Abstract
Background and aims
RootScan is a program for semi-automated image analysis of anatomical traits in root cross-sections.
Methods
RootScan uses pixel thresholds to separate the cross-section from its background and to divide it into tissue regions. Area measurements and object counts are performed within various regions of interest. A graphical user interface permits the user to see which regions are selected, to edit those selections, and to rate and comment on the data. The structure of the program allows for organized workflow and increased data collection efficiency.
Results
The program collects data on more than 20 variables per image including areas of the cross-section, stele, cortex, aerenchyma lacunae, xylem vessels, and counts of cortical cells and cell files. An increased rate of data collection allows collection of four times more variables in less time than is possible with current methods. Correlation analysis shows that RootScan data is equal or greater in accuracy than data collected with Photoshop.
Conclusions
Compared with currently available tools, this software offers considerable improvements in the amount and quality of data, ease of use, and time needed for data collection. RootScan permits phenotypic scoring of physiologically and agronomically important traits on a large number of genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phenotypic profiling is a means of quantifying the expression of plant traits, requiring accurate, rapid, and economical measurement of traits of many samples in a reasonable amount of time. Root anatomical traits have important effects on plant function, including acquisition of nutrients and water from the soil, resource transport within the plant, and the costs and benefits associated with root growth. Traits such as root cortical aerenchyma (RCA) influence the metabolic costs of tissue maintenance by eliminating living cells, thereby permitting enhanced resource acquisition through increased growth (Fan et al. 2003; Zhu et al. 2010; Postma and Lynch 2011a, b). Microscopic images can be used to acquire useful information, including dimension, location and distribution of anatomical features. However, accurate measurements from a large number of images can be time-consuming and laborious.
Despite the central role of the root system in plant function, root traits are underutilized in plant breeding (Gregory et al. 2009). The ability to make accurate, rapid and reliable measurements of anatomical traits would allow for their use as direct phenotypic selection criteria or to identify molecular markers. The anatomical traits measured in RootScan have potential for breeding increased tolerance to edaphic stresses, and include characteristics that may reduce the number of respiring cells, change hydraulic conductance in the xylem vessels, and shorten the radial pathway for transport of soil resources into the vascular tissue (Drew and Saker 1986; Drew and Fourcy 1986; Fan et al. 2007; Tombesi et al. 2010; Zhu et al. 2010). Decreased xylem vessel area has been used as an approach to breeding plants with greater water use efficiency (Richards and Passioura 1989). Traits affecting root cost and maintenance include the density and number of cells, the amount of cell wall area, and the relative proportion of living and non-living area (Justin and Armstrong 1987; Lynch and Ho 2005). Areas of carbon density reflect greater initial investment during growth, and therefore influence construction cost on a whole root or system level. The relative proportion of the stele and cortex occupying the cross-section can vary by genotype, root class, and longitudinal position along the root axis (Justin and Armstrong 1987). Finally, factors such as cell size, packing and the number of cortical cell files could influence the initial carbon investment in a root, and the transport of resources through the apoplast and symplast.
A variety of software packages are used for image processing and data collection in plant biology. Software packages such as Photoshop offer flexibility and user-friendly processing tools. Manual selection of objects within images can be performed using standard image processing packages, and accompanying pixel counts can be converted to area measurements based on micrometer calibration. Photoshop has been used for image analysis of root morphological traits in soybean (Wright et al. 1999), measurement of root cortical cell size (Longstreth and Borkhsenious 2000), processing images before or after root diameter analyses with MacRhizo (Zobel 2003), and assessment of reactive oxygen species damage in root and hyphal symbionts (Fester and Hause 2005). However, such programs are not designed for automated data extraction and are cumbersome when working with more than a handful of traits in a few dozen images. The amount of time spent on each step of data collection becomes especially important with the large number of samples required in phenotypic profiling for genetic analyses.
In contrast to Photoshop, software such as Image J, ImageMaster and IBAS2000 are specifically designed for analysis of biological images, meaning they perform well despite morphological and anatomical variation among samples. Generic tools and pull down menus are user-friendly and provide some adaptability of function. Plug-in modules are available for these programs which allow macros-directed batch analysis of images. However, a major limitation of these programs is that they are not designed for semi-automated processing of a large number of images, and therefore users are not involved in data quality evaluation. In contrast, software that is completely user-driven may generate fatigue and inconsistencies among data collectors, or even the same person among data collection sessions.
Here, RootScan is described, a tool for high-throughput profiling of root anatomical phenotypes from images of root cross-sections. RootScan was developed to analyze traits relevant to soil resource acquisition, with potential applications in other areas of plant biology. This semi-automated image analysis software avoids the time-consuming manual measurements commonly required in programs such as Photoshop or Image J, and is well suited to screening large numbers of images that are biologically variable. To our knowledge, no other programs exist for semi-automated image analysis designed for high-throughput evaluation of anatomical features of root cross-sections. Examples are offered of the utility of RootScan for cross-sectional images of maize, rice and bean roots.
Materials and methods
Genotypes and growth conditions
Seventy genotypes of maize (Zea mays L. ssp. mays) were randomly selected from the recombinant inbred population Intermated B73xMo17 (IBM) for image analysis. Plants were grown in a greenhouse located on the campus of The Pennsylvania State University in University Park, PA (40°48′N, 77°51′W), from May-August 2008. The nutrient solution consisted of the following (in μM): NO3 (2211), NH4 (777), CH4N20 (398), P (411), K (1858), Ca (1454.70), Mg (960), B (16), Cu (0.33), Zn (7), Mn (8), Mo (0.85), Fe-EDTA (16). Environmental data were collected hourly in the greenhouse using a HOBO U10-003 datalogger (Onset Corporation, Pocasset, MA, USA). Mean ambient temperature was 26.5°C ± 5.9 (day)/21.3°C ± 2.4 (night), and mean relative humidity level was 57% ± 12.2. Maximum photosynthetic flux density was 1200 μmol photons m−2 s−1.
Maize root systems were harvested after 28 days (V6 stage), and preserved in 75% ethanol, and stored at 4°C until the time of analysis. A 4-cm tissue segment was collected 5 cm from the base of a second whorl crown root for hand sectioning. Additional test samples were obtained from field-grown maize. Common bean (Phaseolus vulgaris L.) genotype DOR364 and rice (Oryza sativa L. ‘Dular’) plants were grown for 25 days in silica sand mixed with phosphorus-doped alumina (5.0 g Al-P/l sand) to maintain P availability at 1 μM (low P), and 300 μM (high P). At harvest, a 4-cm segment was collected from the base of a representative basal root of each bean plant or from a second-whorl crown root for rice.
Sample sectioning
Preserved, unembedded tissue was sectioned by hand using Teflon-coated double-edged stainless steel blades (Electron Microscopy Sciences, Hatfield, PA, USA) and wet mount slides were immediately prepared. Section thickness was between 30 μm and 50 μm. Dilute toluidine blue (0.05%) was used to stain bean cross-sections for enhanced contrast. Sections were examined on a Diaphot inverted light microscope (Nikon, Chiyoda-ku, Japan), at 4× magnification with an additional 0.7× adapter, for a combined magnification of 2.8×. This allowed larger sections to be viewed in their entirety. The three best sections on a slide were selected as subsamples for image capture. Selection of particular cross-sections was based on tissue integrity, and the relative perpendicularity of sectioning (i.e. uniform thickness across the section). Each section was imaged separately. Prior to image capture, sections were centered in the frame such that no part of the section, its lateral roots or root hairs touched the image frame. Lower resolution images, used to create and test the program, were captured with a black and white XC-77 CCD Video Camera Module (Hamamatsu, Iwata-City, Japan). Analog output was converted by the image capture software ImageMaster 5.0 (Photon Technology International, Birmingham, NJ, USA) to 8-bit images with a pixel dimension of 640 × 480. High resolution images, used later for further validation, testing, and tests of speed, were captured with a Nikon DS-Fi1 digital camera with the Nikon Elements capture software set at high resolution capture. These 8-bit high-resolution images have a pixel dimension of 2560 horizontal × 1920 vertical pixels. Images used in this paper were at the lower resolution unless otherwise noted.
RootScan and photoshop measurements
RootScan was written in MatLab 7.6 2008a (The MathWorks Company, Natick, MA, USA) to generate primary measurements based on number of pixels, which were used to calculate secondary measurements (Table 1). Analysis was carried out on a workstation equipped with 40 MB of random access memory (RAM) and two 2.33 GHz quad-core Harpertown central processing units (CPU) and 8 GB DDR2 ECC memory (Intel, Santa Clara, CA, USA). Primary measurements included area and count variables. The following area measurements were made via pixel-counting: areas of the total cross-section, cross-section cell wall, aerenchyma lacunae, total stele, stele cell wall, xylem vessels, and lateral root (if present). Mean cell size was calculated via pixel counting, with three separate measurements made for the outer, middle and inner cortex. Count data included number of cortical cells, cortical cell files, and aerenchyma lacunae. Some of these primary measurements were used to calculate secondary measurements in RootScan: area of the cortex ([cross-section area]-[stele area]), percent aerenchyma (total aerenchyma area/cortex area), cortical cell area ([cortical area]-[aerenchyma area]), percent cortical cell area ([cortical cell area]/[total cross-sectional area]), and ratios of stele to cross section area and stele to cortex area. Manual measurements of a subset of variables were made using Photoshop 9.0.2 (Adobe, San Jose, CA, USA) for comparison with those made with RootScan. Using the ‘magic wand’ and ‘lasso’ selection tools in Photoshop, primary area measurements were made by selecting objects of interest (stele, cross-section, aerenchyma lacunae, xylem vessels) and recording the pixel area from the histogram display. The cross-section and stele were selected by drawing a circle with the lasso tool, while the aerenchyma were autoselected with the selection icon tool, which depends on finding the borders. Cortex was calculated as the difference between cross-section and stele areas and the percent aerenchyma was calculated from arenchyma area and cortical area. In both programs, pixel values were converted to mm2, based on micrometer calibration (204 pixels/linear mm).
Data accuracy in RootScan was examined by simulating various image quality problems by altering a single image in Photoshop and comparing data collected from the original and altered images. Using standard Photoshop tools, image quality was decreased on a relative scale of 0 (original image) to −5 for each of four issues: focus, exposure, shearing, and section thickness. For altering focus, the filter “Gaussian blur” was used at 2,3,4, and 5 steps. For exposure, adjustments were made to the image using the brightness/contrast adjustment, and scaling the lightness slider at steps of 25, i.e. lightness at 25, 50, 75, 100, and 125, and darker at −125, −100, −75, −50, and −25. Shearing was simulated using the brush stroke filter on angled stroke, scaling stroke length in intervals of 3 from 6 to 18 and setting direction balance = 75. Uneven section thickness was simulated with the “burn” tool using exposure at 50% and swiping from 1 to 5 times.
Statistical analysis
Statistical analysis was performed using the R Program, version 2.9.2 (R Development Core Team 2010). Data from images of the three subsample sections were pooled into mean values by trait. Validation of the data collection methods used in RootScan was performed for multiple users, images, and repeated analysis of the same image. Measurement consistency between RootScan and Photoshop was evaluated by analyzing the same image 10 times in each program by the same user. Measurement precision in RootScan was evaluated by having five users analyze the same image. Users had differing levels of experience with image analysis and with RootScan. RootScan’s measurements were also compared to a corresponding set of Photoshop measurements. Resulting data were evaluated using descriptive statistics and Pearson correlation analysis.
Results
Program overview
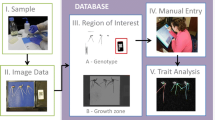
RootScan’s single module graphical user interface (GUI) contains features to monitor data collection in real time. Buttons for each of the major steps allow the user to repeat or skip steps to correct mistakes made by the user or by the program. An overview of the major steps is shown in Fig. 1. As each step is finished, a color-coded miniature image appears in the lower right area of the GUI, showing the regions selected for data collection. This miniature image can be inspected and approved by the user before proceeding to the next step. The graphical user interface requires user involvement. A user with minimal training can quickly and correctly identify a given anatomical component, its location, and edges, even in images that are difficult for the software to analyze due to poor image quality. For those images in which the software has difficulty determining boundaries of an area, a user can manually make the correct selection.
Overview of the major steps for identification of anatomic features in RootScan. The cross section is isolated (a, boundary shown by the orange line). The stele is separated from the rest of the image (b, boundary shown by the green line). Aerenchyma lacunae are identified within the cortex image (c, red fill). Within the stele image, metaxylem (yellow fill) and protoxylem (orange fill) are identified (d). Once all the major features are identified (e), the user can add comments regarding the quality of the image. Cells of the cortex can be separated into the desired number of radial bands for counts and sizes in each class (f)
When the program begins, a temporary data matrix is created to store data as it is collected. This feature allows the user to resume an analysis where it was stopped, in the event that data collection is interrupted or paused. RootScan takes 13 direct measurements from each image, and from this information, calculates another nine derived variables (Table 1). The user may also define additional derived variables, based on the direct measurements.
Program operation overview
The RootScan workflow is divided into six steps: cross-section isolation, lateral root selection, stele selection, aerenchyma selection, xylem selection and data quality evaluation. The user may select automatic batch processing for all steps after cross-section isolation. Batch processing reduces user time requirements while grayscale images are converted to their binary equivalents. Then, the analysis can be completed with manual corrections and approval of the remaining steps. In the absence of automated processing, each step would require the user to approve the progression to the next step. Certain common image analysis techniques are repeatedly used throughout the program, including binary conversion, dilation, filling and edge detection (Gonzalez and Woods 1992).
Part I: Isolation of the cross-section and creation of the data collection module
A series of image manipulations are used in succession to isolate the entire cross-section from the background. Negative cross section images were converted to binary images using Otsu’s method (Otsu 1979). This method groups pixels into two categories, foreground (i.e. the cross-section and any debris) and background, and minimizes the intra-class variances of these two groups. Then an image dilation function uses a disk-shaped structural element with a disk radius of 6 pixel units to patch gaps in the cross section perimeter. The resulting image dilation is filled to remove any holes in foreground objects. The result should be a solid cross-section image (or a solid stele region when this is applied to the stele segmentation step) and surrounding debris. The target region (i.e. cross-section or stele) and debris are evaluated using a label matrix and all debris is removed from the image.
Upon completion of these steps, an erosion step (using the same disk-shaped structural element) is used to shrink the cross-section or stele perimeter back to its original position prior to dilation. The outer boundary of the cross-section is the epidermis, and root hairs are truncated during this step. At the conclusion of this step, cross-sectional area is calculated by pixel number.
Part II: Lateral detection and segmentation
Any lateral roots present in the cross-section may be removed from the image (Online resource 1). While lateral roots occupy a portion of the cross-section and are included in the total cross-sectional area calculation, the purpose of this step is to exclude them from the cortical area measurement. To do this, the user draws a polygon around the lateral root region. The pixels of lateral root area within the bounds of the cross-section are counted as lateral root area, and recorded in the data output. The pixels outside the polygon are the stele and cortex regions, which are passed on to the aerenchyma and stele detection algorithms as a separate image without the area occupied by the lateral root segment.
Part III: Stele detection and segmentation
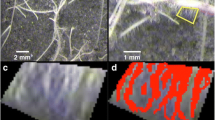
This part of the program separates the stele from the cortex. Since the stele and cortex typically have contrasting pixel values (light intensity contrast), this characteristic is used in the separation of these two regions of the image (Fig. 2). Low contrast in cortical regions adjacent to stele regions presents a problem because separation of tissue regions is based on existence of a high contrast border between the stele and cortex. To place focus on the stele region, a spatial weights matrix (Fig. 2b) (Haining 2003) is applied to the image negative (Fig. 2a) (using image multiplication) that allows emphasis of stele pixels and de-emphasis of cortex pixels based on radial proximity to the centroid of the image, which is assumed to approximate the centroid of the stele. From this attenuated cross-sectional image (Fig. 2c), a temporary threshold is calculated from a pixel density map for elimination of the additional lighter cortical regions. Maximum tissue pixel density usually occurs at the boundary between the stele and cortex. This method calculates the neighboring pixel density inside a square 20 × 20 pixel sampling box for tissue pixels retained in the truncated cross-section image (Fig. 2c) by taking the sum of all pixels contained within each pixels sampling box. The result is another map showing regions of low and high pixel density (Fig. 2d). In Fig. 2d, the lighter areas indicate greater pixel densities (more tissue per unit area) while darker areas indicate lower pixel densities (less tissue per unit area).
RootScan image processing for stele separation. After cross-section isolation, negative used for spatial weights matrix application (a), spatial weights matrix adjusted for cross-section asymmetry bias in the bandwidth calculation (b), truncated cross-section image result after applying spatial weights matrix (c), result from the local pixel density calculation used to calculate the threshold for binary conversion (d), initial binary image showing stele and cortical segments (e), binary image showing only stele region (f), dilated binary image (g), filled dilated binary image (h), eroded filled image (i), stele perimeter image (j), smoothed solid stele image result from applying median smoother (k), final stele negative image (l)
The values from the pixel density map are passed into a modified version of Otsu’s intra-class variance minimization method for binary conversion. All density map elements greater than the calculated threshold are considered stele/cortical boundary or noise pixels while everything else is background. The resulting binary image (Fig. 2e) shows the stele and occasionally a few small cortex cell wall regions. These cortex cell wall regions are eliminated by selection of the largest object (Fig. 2f), and any holes in the stele perimeter are patched by dilation (Fig. 2g) using the disk-shaped structural element. Once the perimeter is dilated it is filled to create a solid stele image (Fig. 2h). The solid stele image is eroded back to it original pre-dilated size (Fig. 2i) using the same structural element. In the final step, a 1-dimensional median filter is applied to the stele perimeter, smoothing the perimeter line by averaging neighboring radial values. The resulting smoothed perimeter is dilated with a 25 pixel disk-shaped structural element to make the stele boundary continuous (Fig. 2j) and filled using the “holes” method (Fig. 2k), then the dilation is reversed to return the stele to its original size. The grayscale equivalent is shown in Fig. 2l.
If the program fails to separate the stele region from the cortex region correctly, the user is allowed to edit the selection. Inaccurate stele and cortex segmentation can be attributed to poor image quality and low contrast between cortex and stele regions for a particular image. If RootScan has selected the wrong area as the stele, the user may draw a multi-sided polygon around the stele region, as with lateral root selection. The stele area is then recalculated from this polygon, and this value replaces the initial incorrect calculation.
The results from the stele selection (Fig. 2l) are passed on to the xylem vessel selection algorithm and the cortex region is passed on to the aerenchyma detection algorithm as a separate image. Within each image, each object receives a unique integer label to distinguish it from other objects. An object is an area of white pixels surrounded by a high contrast edge. The result is a label matrix that is used for indexing and defining each object attribute in the label matrix. Objects label indexes are matched with their object areas, x and y centroid coordinates, perimeters, major axis lengths using the “regionprops” function of Matlab.
Part IV: Cortex data collection
Collection of area and count data in the cortex begins with identification and labeling of objects within the cortex. This is followed by a series of tests that distinguishes aerenchyma lacunae from cells. All cells and lacunae are considered objects. The lengths of major and minor axes are used to distinguish lacunae from cells. In addition, aerenchyma lacunae occur in the middle portion of the cortex, therefore RootScan excludes the inner and outer 20% of the cortex during identification of aerenchyma lacunae. Within that zone, an object is considered to be a lacuna if the major axis length is greater than the sum of the median and standard deviation of all major axis lengths. After aerenchyma detection, the GUI displays the selected regions highlighted in red. This allows the user to select any cortex object as a lacuna that was not selected by RootScan, or to de-select any cortex objects falsely classified as aerenchyma. At the conclusion of Part IV, all cortex-related data collection is complete.
Part V: Xylem vessel detection
The image resulting from the stele selection in Part III reappears in the GUI window for metaxylem vessel detection (Fig. 2l). During this step, stele and xylem data are collected. Late metaxylem vessels are identified within the stele based on their size, since they are generally larger than any other object in the stele. Metaxylem vessels are distinguished from other objects by using the maximum area difference in a ranked list of stele object areas, so that objects with areas above the step of maximum difference are defined as metaxylem, excluding any large erroneous objects that might appear in the stele center. Large objects sometimes are detected near the center of the image as a result of blurring of pith cells in low-contrast images, but since xylem do not form in this position, these are automatically eliminated. In high-resolution images, protoxylem vessels can also be identified by the program, and selections corrected by the user. As with aerenchyma selection, the GUI allows the user to edit the xylem vessels that are automatically selected by the program by selecting or deselecting objects.
Part VI: Data quality evaluation and output
Following completion of Part V, the user is presented with a color-coded image that highlights the regions of interest selected during data collection. At this time, the user may return to any previous step to make corrections. Accompanying the final highlighted image is a basic dialog box that allows the user to rank the image data and record brief comments. The data quality ranking and comments are recorded in the data output. Pixel measurements are converted to mm2 based on the calibration value entered by the user. Examples of resulting values for cross-sections from four maize genotypes with extreme values for cross-sectional area, aerenchyma area, and other anatomical traits are shown are Table 2.
Validation of RootScan
Comparisons of efficiency, accuracy and precision were made between RootScan and Photoshop. RootScan can produce data on more than 20 variables in each image. Using the “magic wand” and “lasso” tools in Photoshop, five measurements can be made on about 30 images per hour (areas of the cross-section, stele, cortex, aerenchyma lacunae, and percentage of the cortex occupied by aerenchyma).
Tissue area measurements made by RootScan and Photoshop were similar (Table 3). Correlations for area measurements between the two methods were high, with r-values for area of the cross-section, stele, cortex, and aerenchyma of 0.9980, 0.9947, 0.9949, and 0.9719 respectively. Variances for Photoshop measurements were greater than those made in RootScan (Table 3). Variance among users of RootScan was evaluated by calculating coefficients of variation for data collected on the same image by five users, operating the program normally including manual corrections (Table 4). Coefficients of variation for primary variables were generally less than those for secondary variables. Correlation coefficients for area measurements between Photoshop and RootScan were mostly high, in repeated analysis of the same image (Table 3) and in a set of 110 images analyzed in both programs (Table 5). In the larger set of images, a low correlation coefficient was observed for aerenchyma area (Table 5).
To test the effect of degraded image quality on data accuracy, we simulated several types of defects by altering a single, good quality image with Photoshop. The pattern and intensity of the decline in data accuracy depended on the variable in question and type of image defect (Fig. 3). Focus and exposure are factors that should be adjusted prior to image capture, while shearing and section thickness are technical errors occurring during sectioning. Focus refers to the relative sharpness of lines and objects in the image. Exposure refers to the amount of light introduced during image capture. Both low and high exposure images result in low contrast images. Shearing refers to a lack of cross-section perpendicularity. In sheared cross-sections, objects may not be clearly defined, and portions of the cross-section may appear out of focus or compressed. Overly thick sections are those that exceed 50 μm; these sections have a darker appearance in images than thinner cross-sections.
Plot panel showing changes in data accuracy for three variables (aerenchyma area, cortical area and cortical cell count) as image quality declines. Y-axis values depict a relative scale, from 0 (original image) to −5. Data are based on alteration of a single original image, using tools and filters in Photoshop, to simulate four image quality factors (focus, shearing, exposure, and cross-section thickness). Select corresponding images can be seen in Fig. 4
Aerenchyma area was not strongly affected by declining quality for focus and cross-section thickness. However, moderate to extreme shearing caused data inaccuracy. Shearing interferes with image analysis because the edges of the “objects”, in this case aerenchyma lacunae, are so blurred that the program cannot accurately select objects in order to measure them (Figs. 3 and 4). Usually when a section is sheared, the entire image is not affected, as it was in these simulated images. Dark images (high exposure, −2, −3) did not affect image quality, but very light images (over-exposed, −4 and −5) caused overestimation of aerenchyma area (Fig. 3). Overexposed images lack defining cell borders, so that the program would include merged cells as aerenchyma lacunae. Cortical area values declined slightly with increased amounts of shearing and with over-exposure, but the errors were not large (Figs. 3 and 4). Changes in cortical area were observed in thicker sections, and for images that were very out-of-focus (−4, −5). Cortical cell counts declined with decreased focus, while shearing caused a sharp decline in this variable. Thick cross-sections, and images with low exposure caused a moderate decrease in cortical cell count.
Images depicting declining image quality for focus and exposure. A single original image (“0”) was altered using blur and contrast tools in Photoshop. Images of a lower relative quality (“−2” and “−5”) were analyzed in RootScan, and plotted for several variables in Fig. 3
Although it was developed using maize as a model, RootScan can be used to analyze transverse sections of roots from other species, as shown by analysis of color images of common bean (Phaseolus vulgaris L.) and black and white images of rice (Oryza sativa L.) root cross sections (Fig. 5). Verification of these data was done by manual measurement in Photoshop for areas of the stele, cortex, cross-section and aerenchyma. Correlations were high for all variables between manual and semi-automated selection (data not shown).
Image analysis data for cross-sections from bean (Phaseolus vulgaris ‘DOR 364’) and rice (Oryza sativa ‘Dular’) grown under low (1 μM) and high (300 μM) phosphorus. Tissue samples were from the base of a basal root for bean and 5 cm from the apex of a crown root for rice. All variables are in units of mm2, except for count variables (#CC, #CF) and ratio variable (TSA:TCA)
Using RootScan for high-throughput phenotyping
To test the speed of operations for RootScan, a batch of 180 high-resolution images were analyzed by an experienced user and the times for processing each phase recorded (Table 6). Active time was less than 2 min per image (Table 6). From the user’s perspective, the operations are divided into three phases. In phase 1, the user loads the batch of images into RootScan for initial cross-section isolation. The user checks each section to be sure that the outside border is drawn correctly. If there are errors, for example if RootScan has drawn the cross-section border around the root hairs instead of eroding them, the user can alter the selected border by drawing a polygon that excludes the corrected area (Online resource 2). Since the new border is used only if it is inside the RootScan defined border, sections with broken or incomplete borders cannot be processed and should not be included.
Once the cross sections have been satisfactorily isolated, the user can submit the images for batch processing (Phase 2). This step does not require user intervention, and saves labor since the detection of all remaining objects is accomplished during this time.
In phase 3, the user reviews the output to check for errors and make corrections. Objects that have been incorrectly defined, such as aerenchyma lacunae or xylem vessels, can be selected or deselected (Online resource 3). If there are dark areas around the stele, for example due to excessive section thickness or fungal areas near the stele, the stele border may need to be redefined (Online resource 4). There is an opportunity for the user to add comments on problems with the image and to rate the image quality for consideration during statistical analysis. Once the user completes these steps, the data is ready for analysis.
Discussion
We have presented a semi-automated method for high-throughput image analysis of root anatomical phenotypes. Image analysis in RootScan offers several advantages over manual area selection in Photoshop, including analysis of a greater number of images per hour, greater number of traits distinguished, improved format of data output, and more sophisticated methods for handling variation among images. RootScan improves workflow organization and efficiency by automatically loading images, prompting the user at each step, and recording data as it is collected. RootScan measurements include areas, object counts, and mean size by region, as well as derived variables such as ratios, indices, or percentages of an object’s area within a tissue (Table 1). Certain measurements, such as mean cell size, would be impractical to perform manually in Photoshop. Therefore, the Photoshop method is limited to area measurements and any secondary variables that can be derived from area measurements.
Comparison of data collected by the two methods demonstrates that RootScan makes accurate measurements of tissue areas, and has greater accuracy of aerenchyma measurement than Photoshop. Low correlation between aerenchyma measurements by the two programs (Table 3) can be explained by RootScan’s more sophisticated techniques for identifying aerenchyma. With the magic wand tool in Photoshop, the user may be able to visually identify a lacuna, but may be unable to select it to due to low contrast borders. RootScan is designed to account for differences in contrast among images via iterative thresholding, and includes thorough testing to classify objects such as cells, xylem vessels and aerenchyma lacunae. When the program makes errors, the user can still select or deselect objects provided the borders are intact. Overall, this results in more accurate aerenchyma selection in RootScan, compared to Photoshop.
In repeated analysis of the same image by a single user, lower variance was observed with RootScan than with Photoshop (Table 3). The Photoshop method is based entirely on user-selected regions, and variances for repeated measurement of the same image in Photoshop reflect this. By comparison, RootScan makes measurements on automated selections that are corrected by the user only if necessary. In general, greater variances were observed for secondary variables, such as percent aerenchyma, than for primary variables in repeated measurement of the same image. Secondary variables are prone to compounding of errors from primary variables.
As with other image analysis programs, measurement precision and accuracy in RootScan are dependent upon image quality. The user determines image quality during tissue preparation, sectioning and image capture. A high-quality image contains minimal debris, and depicts a high-contrast, thin section (<50 μm) centered in the frame. The method of tissue selection, preparation and sectioning can affect image quality, as does the experience of the researcher. Simulated reductions in image quality show that some variables are more affected by certain image quality issues than others (Figs. 3 and 4). Decreased focus within an image appears to affect the identification of small objects, such as cells, more than identification of larger objects, like aerenchyma lacunae or the perimeter of the cortex (Fig. 3). Shearing affects the data accuracy in several variables because it blurs the edges of large and small objects alike, but may only do so in part of the image (Fig. 3, Online resource 5). Finally, underexposed (very light images) are more problematic than overexposed images due to the fact that perimeters of some objects may completely disappear in underexposed images (Fig. 4). RootScan is capable of analyzing roots of any diameter, provided that the camera and imaging platform can capture the entire cross-section within the image frame. High quality sections of fine roots (<1 mm) may be obtained by embedding in paraffin or resin, and taking sections on a microtome. Roots with a diameter greater than 1 mm may be sectioned without embedding, particularly by experienced researchers. To obtain the thinnest possible sections with unembedded tissues, root segments should be stored in 75–100% alcohol for several weeks to attain a rigid texture.
Automated selection in high contrast images is likely to be consistent in repeated analysis. The test image used for repeated analysis in Photoshop and RootScan was a high contrast image. The cross-section was automatically selected in its entirety each time in RootScan, resulting in a variance of zero for that variable (Table 3). When the border is not correctly defined by the program (e.g. see Online resource 2), user variation would likely increase. Variation for stele and cortex measurements was lower in RootScan than in Photoshop. Automated stele selection in RootScan is determined by contrast between the stele and cortex, and in turn, affects the calculation of stele and cortex areas. Contrast between the stele and cortex is primarily based on tissue density in the stele but is also affected by the thickness of the section. If the contrast is too low, the user may employ a tissue stain to enhance contrast. Tissue staining is optional with RootScan, though users should be consistent in its use within sets of images. If the stele is not correctly selected by RootScan, the user can redraw the border with the polygon selection feature, though this will always introduce some error and takes more time than automated selection. In the test image, some variation occurred in the selection of aerenchyma. High-quality sections and high contrast images increases precision in aerenchyma selection. Among five different users, RootScan’s automated selection features had greater precision than Photoshop. Coefficients of variation for primary variables were generally less than those of secondary variables (Table 4). This result was expected since most of the analysis was carried out by the program rather than the user. A notable exception to smaller coefficients of variation for primary variables was aerenchyma area (CV = 3.52%). Selection of aerenchyma area usually requires some user involvement (Online resource 3), and small differences may occur in the designation of lacunae among users. In general, user intervention reduces precision, and therefore, steps that are not completely user-directed will be less variable in repeated measurement.
RootScan presents an opportunity for further investigation of the value of anatomical traits. The possibility exists for further development and additional applications of this program. Although developed using maize root cross-sections, RootScan can be used with other species, as demonstrated by analysis of images of rice and bean. The primary requirements for data collection are identification of a circular object of interest, the presence of cortex and stele, and high image contrast, therefore use of this software for anatomical analysis of other species is possible. Additional variables of interest may be added such as cross-section, stele or cortex diameter, radial width of aerenchyma region, position of aerenchyma within cross-section, and characteristic shape of lacunae, among others. It is also of interest to develop the ability to separate physiologically important cell types such as the epidermis, pericycle and endodermis as a part of the analysis, if consistent high-resolution images with adequate detail could be obtained. The most recent version of this software is available at the website: http://roots.psu.edu/en/methods.
Conclusion
RootScan is a new tool for high-throughput image analysis of root cross-sectional images, with potential applications in plant breeding, cell biology and agronomy. It improves upon some commonly-used image analysis methods by offering improved workflow organization, greater efficiency and speed, more data per image, and improved accuracy and precision. Possible future development of the program may include addition or flexibility of variables based on species, treatment or specific research objectives.
Abbreviations
- GUI:
-
(graphical user interface)
- RCA:
-
(root cortical aerenchyma)
References
Drew M, Fourcy A (1986) Radial movement of cations across aerenchymatous roots of Zea mays measured by electron probe X-Ray microanalysis. J Exp Bot 37:823–831
Drew MC, Saker LR (1986) Ion transport to the xylem in aerenchymatous roots of Zea mays L. J Exp Bot 37:22–33
Fan MS, Zhu JM, Richards C, Brown KM, Lynch JP (2003) Physiological roles for aerenchyma in phosphorus-stressed roots. Funct Plant Biol 30:493–506
Fan MS, Bai RQ, Zhao XF, Zhang JH (2007) Aerenchyma formed under phosphorus deficiency contributes to the reduced root hydraulic conductivity in maize roots. J Int Plant Biol 49:598–604
Fester T, Hause G (2005) Accumulation of reactive oxygen species in arbuscular mycorrhizal roots. Mycorrhiza 15:373–379
Gonzalez RC, Woods RE (1992) Digital image processing. Addison-Wesley, Reading, MA
Gregory PJ, Bengough AG, Grinev D, Schmidt S, Thomas WTB, Wojciechowski T, Young IM (2009) Root phenomics of crops: opportunities and challenges. Func Plant Biol 36:922–929
Haining RP (2003) Spatial data analysis: theory and practice. Cambridge University Press.
Justin S, Armstrong W (1987) The anatomical characteristics of roots and plant response to soil flooding. New Phytol 106:465–495
Longstreth DJ, Borkhsenious ON (2000) Root cell ultrastructure in developing aerenchyma tissue of three wetland species. Ann Bot 86:641–646
Lynch JP, Ho MD (2005) Rhizoeconomics: carbon costs of phosphorus acquisition. Plant Soil 269:45–56
Otsu N (1979) Threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9:62–66
Postma JA, Lynch JP (2011a) Root cortical aerenchyma enhances growth of Zea mays L. on soils with suboptimal availability of nitrogen, phosphorus and potassium. Plant Physiol 156:1190–1201. doi:10.1104/pp.111.175489
Postma JA, Lynch JP (2011b) Theoretical evidence for the functional benefit of root cortical aerenchyma in soils with low phosphorus availability. Ann Bot 107:829–841
R Development Core Team (2010) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org.
Richards RA, Passioura JB (1989) A breeding program to reduce the diameter of the major xylem vessel in the seminal roots of wheat and its effect on grain yield in rain-fed environments. Aust J Agr Res 40:943–950
Tombesi S, Johnson RS, Day KR, DeJong TM (2010) Relationships between xylem vessel characteristics, calculated axial hydraulic conductance and size-controlling capacity of peach rootstocks. Ann Bot 105:327–331
Wright SR, Jennette MW, Coble HD, Rufty TW (1999) Root morphology of young Glycine max, Senna obtusifolia, and Amaranthus palmeri. Weed Sci 47:706–711
Zhu J, Brown KM, Lynch JP (2010) Root cortical aerenchyma improves the drought tolerance of maize (Zea mays L.). Plant Cell Environ 33:740–749
Zobel RW (2003) Sensitivity analysis of computer-based diameter measurement from digital images. Crop Sci 43:583–5
Acknowledgements
We thank Lauren Gelesh, Johanna Mirenda, Gina Riggio, Andy Evensen, Robert Snyder, and Chinmay Rao for technical assistance. United States Department of Agriculture, National Research Initiative provided funding for this research via grant 207-35100-18365 to JPL and KMB
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Matthias Wissuwa.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1
RootScan screenshots before and after lateral root selection in Zea mays. The user outlines the lateral root segment (a) and it is removed (b). (JPEG 114 kb)
Online resource 2
Example of erroneous automatic cross-section selection. RootScan selected root hairs as part of this rice (Oryza sativa) root cross-section (red line). The user removed them by drawing a polygon that excluded the incorrect areas (blue line). Note that it is not necessary to draw the outline exactly, since the original selection will be used when it is inside the polygon. (JPEG 134 kb)
Online resource 3
Rice root section images during aerenchyma selection and correction. The left image (a) shows that the program missed some aerenchyma lacunae, so the user manually selected the omitted objects by clicking with a crosshair tool. The right image (b) shows the aerenchyma lacunae in red after both RootScan and manual selection. (JPEG 215 kb)
Online resource 4
RootScan incorrectly included part of the cortex of this field-grown maize root during the stele selection step (green line). The user redraws the stele boundary with a polygon tool (blue line). The error was caused by the high-contrast areas in the inner cortex due to fungal infection. (JPEG 124 kb)
Online resource 5
The left image (a) shows a maize root section that has been sheared during sectioning. The cells and lacunae on the upper left are blurred. As a result, the lacunae in that region (image b, in red) are no longer completely visible and the area is reduced. The cells in that area are pixelated and cell counts would be too high. (JPEG 162 kb)
Rights and permissions
About this article
Cite this article
Burton, A.L., Williams, M., Lynch, J.P. et al. RootScan: Software for high-throughput analysis of root anatomical traits. Plant Soil 357, 189–203 (2012). https://doi.org/10.1007/s11104-012-1138-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-012-1138-2