 2007; Bani et al. 2010). Interestingly, our results indicate that non-ultramafic populations of A. .serpyllifolium have a considerable capacity for Ni accumulation: leaf Ni concentrations in populations SE and SN, of limestone origin, ranged from around 500 to around 2000 mg kg−1. Moreover 70% of surviving plants from population SE had shoot Ni concentrations in the pot experiment above the Ni hyperaccumulation threshold of 1 g kg−1. However, it should be noted that around 50% of plants from the SE population died during the first weeks of growth and that the percentage of Ni-hyperaccumulating plants was based only on those plants able to grow on the compost-amended ultramafic soil. Thus, the proportion of plants surpassing the hyperaccumulator threshold in that population in the wild is likely to be considerably lower. Although further evidence is needed to substantiate the hyperaccumulation trait in NU populations (such as a bioconcentration factor > 1 or Ni hypertolerance: Van der Ent et al. 2013), the present result points to the presence of the genetic potential for metal hyperaccumulation even in non-ultramafic populations.
Low soil Ca concentration in comparison to Mg (i.e. soil Ca/Mg ratio < 1) has been widely considered one of the main stress factors limiting plant colonization and establishment on ultramafic soils (Nyberg-Berglund et al. 2004; Brady et al. 2005). Here, amendment of the ultramafic soil with compost increased the soil Ca/Mg ratio (reaching values >1.5, compared with 0.15 in the original ultramafic soil), presumably ameliorating stress conditions to non-ultramafic plants. However, we observed very different responses among population types: non-ultramafic populations showed leaf Ca/Mg ratios similar to those of the soil, whereas in ultramafic populations the corresponding values were three times higher (Fig. 3). O’Dell et al. (2006) suggested that the ability to maintain a high leaf Ca/Mg ratio was the physiological feature distinguishing Californian serpentine shrub species from their non-serpentine congeners. This may also apply to A. serpyllifolium and to other Alyssum species, although it should be noted that considerable variation has been observed in Ca/Mg values in these species, e.g. in A. bertolonii from 0.9 to 4.2 (Galardi et al. 2007), and in A. montanum ssp. montanum from 0.8 to 12.4 (Bani et al. 2010).
By means of the AFLP technique more than 300 anonymous markers were amplified from the A. serpyllifolium genome. The use of plants obtained from a mixed seed pool from each population may have impacted our estimations of genetic diversity and differentiation, due to the possibility of analysing plants coming from the same mother. However, the values of genetic diversity, which were similar for all populations (irrespective of their soil of origin) with the exception of U population L, fell well within the observed range reported by Nybom (2004) in a review of 307 published studies. Sobczyk et al. (2017) published recently an analysis of Alyssum serpyllifolium populations using nuclear microsatellites. These authors provided estimates of within-population genetic diversity highly congruent with our own findings with AFLPs. Moreover Sobczyk et al. (2017) estimated by AMOVA analyses that differences between population types accounted for 5% of molecular variance, whereas the amounts of variance explained by differences between populations and within populations accounted for 21 and 74% of variance, respectively. These results are almost the same we obtained with AFLPs on our five populations (Fig. 4b). Overall, all these evidences seem to indicate that despite the pooling of seeds, our sampled plants are a fair representation of the populations’ genetic variability and structure.
The low genetic diversity inferred in population L (Barazón, Santiso, A Coruña) and its degree of differentiation from the other populations may be related to its geographically isolated position within the species range of A. serpyllifolium: it is a relict population growing in the Atlantic biogeographic region, while A. serpyllifolium as a whole is mainly distributed in the Mediterranean biogeographic region. Moreover, the L population is found in the northwestern edge of the species distribution, more than 100 km from the nearest A. serpyllifolium population. A similar effect (increased genetic differentiation) was observed by Adamidis et al. (2014) in an isolated population of the serpentine endemic Alyssum lesbiacum.
The ‘abundant centre’ model holds that peripheral populations of a certain species have a lower genetic diversity and a greater degree of genetic differentiation than central populations as a consequence of a lower effective size and a greater degree of geographic isolation. It seems that this model (present in more than 60% of species: Eckert et al. 2008) can also be applied to A. serpyllifolium, although the number of populations sampled here is limited. In spite of its isolation and lower genetic diversity, the L population seems to be well conserved, being composed of several thousands of plants and without any clear signs of population decline (personal observation C. Quintela-Sabarís). However, land-use change in the area, together with the unpredictable effects of climate change, would justify monitoring the population in the following years so as to ensure its conservation.
Metal hyperaccumulation and tolerance to ultramafic soils are genetically complex traits that involve many component physiological processes. The exploratory genome scan with AFLP markers and its analysis using two approaches (a population-genomics outlier approach and a ‘population free’ phenotype to band distribution regression approach) was developed here to take into account this complexity. Together, outlier and regression analyses provided 34 AFLP loci with potentially adaptive value. AMOVA analysis of neutral loci assigned 0% of genetic variance to differences between ultramafic and non-ultramafic populations, so the set of potentially adaptive loci account for all the genetic variability due to adaptation to different soil type.
The outlier approach, based on comparisons of U vs NU populations, led to the identification of nine outlier loci with potentially adaptive value. Interestingly, eight of these loci were identified in comparisons between populations from the S of the Iberian Peninsula, whereas only one outlier was detected in populations from the NW. We think that this imbalance between regions is a result of differences in serpentine adaptation and Ni accumulation that exist between NU populations. As discussed above, a significant percentage of plants from the SE population possessed the capacity for Ni hyperaccumulation, whereas all plants from the population SN were non-accumulators. Moreover, if we consider that (in our experiment with all the populations growing in a common garden) the leaf ionome may represent a proxy for genetically-based serpentine adaptation and Ni accumulation, plants from the SE population were closer to ultramafic populations than plants from the SN population. As a result, we may expect that population SE has a higher probability than population SN of sharing potentially adaptive AFLP loci with ultramafic populations, making the detection of outlier loci more difficult.
In an AFLP study evaluating the adaptation of Arabidopsis halleri to metalliferous soils, Meyer et al. (2009) found two types of outlier loci: those selected in all the metallicolous populations (considered as strong candidates for general adaptation to metal-rich environments), and those selected only in one of the metallicolous populations. This second group of markers were considered the result of site-specific selective pressures. In our study, none of the identified outlier loci was selected in more than one ultramafic population. According to Meyer et al. (2009), this may be the result of different selective forces in each ultramafic area. Indeed, Cabello-Conejo (2015) showed significant differences in several soil properties (including pH, C/N ratio, exchangeable Ca, Ca/Mg ratio, CEC, total and extractable Ni concentration) amongst the U populations included in our study. Thus, we can assume that (at least due to soil conditions) the studied U populations have experienced different selective pressures that may have resulted in different detected outliers.
‘Population free’ regression analysis of the leaf ionome on band distribution was more powerful for detecting potential adaptive loci than the outlier approach: this identified 26 bands with significant regression over one or more elements of the leaf ionome (one of which was already identified by the outlier approach). A second advantage of this method is that it allowed assessment of the ionome variables with more significant regressions, which can indicate or confirm those factors previously identified that act as the basis of serpentine adaptation. Significant regressions were found exclusively for elements that have been implicated in the serpentine syndrome, namely Ni (toxicity), Ca, Mg, Ca/Mg (nutrient imbalance) and P (macronutrient deficiency) (Brady et al. 2005; Kazakou et al. 2008). Similar results were obtained by Turner et al. (2010) when studying serpentine adaptation in the non-hyperaccumulator Arabidopsis lyrata: using NGS techniques, these authors found that candidate loci for serpentine adaptation were mostly related to heavy-metal detoxification and Ca and Mg transport.
As stated above, some of the identified loci were related to two or more nutrient elements, but not to Ni. This fact may suggest that serpentine adaptation (assessed as the ability to maintain leaf elemental homeostasis) and Ni hyperaccumulation are independent traits, i.e. expressed by different genes, controlled by different mechanisms. Indeed, Ni-hyperaccumulators are only a small subset of serpentine adapted populations (Brady et al. 2005). Several studies have shown that metal tolerance and hyperaccumulation are independent mechanisms (for a review, see Maestri et al. 2010), but few have specifically related serpentine adaptation (which involves other stresses, in addition to toxic metals) and Ni hyperaccumulation (Ghasemi and Ghaderian 2009; Kazakou et al. 2010; Cabello-Conejo 2015).
Based on a broader molecular analysis of the tribe Alysseae, Flynn (2012) inferred that the putative ancestral species of Alyssum section Odontarrhena was a Ni hyperaccumulator and, with high probability, endemic to serpentine substrates. However, there is a lack of genetic data at a finer taxonomic level that allows us to determine whether Ni hyperaccumulation and serpentine adaptation are ancestral (i.e. non-ultramafic populations are derived from ultramafic ones) or recently recovered (i.e. ultramafic populations have evolved from non-ultramafic ones) in A. serpyllifolium.
The present study has revealed that the Ni-hyperaccumulation trait is present in non-ultramafic populations of A. serpyllifolium, although a wider population sampling and analysis would be necessary to estimate more accurately the degree of conservation of this trait (i.e. the percentage of Ni hyperaccumulators in non-ultramafic populations). Conversely, the ability to maintain high Ca/Mg ratios seems to be restricted to ultramafic populations. However, the frequent presence of A. serpyllifolium on dolomite-derived soils (Mota et al. 2008), where the tolerance of high Mg concentrations is also a beneficial trait, may indicate that this trait may also be present in such non-ultramafic populations.
Although more analyses are needed to confirm the adaptive role of the identified loci, it can be inferred that regulation of leaf concentrations of Ni, Ca, Mg and P are amongst the main factors explaining the genetic differences between ultramafic and non-ultramafic populations within this species. This provides a first step towards the identification of mechanisms involved in serpentine adaptation and Ni hyperaccumulation in Alyssum section Odontarrhena.
References
Adamidis GC, Dimitrakopoulos PG, Manolis A, Papageorgiou AC (2014) Genetic diversity and population structure of the serpentine endemic Ni hyperaccumulator Alyssum lesbiacum. Plant Syst Evol 300:2051–2060
Álvarez-López V 2016 Plant-microbe-soil interactions and their role in phytotechnologies applied to trace metal-rich soils. PhD Thesis, CSIC-USC, Santiago de Compostela 356 pp.
Álvarez-López V, Prieto-Fernández A, Becerra-Castro C, Monterroso C and Kidd P S 2015 Rhizobacterial communities associated with the flora of three serpentine outcrops of the Iberian Peninsula. Plant Soil, 1–20
Antao T, Beaumont MA (2011) Mcheza: A workbench to detect selection using dominant markers. Bioinformatics 27:1717–1718
Baker AJM (1981) Accumulators and excluders – strategies in the response of plants to heavy metals. J Plant Nutr 3:643–654
Ball PW, Dudley TR (1993) Alyssum. In: Flora Europaea Vol. I, 2nd edition. Ed. T G Tutin et al. Cambridge University Press, Cambridge, pp 359–369
Bani A, Pavlova D, Echevarria G, Mullaj A, Reeves RD, Morel JL, Sulçe S (2010) Nickel hyperaccumulation by the species of Alyssum and Thlaspi (Brassicaceae) from the ultramafic soils of the Balkans. Bot Serb 34:3–14
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. P Roy Soc B-Biol Sci 263:1619–1626
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300
Bonin A, Bellemain E, Eidesen PB, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273
Bonin A, Ehrich D, Manel S (2007) Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionist. Mol Ecol 16:3737–3758
Brady KU, Kruckeberg AR, Bradshaw HD Jr (2005) Evolutionary ecology of plant adaptation to serpentine soils. Annu Rev Ecol Evol Syst 36:243–266
Brooks RR (1987) Serpentine and its Vegetation: a Multidisciplinary Approach. Dioscorides Press, Oregon, 454 pp
Brooks RR, Shaw S, Asensi Marfil A (1981) Some observations on the ecology, metal uptake and nickel tolerance of Alyssum serpyllifolium subspecies from the Iberian peninsula. Vegetatio 45:183–188
Cabello-Conejo M I 2015 Nickel hyperaccumulating plants: strategies to improve phytoextraction and a characterisation of Alyssum endemic to the Iberian Peninsula. PhD Thesis, CSIC-USC, Santiago de Compostela. 204 pp.
Cecchi L, Colzi I, Coppi A, Gonnelli C, Selvi F (2013) Diversity and biogeography of Ni hyperaccumulators of Alyssum section Odontarrhena (Brassicaceae) in the central western Mediterranean: evidence from karyology, morphology and DNA sequence data. Bot J Linn Soc 173:269–289
Centofanti T, Sayers Z, Cabello-Conejo MI, Kidd PS, Nishizawa NK, Kakei Y, Davis AP, Sicher RC, Chaney RL (2013) Xylem exudate composition and root-to-shoot nickel translocation in Alyssum species. Plant Soil 373:59–75
Coart E, Van Glabeke S, Petit RJ, Van Bockstaele E, Roldan-Ruiz I (2005) Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.) Conserv Genet 6:259–273
Deng THB, Tang YT, van der Ent A, Sterckeman T, Echevarria G, Morel JL, Qiu RL (2016) Nickel translocation via the phloem in the hyperaccumulator Noccaea caerulescens (Brassicaceae). Plant Soil 404:35–45
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species' geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol 17:1170–1188
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Flynn TA (2012) Evolution of nickel hyperaccumulation in Alyssum L. D.Phil. thesis. University of Oxford, Oxford, 232 pp
Galardi F, Mengoni A, Pucci S, Barletti L, Massi L, Barzanti R, Gabbrielli R, Gonnelli C (2007) Intra-specific differences in mineral element composition in the Ni-hyperaccumulator Alyssum bertolonii: a survey of populations in nature. Environ Exp Bot 60:50–56
Gao J, Sun L, Yang X, Liu JX (2013) Transcriptomic analysis of cadmium stress response in the heavy metal hyperaccumulator Sedum alfredii Hance. PLoS One 8(6):e64643
Gerth A, Merten D, Baumbach H (2011) Verbreitung, Vergesellschaftung und genetische Populationsdifferenzierung des Berg-Steinkrautes (Alyssum montanum L.) auf Schwermetallstandorten im östlichen Harzvorland. Hercynia 44:73–92
Ghasemi R, Ghaderian SM (2009) Responses of two populations of an Iranian nickel-hyperaccumulating serpentine plant, Alyssum inflatum Nyar., to substrate Ca/Mg quotient and nickel. Environ Exp Bot 67:260–268
Halimaa P, Blande D, Aarts MGM, Tuomainen M, Tervahauta A, Kärenlampi S (2014) Comparative transcriptome analysis of the metal hyperaccumulator Noccaea caerulescens. Front Plant Sci 5:213. doi:10.3389/fpls.2014.00213
Herrera CM, Bazaga P (2009) Quantifying the genetic component of phenotypic variation in unpedigreed wild plants: tailoring genomic scan for within-population use. Mol Ecol 18:2602–2614
Holderegger R, Herrmann D, Poncet B, Gugerli F, Thuiller W, Taberlet P, Gielly L, Rioux D, Brodbeck S, Aubert S, Manel S (2008) Land ahead: using genome scans to identify molecular markers of adaptive relevance. Plant Ecol Divers 1:273–283
Kazakou E, Dimitrakopoulos PG, Baker AJM, Reeves RD, Troumbis AY (2008) Hypotheses, mechanisms and trade-offs of tolerance and adaptation to serpentine soils: from species to ecosystem level. Biol Rev 83:495–508
Kazakou E, Adamidis GC, Baker AJM, Reeves RD, Godino M, Dimitrakopoulos PG (2010) Species adaptation in serpentine soils in Lesbos Island (Greece): metal hyperaccumulation and tolerance. Plant Soil 332:369–385
Kidd P S, Cabello-Conejo M I, Monterroso C, Becerra-Castro C, Álvarez-López V, Acea M J and Prieto-Fernández A 2011 Nickel bioaccumulation in different populations of Alyssum pintodasilvae and Alyssum malacitanum: Application in phytoextraction. XI ICOBTE Conference.
Kidd PS, Mench M, Álvarez-López V, Bert V, Dimitriou I, Friesl-Hanl W, Herzig R, Janssen JO, Kolbas A, Müller I, Neu S, Renella G, Ruttens A, Vangronsveld J, Puschenreiter M (2015) Agronomic practices for improving gentle remediation of trace element-contaminated soils. Int J Theor Phys 17:1005–1037
Krämer U (2010) Metal hyperaccumulation in plants. Annu Rev Plant Biol 61:517–534
Kruckeberg A R 2002 The influences of lithology on plant life. In Geology and Plant Life: The Effects of Landforms and Rock Type on Plants pp. 160–181. Seattle/London: Univ. Wash. Press. 362 pp.
Küpfer P, Nieto-Feliner G (1993) Alyssum. In: Flora Iberica Vol. IV. Ed. S Castroviejo et al. Real Jardín Botánico. CSIC, Madrid, pp 167–184
Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H (2014) UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 20:1983–1992
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Maestri E, Marmiroli M, Visioli G, Marmiroli N (2010) Metal tolerance and hyperaccumulation: costs and trade-offs between traits and environment. Environ Exp Bot 68:1–13
Mengoni A, Cecchi L, Gonnelli C (2012) Nickel hyperaccumulating plants and Alyssum bertolonii: model systems for studying biogeochemical interactions in serpentine soils. In: Bio-Geo Interactions in Metal-Contaminated Soils. Soil Biology 31. Ed. E Kothe and A Varma. Springer, Berlin, pp 279–296
Merlot S, Hannibal L, Martins S, Martinelli L, Amir H, Lebrun M, Thomine S (2014) The metal transporter PgIREG1 from the hyperaccumulator Psychotria gabriellae is a candidate gene for nickel tolerance and accumulation. J Exp Bot 65:1551–1564
Meyer CL, Vitalis R, Saumitou-Laprade P, Castric V (2009) Genomic pattern of adaptive divergence in Arabidopsis halleri, a model species for tolerance to heavy metal. Mol Ecol 18:2050–2062
Morrison RS, Brooks RR, Reeves RD (1980) Nickel uptake by Alyssum species. Plant Sci Lett 17:451–457
Mota JF, Medina-Cazorla JM, Navarro FB, Pérez-García FJ, Pérez-Latorre A, Sánchez-Gómez P, Torres JA, Benavente A, Blanca G, Gil C, Lorite J, Merlo ME (2008) Dolomite flora of the Baetic ranges glades (South Spain). Flora 203:359–375
Nkrumah PN, Baker AJM, Chaney RL, Erskine PD, Echevarria G, Morel JL, van der Ent A (2016) Current status and challenges in developing nickel phytomining: an agronomic perspective. Plant Soil 406:55–69
Nyberg-Berglund AB, Dahlgren S, Westerbergh A (2004) Evidence for parallel evolution and site-specific selection of serpentine tolerance in Cerastium alpinum during the colonization of Scandinavia. New Phytol 161:199–209
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
O’Dell RE, James JJ, Richards JH (2006) Congeneric serpentine and nonserpentine shrubs differ more in leaf Ca: Mg than in tolerance of low N, low P, or heavy metals. Plant Soil 280:49–64
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pollard AJ, Reeves RD, Baker AJM (2014) Facultative hyperaccumulation of heavy metals and metalloids. Plant Sci 217:8–17
Quintela-Sabarís C, Ribeiro MM, Poncet B, Costa R, Castro-Fernández D, Fraga MI (2012) AFLP analysis of the pseudometallophyte Cistus ladanifer: comparison with cpSSRs and exploratory genome scan to investigate loci associated to soil variables. Plant Soil 359:397–413
Sobczyk MK, Smith JAC, Pollard AJ, Filatov DA (2017) Evolution of nickel hyperaccumulation and serpentine adaptation in the Alyssum serpyllifolium species complex. Heredity 118:31–41
Spaniel S, Marhold K, Filova B, Zozomova-Lihova J (2011a) Genetic and morphological variation in the diploid-polyploid Alyssum montanum in Central Europe: taxonomic and evolutionary considerations. Plant Syst Evol 294:1–25
Spaniel S, Marhold K, Passlacqua NG, Zozomova-Lihova J (2011b) Intricate variation patterns in the diploid-polyploid complex of Alyssum montanum-A. repens (Brassicaceae) in the Apennine Peninsula: evidence for long-term persistence and diversification. Am J Bot 98:1887–1904
Spaniel S, Marhold K, Thiv M, Zozomova-Lihova J (2012) A new circumscription of Alyssum montanum ssp. montanum and A. montanum ssp. gmelinii (Brassicaceae) in Central Europe: molecular and morphological evidence. Bot J Linn Soc 169:378–402
Strasburg JL, Sherman NA, Wright KM, Moyle LC, Willis JH, Rieseberg LH (2012) What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos Trans R Soc B 367:364–373
Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV (2010) Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet 42:260–264
van der Ent A, Baker AJM, Reeves RD, Pollard AJ, Schat H (2013) Hyperaccumulators of metal and metalloid trace elements: facts and fiction. Plant Soil 362:319–334
Vekemans X (2002) AFLP-SURV version 1.0. Distributed by the author. Laboratoire de Génétique et Ecologie Végétale. Université Libre de Bruxelles, Belgium
Verbruggen N, Hermans C, Schat H (2009) Molecular mechanisms of metal hyperaccumulation in plants. New Phytol 181:759–776
Vos P, Hagers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgments
This research was funded by the FACCE Surplus project Agronickel (ID71). CQS was awarded a Research Grant from the Deputación da Coruña, and a Postdoctoral contract financed by the French National Research Agency through the national program “Investissements d’avenir” with the reference ANR-10-LABX-21-01 / LABEX RESSOURCES21.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Antony Van der Ent.
Rights and permissions
About this article
Cite this article
Quintela-Sabarís, C., Marchand, L., Smith, J.C. et al. Using AFLP genome scanning to explore serpentine adaptation and nickel hyperaccumulation in Alyssum serpyllifolium . Plant Soil 416, 391–408 (2017). https://doi.org/10.1007/s11104-017-3224-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3224-y