Abstract
Key message
NtCycB2 negatively regulates the initiation of tobacco long stalk glandular trichomes and influences the expression of diterpenoid biosynthesis- and environmental stress resistance-related genes.
Abstract
Many asterid plants possess multicellular trichomes on their surface, both glandular and non-glandular. The CycB2 gene plays a key role in multicellular trichome initiation, but has distinct effects on different types of trichomes; its mechanisms remain unknown. In tomato (Solanum lycopersicum), SlCycB2 negatively regulates non-glandular trichome formation, but its effects on glandular trichomes are ambiguous. In this study, we cloned the SlCycB2 homolog of Nicotiana tabacum, NtCycB2, and analyzed its effect on three types of trichomes, long stalk glandular trichomes (LGT), short stalk glandular trichomes (SGT), and non-glandular trichomes (NGT). Knocking out NtCycB2 (NtCycB2-KO) promoted LGT formation, while overexpression of NtCycB2 (NtCycB2-OE) decreased LGT density. SGT and NGT were not significantly influenced in either NtCycB2-KO or NtCycB2-OE plants, indicating that NtCycB2 regulated only LGT formation in tobacco. In addition, compared with NtCycB2-OE and control plants, NtCycB2-KO plants produced more trichome exudates, including diterpenoids and sugar esters, and exhibited stronger aphid resistance. To further elucidate the function of NtCycB2, RNA-Seq analysis of the NtCycB2-KO, NtCycB2-OE, and control plants was conducted. 2,552 and 1,933 differentially expressed genes (DEGs) were found in NtCycB2-KO and NtCycB2-OE plants, respectively. Gene Ontology analysis of the common DEGs revealed that ion transport, carbohydrate and amino acid metabolism, photosynthesis, and transcription regulation processes were significantly enriched. Among these DEGs, diterpenoid biosynthesis genes were upregulated in NtCycB2-KO plants and downregulated in NtCycB2-OE plants. Two MYB transcription factors and several stress resistance-related genes were also identified, suggesting they may participate in regulating LGT formation and aphid resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant trichomes are hair-like tissues that develop from aerial epidermal cells and can be found on many plant species. Trichomes vary in shape, size, metabolism, and secretion ability, and may be unicellular or multicellular, glandular or non-glandular (Huchelmann et al. 2017). Trichomes play an important role in response to biotic stress, as they act as a physical barrier against herbivores. Moreover, glandular trichomes can synthesize, store, and secrete large amounts of exudates, including alkaloids, polysaccharides, terpenoids, polyphenols, organic acids, and defensive proteins. In turn, these exudates can entrap or poison herbivores and prevent pathogen infection (Huchelmann et al. 2017; Alain 2018). In addition, trichomes provide protection against abiotic stressors, such as UV–B radiation, drought, and high temperatures (Karabourniotis et al. 1995, 2019).
As trichomes are easily accessed, and their development is precisely controlled, they are considered an excellent model for studying the molecular mechanisms involved in controlling plant cell division and differentiation (Yang and Ye 2013). In Arabidopsis, which is covered by unicellular non-glandular trichomes, the regulatory network controlling trichome initiation and development has been extensively studied, and many regulatory genes have been identified. These genes include GLABRA1 (GL1), which encodes an R2R3 MYB transcription factor, GLABRA3 (GL3) and its homolog ENHANCER OF GL3 (EGL3), which encode basic helix-loop-helix (bHLH) transcription factors, and TRANSPARENT TESTA GLABRA1 (TTG1), which encodes a WD-40 repeat protein. These proteins are able to form a MYB-bHLH-WD-repeat complex (Zhao et al. 2008), which activates the expression of GLABRA2 (GL2), a gene encoding a homeodomain transcription factor that promotes trichome initiation (Rerie et al. 1994). In addition to these positive regulatory genes, negative regulatory genes have also been identified, including TRIPTYCHON (TRY), CAPRICE (CPC), ENHANCER OF TRY AND CPC 1 (ETC1), ETC2, TRICHOMELESS1 (TCL1), TCL2, and CPL3, which encode R3 MYB transcription factors that compete with GL1 for binding to GL3/EGL3 to form an inactive complex (Ishida et al. 2008).
In contrast to Arabidopsis with its unicellular non-glandular trichomes, many asterid plants possess multicellular glandular trichomes (Yang et al. 2015). However, overexpression of Arabidopsis GL1 in tobacco plants does not influence glandular trichome formation (Payne et al. 1999), suggesting that the molecular mechanism involved in controlling glandular trichome development may differ from that in Arabidopsis. Our current knowledge regarding this mechanism is limited; however, advances have been made with the discovery of some vital transcription factors (Chalvin et al. 2020). For instance, the R2R3 MYB transcription factor MIXTA has been found to positively regulate glandular trichome initiation in Artemisia annua (Shi et al. 2018) and tomato (Solanum lycopersicum) (Ewas et al. 2016). Another MYB protein in A. annua, AaMYB1, also promotes glandular trichome development (Matías-Hernández et al. 2017). In tomato (S. lycopersicum), the C2H2 zinc-finger gene Hair (Chang et al. 2018) and bHLH gene SlMYC1 (Xu et al. 2018) have also been shown to positively regulate glandular trichome formation.
HD-ZIP IV transcription factors are also important regulators of glandular trichome development. For instance, AaHD1 and AaHD8 are two HD-ZIP IV proteins in A. annua that play important roles in glandular trichome development (Yan et al. 2017, 2018), while the AaHD8 homolog SlCD2 promotes glandular trichome initiation in tomato (S. lycopersicum) (Nadakuduti et al. 2012). Woolly (Wo) is another HD-ZIP IV transcription factor gene that controls glandular trichome formation in tomato (S. lycopersicum) (Yang et al. 2011). Microarray analysis has shown that Wo upregulates the expression of SlCycB2, a B-type cyclin gene, and direct protein–protein interactions are formed between Wo and SlCycB2 (Yang et al. 2011). Overexpression of SlCycB2 results in significantly decreased numbers of both glandular and non-glandular trichomes. Meanwhile, RNA interference (RNAi) of SlCycB2 leads to increased density of non-glandular type III and V trichomes and decreased density of glandular type I trichomes. However, glandular type IV and VI trichome density are not changed by RNAi of SlCycB2 (Gao et al. 2017). These findings suggest that SlCycB2 is a key regulatory gene in tomato trichome development, but also that it has different effects on the various types of trichomes. Specifically, SlCycB2 negatively regulates non-glandular trichome formation, but has ambiguous effects on glandular trichomes.
In our previous study, we cloned the homologs of Wo and SlCycB2 in Nicotiana benthamiana and found that NbWo and NbCycB2 are able to control trichome development. Similar to that in tomato, NbWo could positively regulate trichome formation; however, unlike that in tomato, NbCycB2 could negatively regulate glandular trichome formation. We further demonstrated that NbWo is able to directly bind the NbCycB2 promoter and promote its expression. On the other hand, NbCycB2 represses the activity of NbWo through protein-protein interactions, thereby negatively regulating trichome formation (Wu et al. 2020).
Although the functions of Wo and CycB2 in controlling glandular trichome initiation have been characterized and the relationships between them identified, this cannot explain the reason for CycB2 having different effects on different types of trichomes, and thus, further information is still needed.
Taking this in consideration, we chose Nicotiana tabacum as a model system as it contains only three types of trichomes (Cui et al. 2011), being much simpler than tomato, which contains seven types (McDowell et al. 2011). We cloned NtCycB2, the homolog of CycB2 in N. tabacum, and then constructed its overexpression (NtCycB2-OE) and knockout (NtCycB2-KO) lines. The effects of NtCycB2 on trichome density and exudate content were analyzed by comparing the wild-type, overexpression, and knockout lines. We also conducted transcriptome analysis of NtCycB2-KO, NtCycB2-OE, and control plants to further explore the function of NtCycB2. Finally, the newly gained information and its implications for understanding the function of NtCycB2 and the molecular mechanisms of multicellular glandular trichome formation were discussed.
Materials and methods
Plant materials and growth conditions
Tobacco (Nicotiana tabacum) cultivar K326 was used as the wild type, plants were grown under greenhouse conditions of 16 h light/8 h dark and 25 °C/20 °C day/night temperature.
Sequence analysis
The coding sequence (CDS) of SlCycB2 (Gao et al. 2017) and NbCycB2 (Wu et al. 2020) were used as queries to blast against the reference genome sequence of N. tabacum at Sol Genomics Network (SGN) database (https://solgenomics.net/), and e-value < 1e−10 was set as the query threshold. The CDS and amino acid sequence of the putative NtCycB2 genes were downloaded; multiple sequence alignments were carried out using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/) using default parameters.
Vector construction and transformation of N. tabacum
The CRISPR/Cas9-mediated targeted mutagenesis was used to knockout NtCycB2, with the methods as described before (Gao et al. 2015), the targeting sequence (GTGATCAACCACCCACAAG) and its reverse complement oligomer were chemically synthesized and annealed, the dimers obtained were then inserted into the pORE-Cas9 binary vector (Gao et al. 2015). For NtCycB2 overexpression, the full-length CDS were amplified (Primers are listed in Table S1) from the cDNA of N. tabacum leaves, and inserted into the pCXSN vector under the control of CaMV 35 S promoter (Chen et al. 2009).
The constructs obtained above were separately transformed into the competent cells of Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method. Leaf discs from 6-week-old N. tabacum were infected by A. tumefaciens strain GV3101 harboring the NtCycB2 knockout and overexpression vectors respectively using the methods described before (Gao et al. 2015). The leaf discs infected were then plated onto the regeneration medium (MS medium contains 3% (w/v) sucrose, 1 mg/L 6-Benzylaminopurine, 0.15 mg/L 1-Naphthaleneacetic acid and 50 mg/L Cefotaxime sodium, 8 mg/L hygromycin, pH 5.6). The hygromycin resistant seedlings were obtained, the genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Germany) and used for further examination.
For the screening of NtCycB2-OE plants, PCR amplification of the hygromycin resistant gene from the genomic DNA was performed for preliminary screening (primers used were shown in Table S1), and the positive singletons were further confirmed by qRT-PCR measurement of NtCycB2 expression level.
For the screening of NtCycB2-KO plants, the NtCycB2 gene was amplified by PCR from genomic DNA (primers used were shown in Table S1), the PCR products were Sanger sequenced to detect the mutations.
Observation of trichome morphology
At the flowering stage, the newly emerged leaves (5 cm in length) of NtCycB2-KO, NtCycB2-OE, and control plants were collected and stained with 2% rhodamine B for 30 min, after washed away the unbound dye, the trichome was observed using a VHX-5000 microscope (Keyence, Japan). The trichome density was counted and shown as trichome number per mm2 leaf area.
Analysis of trichome exudates
At the flowering stage, the trichome exudates of NtCycB2-KO, NtCycB2-OE and control plants were analyzed. Five healthy plants were selected from each line, and their middle leaves (10th to 12th leaf from the stem base) were collected. Leaf disks of 10 cm in diameter were cut out along both sides of the midrib, a total of 60 disks were obtained for each line, and divided into three repeats. Trichome exudates were extracted using the method described before (Huang et al. 2018). Briefly, leaf disks were immersed in 500 ml of dichloromethane for eight times of 2 s each. After that, 1 ml of internal standard solution (mixture of 2.020 mg/ml sucrose octaacetate and 2.542 mg/ml n-heptadecane alcohol) was added into each extract, well mixed and concentrated using a rotary evaporator, and finally dried in a nitrogen concentration. The extracts were then subjected to silylation treatment by adding 500 µl mixture of N,N-Dimethylformamide and N,O-Bis (trimethylsilyl) trifluoroacetamide (1:1, v:v), and incubated in 75 °C for 60 min.
The solutions obtained above were then analyzed by gas chromatography-mass spectrometry (GC-MS) using a TRACE GC ULTRA-DSQ II (Thermo Fisher Scientific, US) system coupled with a DB-5MS column (30 m × 0.25 mm) (Agilent Technologies, US). Helium was used as the carrier gas, the column flow was set as 0.8 ml/min. For each run, a volume of 1 µl sample was analyzed, with the split ratio set as 1:15. The injector temperature was 250 °C, and the oven was programmed to begin at 40 °C for 2 min, then 6 °C per min to 180 °C, hold for 2 min; the temperature then increased by 2 °C per min until it reached to 280 °C, and maintained at this temperature for 20 min. The MS transfer line temperature was 250 °C, and MS conditions were 50–650 AMU scanning range, and the ion source 230 °C. The MS profiles were used to identify components based on the NIST12 spectral library (https://www.nist.gov/srd). For each component, the area under the corresponding GC chromatogram peak was integrated, and compared to that of the internal standard, so that their amounts were calculated.
Aphid bioassay
Tobacco aphids (Myzus nicotianae) resistance of NtCycB2-KO, NtCycB2-OE, and control plants were compared at the 8-leaf stage. Four healthy plants for each line were selected, and ten aphid larvae were inoculated on the leaves for each plant. Each plant was confined to a cage assembled with an insect proof net, and grown in a chamber with conditions of 25 °C, 70% relative humidity, and 16:8 h light/dark photoperiod. The aphid number was calculated every week for a nine-week period.
RNA extraction
Different plant tissues were harvested and frozen immediately using liquid nitrogen. Total RNA was isolated using TRIzol reagent (Invitrogen, US). The integrity of the RNA samples was examined by 1% agarose gel electrophoresis. The quality and quantity of these RNA samples were further determined by measuring the relative absorbance at 260/280 and 260/230 nm using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, US).
RNA-Seq analysis
The RNA-Seq libraries were produced and sequenced in Shanghai OE Biotech Co., Ltd (China) using the Illumina HiSeqTM 2000 sequencing platform. 150 bp paired-end clean reads were generated from the raw data by removing the adaptors and low-quality reads. All the clean reads were then mapped to the N. tabacum reference genome (ftp://ftp.solgenomics.net/genomes/Nicotiana_tabacum/edwards_et_al_2017/assembly/Nitab-v4.5_genome_Scf_Edwards2017.fasta) (Edwards et al. 2017) using hisat2 software (Kim et al. 2015). Gene expression levels were calculated as FPKM, and differentially expressed genes (DEGs) were identified using the DESeq R package (Anders and Huber 2012), P-value < 0.05, fold change < 0.5 or > 2 were set as the thresholds for significantly differential expression. To analysis the function of DEGs, Gene Ontology (GO) functional enrichment analysis was performed using topGO (Alexa and Rahnenfuhrer 2010) software.
Quantitative real time PCR (qRT-PCR)
First-strand cDNA was synthesized from 2 µg of total RNA using Goscritp reverse transcriptase (Promega, US). qRT-PCR reaction mixture contains 10 µl of SYBR Green I Master Mix (TOYOBO, Japan), 0.5 µl of forward primers (10 µM), 0.5 µl of reverse primers (10 µM), 1 µl of cDNA template and 8 µl of distilled water. qRT-PCR was performed on the Roche Light Cycler 480 system (Roche, Switzerland) with the parameters set as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, annealing temperature for 30 s, and 72 °C for 20 s. A melting curve was established by slow heating from 60 to 95 °C within 20 min. Relative gene expression levels were calculated using 2−ΔΔCt, with three technical replicates for each sample and the results were shown as means ± standard deviation (SD). All the primers used are listed in Table S1.
Statistical analysis
For the experiments described above, three separate tests were carried out for each sample. The data obtained were subjected to one-way analysis of variance (ANOVA) using IBM SPSS Statistics 19 software (IBM, New York, USA), followed by the least significant difference multiple comparison test. Statistical significance of the difference between data obtained from different materials was considered at P < 0.05.
Results
NtCycB2 negatively regulated tobacco long stalk glandular trichome (LGT) initiation
Two loci, Nitab4.5_0014697g0010.1 and Nitab4.5_0012062g0010.1, were identified as putative homologs of CycB2 in the N. tabacum reference genome (Edwards et al. 2017) and were arbitrarily designated as NtCycB2-1 and NtCycB2-2, respectively. Both contained a 333-bp open reading frame (Fig. S1a), and no introns were found in their genomic sequence. Their protein sequences were identical to each other (Fig. S1b) and showed 96.4% and 71.1% identity to NbCycB2 and SlCycB2 protein sequences, respectively.
The expression pattern of NtCycB2 was examined in different plant tissues, including roots, stems, leaves, flowers, and trichomes separated from leaves. NtCycB2 showed the highest expression in trichomes and the lowest in roots, while its expression in flowers was higher than that in leaves and stems. When comparing NtCycB2-1 and NtCycB2-2, we found that NtCycB2-1 was expressed at higher levels than that of NtCycB2-2 in all tissues except the roots (Fig. S1c).
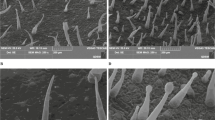
We then constructed the NtCycB2 knockout line NtCycB2-KO using a CRISPR-Cas9 system. Sanger sequencing showed that a 7-bp deletion was induced for NtCycB2-1, and a 5-bp deletion coupled with an A/T bp insertion was found for NtCycB2-2 (Fig. 1a). The NtCycB2-1 coding sequence was used to construct the overexpression line NtCycB2-OE. A pair of conserved primers (Table S1) was designed to estimate the total NtCycB2-1 and NtCycB2-2 expression. Based on qRT-PCR analysis, there was a 58-fold higher expression level of NtCycB2 in NtCycB2-OE than that in control plants (Fig. 1b). Three types of trichomes, long stalk glandular trichomes (LGT), short stalk glandular trichomes (SGT), and non-glandular trichomes (NGT) were found on the leaf surface of wild-type plants (Fig. 1d). Compared to that of the control plants, significantly more trichomes were observed on the leaves and stems of NtCycB2-KO plants (Fig. 1e). In contrast, a hairless phenotype was observed in NtCycB2-OE plants (Fig. 1f). The densities of LGT, SGT, and NGT were individually calculated to elucidate detailed changes. The results showed that LGT density in NtCycB2-KO plants was two-fold higher than that of the control plants, whereas in NtCycB2-OE plants was 15% of that in control plants. However, no significant changes were found for SGT and NGT densities in either NtCycB2-KO or NtCycB2-OE plants compared to those of the control plants (Fig. 1c). We also examined the flower phenotypes, since NtCycB2 showed a higher expression in flowers than that in leaves, as shown in Fig. S2, flowers of both NtCycB2-KO and NtCycB2-OE plants were well developed. LGT density was increased in NtCycB2-KO flowers but decreased in NtCycB2-OE flowers, which is similar to that in leaves. These results suggest that NtCycB2 negatively regulated the formation of LGT, but had no effect on that of SGT or NGT.
Construction of NtCycB2 knockout and overexpression lines and observation of leaf trichome. a Targeted mutagenesis of NtCycB2 gene using CRISPR-Cas9 system. DNA insertions and deletions are shown in red letters and dashes. PAM, the protospacer adjacent motif. b Relative expression of NtCycB2 in NtCycB2-KO, NtCycB2-OE and control plants measured by qRT-PCR, the columns and bars represent the means and standard errors (n = 3) respectively. The relative expression levels in different plants were calculated by setting the expression value in control plants as 1. c Statistics of the leaf trichome density. Representative phenotype pictures for d wild type, e NtCycB2-KO and f NtCycB2-OE are shown here
NtCycB2 affected the accumulation of leaf surface chemical components
We analyzed the trichome exudates of NtCycB2-KO, NtCycB2-OE, and control plants. The main components of tobacco glandular trichomes are generally diterpenoids and sugar esters (Huang et al. 2018). GC-MS analysis showed that the quantity of both diterpenoids and sugar esters in NtCycB2-KO plants was significantly increased, specifically 1.76-fold for diterpenoids and 1.36-fold for sugar esters, compared to that in control plants (Fig. 2b, c). Conversely, the quantity of diterpenoids and sugar esters in NtCycB2-OE plants was dramatically decreased to only 7.1% for diterpenoids and 24.4% for sugar esters relative to that in wild-type plants (Fig. 2b, c).
GC-MS analysis of glandular trichome exudates. a GC-MS profiles of trichome exudates on control, NtCycB2-KO and NtCycB2-OE plants. b Statistics of diterpenoids content. c Statistics of sugar ester content. The columns and bars represent the means and standard errors (n = 3) respectively. The means marked by different letters are statistically significant (P < 0.05)
NtCycB2 affected tobacco resistance to aphids
We investigated aphid resistance differences between NtCycB2-KO, NtCycB2-OE, and control plants by comparing aphid survival and reproduction. Ten aphids were transferred to each tobacco plant, and the aphid number was determined weekly. Our results showed that the aphid number for NtCycB2-OE plants was much higher than that for control plants throughout the experimental period (Fig. 3b, d). In contrast, NtCycB2-KO plants showed fewer aphids than that of the control plants (Fig. 3c, d). The aphid number reached its highest point five weeks after the initial infestation, and the average aphid number for NtCycB2-OE plants was 3.5-fold higher than that for control plants. In contrast, the aphid number for the NtCycB2-KO plants was reduced to only 25% of that for wild-type plants (Fig. 3d). These results suggest that aphid resistance was increased in NtCycB2-KO plants and decreased in NtCycB2-OE plants.
NtCycB2 manipulation triggered gene expression profile changes
We performed a comparative transcriptome analysis of NtCycB2-KO, NtCycB2-OE, and control plants. Three independent biological replicates were used for each line. Approximately 47 million paired-end clean reads of 150 bp in length were generated for each sample. Approximately 97% of the clean reads were successfully mapped to the N. tabacum reference genome (Edwards et al. 2017), and approximately 87% of the reads were uniquely mapped (Table S2). Sample cluster analysis based on gene expression showed that the biological replicates were more highly correlated within samples than between samples (Fig. S3a). These results suggest that the quality of the sequence data was sufficient for subsequent gene expression analysis.
Gene expression levels were compared between different samples, and differentially expressed genes (DEGs) were defined as fold change < 0.5 or > 2. Statistical significance was set at P-value < 0.05. Compared with that of the control plants, 1,235 DEGs were upregulated and 1,317 DEGs were downregulated in NtCycB2-KO plants, while 1,063 DEGs were upregulated and 870 DEGs were downregulated in NtCycB2-OE plants. To further confirm the RNA-Seq results, 17 DEGs were randomly selected, and their expression were validated using qRT-PCR (genes and primers are listed in Table S1). The results showed that gene expression levels measured by qRT-PCR and RNA-Seq had a strong correlation (R2 = 0.71) (Fig. S3b), indicating the high quality of our RNA-Seq results.
Among the 2,552 total DEGs in NtCycB2-KO plants and 1,933 total DEGs in NtCycB2-OE plants, 695 were shared by both plant lines (Fig. 4a). We performed Gene Ontology (GO) enrichment of these common DEGs to evaluate their functional categories. The GO terms of the enriched biological processes are listed in Table S5. The top 20 most significant GO terms are shown in Fig. 4b and are mainly involved in ion transport, carbohydrate and amino acid metabolism, photosynthesis, and transcription regulation processes.
RNA-Seq analysis of control, NtCycB2-KO and NtCycB2-OE plants. a Venn diagram showing common and exclusive DEGs between NtCycB2-KO and NtCycB2-OE. b Top 20 most significant enriched GO terms of the common 695 DEGs between NtCycB2-KO and NtCycB2-OE. c Venn diagram showing common and exclusive DEGs that up regulated in NtCycB2-KO and down regulated in NtCycB2-OE. d Venn diagram showing common and exclusive DEGs that down regulated in NtCycB2-KO and up regulated in NtCycB2-OE. e Gene numbers of secondary metabolites biosynthesis and resistance response among the 25 DEGs that up regulated in NtCycB2-KO and down regulated in NtCycB2-OE. f Gene numbers of secondary metabolites biosynthesis and resistance response among the 13 DEGs that down regulated in NtCycB2-KO and up regulated in NtCycB2-OE
We speculated that the DEGs showing opposite changes in NtCycB2-KO and NtCycB2-OE plants were functionally associated with NtCycB2. As shown in the Venn diagram (Fig. 4c), 25 DEGs were upregulated in NtCycB2-KO plants and downregulated in NtCycB2-OE plants, while only 13 DEGs were downregulated in NtCycB2-KO plants and upregulated in NtCycB2-OE plants (Fig. 4d), and they were listed in Table S6. Among the group of 25 DEGs in Fig. 4c and 13 were related to secondary metabolite biosynthesis and transport, while five were related to abiotic or biotic stress resistance (Fig. 4e, Table S6). Among the group of 13 DEGs in Fig. 4d, three were related to secondary metabolite biosynthesis and four were related to abiotic or biotic stress resistance (Fig. 4f, Table S6). These results indicated that in addition to trichome formation, NtCycB2 may also regulate metabolic processes and environmental stress resistance.
Interestingly, among the group of 25 DEGs in Fig. 4c, two transcription factor genes were identified, namely Nitab4.5_0004186g0020.1 and Nitab4.5_0005769g0080.1, both belonging to the MYB family. Expression of Nitab4.5_0004186g0020.1 and Nitab4.5_0005769g0080.1 was confirmed using qRT-PCR (Fig. S4), indicating that they may work in conjunction with NtCycB2 to control the formation of glandular trichomes.
NtCycB2 affected the expression of diterpenoid biosynthesis-related genes
Changes in the amounts of trichome exudate and the DEGs related to secondary metabolite biosynthesis prompted us to compare the expression levels of diterpenoid biosynthesis genes. In tobacco glandular trichomes, diterpenoid biosynthesis is catalyzed by four enzymes, cembratrien-ol synthases (CBTS), cytochrome P450 (CYP71D16), copalyl diphosphate synthases (CPS2), and abienol synthase (ABS) (Wang and Wagner 2003; Sallaud et al. 2012). Based on the fragments per kilobase of transcript per million mapped reads (FPKM) values of these four gene families, we found that most of them were upregulated in NtCycB2-KO plants, but downregulated in NtCycB2-OE plants (Fig. 5). To further confirm these results, one member from each gene family was randomly selected, and their expression analyzed using qRT-PCR. Although some quantitative differences existed at the expression level, the qRT-PCR results revealed similar expression patterns to those of the RNA-Seq data (Fig. S4).
Homologs of known trichome development-related genes were not regulated by NtCycB2
Many trichome development regulatory genes have been identified, such as GL1, GL3/EGL3, and TTG1 in Arabidopsis (Pattanaik et al. 2014), Wo and SlCycB2 in tomato (Yang et al. 2011; Gao et al. 2017), and AaMIXTA and AaHD1 in A. annua (Yan et al. 2017; Shi et al. 2018). We compared the expression levels of tobacco homologs of 23 known trichome development-related genes (Table S7). Although the expression of some genes changed, these expression changes were only significant for a small number of genes, and changes in expression were not consistent with changes in trichome density. This suggested that these genes may not be associated with NtCycB2.
Discussion
Trichomes, especially glandular trichomes, can protect plants from damage by various biotic and abiotic stressors (Glas et al. 2012), thus increasing trichome density is an efficient way to improve plant resistance. Therefore, a deep study of the molecular mechanisms controlling multicellular glandular trichome formation will aid in increasing trichome density. CycB2 encodes a putative B-type cyclin and is a key gene regulating trichome initiation in tomato and N. benthamiana (Gao et al. 2017; Wu et al. 2020), but the detailed mechanism remains unclear.
In this study, we cloned NtCycB2, the homolog of CycB2 in N. tabacum. We then analyzed the role of NtCycB2 in the formation of different types of trichomes, biosynthesis of trichome exudates, and aphid resistance by generating overexpression and knockout tobacco lines. Furthermore, we identified genes through RNA-Seq analysis that may be regulated by NtCycB2. Our results provide new information regarding the function of NtCycB2.
NtCycB2 has different roles in glandular and non-glandular trichome formation
Previous studies have shown that suppression of CycB2 increases trichome density in both tomato and N. benthamiana, while overexpression of CycB2 causes a hairless phenotype (Gao et al. 2017; Wu et al. 2020). We obtained similar results in our study, but differences still exist in these similar appearances. In tomato plants, which are covered by seven types of trichomes (Glas et al. 2012), the non-glandular trichome (type III and V) numbers are dramatically decreased in SlCycB2 overexpression plants, but significantly increased in SlCycB2 RNAi lines, suggesting that non-glandular trichomes formation is negatively regulated by SlCycB2. Regarding glandular trichomes (type I, IV, VI, and VII), type I are almost absent in both SlCycB2 overexpression and RNAi plants, whereas type IV and VI glandular trichome density is decreased in SlCycB2 overexpression lines but not changed in SlCycB2 RNAi plants (Gao et al. 2017). This makes it difficult to define the role of SlCycB2 in glandular trichome formation. In contrast, CycB2 in N. benthamiana has been shown to be a negative regulator of glandular trichomes, as their density decreases in NbCycB2 overexpression lines and increases in NbCycB2 RNAi lines (Wu et al. 2020). However, N. benthamiana does not possess non-glandular trichomes and thus, it was not clear whether CycB2 has any effects on non-glandular trichomes in tobacco plants. In our study, NtCycB2 negatively regulated LGT formation but had no effect on SGT or NGT formation (Fig. 1). These inconsistent results indicate that CycB2 has different roles in the formation of glandular and non-glandular trichomes and that the function of CycB2 in trichome formation may differ among Solanaceae species.
Previous studies have also found that the function of some trichome regulatory genes is conserved among different species, such as MIXTA in A. annua (Shi et al. 2018) and tomato plants (Ewas et al. 2016), and AaHD8 in A. annua (Yan et al. 2018) and its homolog SlCD2 in tomato plants (Nadakuduti et al. 2012). However, few studies have compared their roles in different trichome types among species. Here, we found that even though CycB2 decreased trichome density in tomato plants, N. benthamiana, and N. tabacum, it had different impacts on glandular and non-glandular trichomes. This finding expands our knowledge about multicellular trichome formation.
NtCycB2 negatively regulates the accumulation of leaf surface chemical components and aphid resistance
Trichomes play an important role in response to biotic stress, as they not only act as a physical barrier against herbivores, but, due to their exudates, also provide chemical protection (Huchelmann et al. 2017; Alain 2018). Consistent with this, our study showed that knocking out of NtCycB2 significantly increased aphid resistance (Fig. 3). Based on our results, we speculate that NtCycB2 regulated tobacco aphid resistance in three ways.
First, NtCycB2 was able to dramatically influence trichome density, which directly enhanced the physical barrier. Furthermore, more glandular trichomes meant more “factories” for the biosynthesis of exudates, which further strengthened aphid resistance.
Second, NtCycB2 negatively regulated the expression of trichome exudate biosynthesis genes, as they were upregulated in NtCycB2-KO plants and downregulated in NtCycB2-OE plants (Fig. 5). Consistent with this, NtCycB2 showed a higher expression level in trichomes than that in other tissues we examined (Fig. S1c). Thus, changes in the accumulation of leaf surface chemical components may have been partially due to changes in the expression of these genes. It has also been found in previous studies that some trichome formation-related transcription factors have a dual function. In addition to controlling trichome initiation, these transcription factors are able to regulate trichome metabolism. For example, the AP2 transcription factor TAR1 of A. annua regulates the development of trichomes and biosynthesis of artemisinin (Tan et al. 2015), and the transcription factor SlMYC1 of tomato plants regulates type VI glandular trichome formation and terpenoid biosynthesis (Xu et al. 2018). A recent study showed that SlWo in tomato plants can directly bind the promoter of terpene synthase genes and activate their expression (Hua et al. 2021), indicating that the CycB2-Wo pathway (Wu et al. 2020) may also participate in tobacco trichome secondary metabolite biosynthesis.
Third, NtCycB2 regulated the expression of other resistance-related genes. Among the DEGs that exhibited opposite changes in NtCycB2-KO and NtCycB2-OE plants, many were involved in abiotic/biotic stress resistance (Table S6), which provided additional protection against aphid invasion. Similar to our results, SlCycB2 in tomato plants regulates the expression of defense-related genes, such as protein inhibitors (Gao et al. 2017).
NtCycB2 may regulate trichome formation through pathways that different from those of known genes
NbCycB2 has been found to influence trichome formation through the CycB2-Wo feedback loop by suppressing the activity of the NbWo protein in N. benthamiana (Wu et al. 2020). However, it is unknown whether other genes are involved in this process. Therefore, further investigation is needed.
Trichome formation regulating genes have been identified in many species, such as Arabidopsis (Pattanaik et al. 2014), cotton (Yang and Ye 2013), A. annua, and tomato (Chalvin et al. 2020), and previous studies suggest that their function may be conserved in different plant species (Payne et al. 1999; Chang et al. 2018). We identified the homologs of 23 known trichome-related genes in tobacco (Table S7), and their expression in NtCycB2-KO, NtCycB2-OE, and control plants were compare using RNA-Seq data. However, none showed an expression pattern related with the changes in glandular trichome density. Similarly, in a previous study, the expression of known trichome-related genes showed no significant changes when tomato Wo was ectopically expressed in N. tabacum (Yang et al. 2015). This indicates that the expression of these genes is not regulated by NtCycB2 and that they may not participate in NtCycB2-regulated trichome formation pathways.
Interestingly, two transcription factor genes were identified among the DEGs that were upregulated in NtCycB2-KO plants and downregulated in NtCycB2-OE plants, Nitab4.5_0004186g0020.1 and Nitab4.5_0005769g0080.1 (Table S6).
Nitab4.5_0004186g0020.1 is a RADIALIS-like protein, which is a member of MYB transcription factor family. A previous study showed that the RADIALIS-like gene GbRL1 of cotton is strongly expressed in ovules and cotton fiber during the initiation stage. The authors of that study inferred that GbRL1 may play a role in fiber initiation (Zhang et al. 2011), which is a process similar to trichome formation. Meanwhile, Nitab4.5_0005769g0080.1 is a homolog of the Arabidopsis MYB108 protein. In Arabidopsis, MYB108 is a jasmonic acid (JA)-inducible transcription factor that plays an important role in stamen development (Mandaokar and Browse 2009) and biotic stress responses (Mengiste et al. 2003). In many plant species, it has been found that JA plays an important role in trichome development (Boughton et al. 2005; Qi et al. 2011; Yan et al. 2017), suggesting that this gene may also regulate glandular trichome development in tobacco. We hypothesize that these two transcription factors may regulate tobacco glandular trichome development, but this requires further study.
Data Availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files). The raw filtered RNA-seq data used in this article are available in the NCBI Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra/) under BioProject Accession: PRJNA622901.
Abbreviations
- DEG:
-
Differentially expressed gene
- GC–MS:
-
Gas chromatography–mass spectrometry
- LGT:
-
Long stalk glandular trichomes
- NGT:
-
Non-glandular trichomes
- qRT-PCR:
-
Quantitative real time PCR
- SGT:
-
Short stalk glandular trichomes
References
Alain T (2018) Harnessing plant trichome biochemistry for the production of useful compounds. In: Kermode AR, Jiang L (eds) Molecular pharming: applications, challenges, and emerging areas. Wiley, Hoboken, pp 353–382
Alexa A, Rahnenfuhrer J (2010) topGO: enrichment analysis for gene ontology. Bioconductor R package. http://www.bioconductor.org/packages/2.6/bioc/vignettes/topGO/inst/doc/topGO.pdf. Accessed 4 June 2010.
Anders S, Huber W (2012) Differential expression of RNA-Seq data at the gene level–the DESeq package. Eur Mol Biol Lab. https://www.genomatix.de/online_help/help_regionminer/DESeq_1.10.1.pdf. Accessed 12 Sept 2012.
Boughton AJ, Hoover K, Felton GW (2005) Methyl jasmonate application induces increased densities of glandular trichomes on tomato, Lycopersicon esculentum. J Chem Ecol 31(9):2211–2216. https://doi.org/10.1007/s10886-005-6228-7
Chalvin C, Drevensek S, Dron M, Bendahmane A, Boualem A (2020) Genetic control of glandular trichome development. Trends Plant Sci 25(5):477–487. https://doi.org/10.1016/j.tplants.2019.12.025
Chang J, Yu T, Yang Q, Li C, Xiong C, Gao S, Xie Q, Zheng F, Li H, Tian Z, Yang C, Ye Z (2018) Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J 96(1):90–102. https://doi.org/10.1111/tpj.14018
Chen S, Songkumarn P, Liu J, Wang G-L (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150(3):1111–1121. https://doi.org/10.1104/pp.109.137125
Cui H, Zhang S-T, Yang H-J, Ji H, Wang X-J (2011) Gene expression profile analysis of tobacco leaf trichomes. BMC Plant Biol 11(1):76. https://doi.org/10.1186/1471-2229-11-76
Edwards KD, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans AD, Bombarely A, Allen F, Hurst R, White B, Kernodle SP, Bromley JR, Sanchez-Tamburrino JP, Lewis RS, Mueller LA (2017) A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom 18:448. https://doi.org/10.1186/s12864-017-3791-6
Ewas M, Gao Y, Wang S, Liu X, Zhang H, Nishawy EME, Ali F, Shahzad R, Ziaf K, Subthain H, Martin C, Luo J (2016) Manipulation of SlMXl for enhanced carotenoids accumulation and drought resistance in tomato. Sci Bull 61(18):1413–1418. https://doi.org/10.1007/s11434-016-1108-9
Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87(1):99–110. https://doi.org/10.1007/s11103-014-0263-0
Gao S, Gao Y, Xiong C, Yu G, Chang J, Yang Q, Yang C, Ye Z (2017) The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci 262:103–114. https://doi.org/10.1016/j.plantsci.2017.05.006
Glas J, Schimmel B, Alba J, Escobar-Bravo R, Schuurink R, Kant M (2012) Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int J Mol Sci 13(12):17077–17103. https://doi.org/10.3390/ijms131217077
Hua B, Chang J, Wu M, Xu Z, Zhang F, Yang M, Xu H, Wang L-J, Chen X-Y, Wu S (2021) Mediation of JA signalling in glandular trichomes by the woolly/SlMYC1 regulatory module improves pest resistance in tomato. Plant Biotechnol J 19(2):375–393. https://doi.org/10.1111/pbi.13473
Huang M, Zhang H, Wang Z, Niu D, Li Y, Cui H (2018) Comparative studies of leaf surface chemical biosynthesis in different tobacco cultivars. Acta Physiol Plant 40(4):67. https://doi.org/10.1007/s11738-018-2642-7
Huchelmann A, Boutry M, Hachez C (2017) Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol 175(1):6–22. https://doi.org/10.1104/pp.17.00727
Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59(1):365–386. https://doi.org/10.1146/annurev.arplant.59.032607.092949
Karabourniotis G, Kotsabassidis D, Manetas Y (1995) Trichome density and its protective potential against ultraviolet-B radiation damage during leaf development. Can J Bot 73(3):376–383. https://doi.org/10.1139/b95-039
Karabourniotis G, Liakopoulos G, Nikolopoulos D, Bresta P (2019) Protective and defensive roles of non-glandular trichomes against multiple stresses: structure–function coordination. J For Res 31(1):1–12. https://doi.org/10.1007/s11676-019-01034-4
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. https://doi.org/10.1038/NMETH.3317
Mandaokar A, Browse J (2009) MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol 149(2):851–862. https://doi.org/10.1104/pp.108.132597
Matías-Hernández L, Jiang W, Yang K, Tang K, Brodelius PE, Pelaz S (2017) AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J 90(3):520–534. https://doi.org/10.1111/tpj.13509
McDowell ET, Kapteyn J, Schmidt A, Li C, Kang JH, Descour A, Shi F, Larson M, Schilmiller A, An L, Jones AD, Pichersky E, Soderlund CA, Gang DR (2011) Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol 155(1):524–539. https://doi.org/10.1104/pp.110.167114
Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15(11):2551–2565. https://doi.org/10.1105/tpc.014167
Nadakuduti SS, Pollard M, Kosma DK, Allen C, Ohlrogge JB, Barry CS (2012) Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiol 159(3):945–960. https://doi.org/10.1104/pp.112.198374
Pattanaik S, Patra B, Singh SK, Yuan L (2014) An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front Plant Sci 5:259. https://doi.org/10.3389/fpls.2014.00259
Payne T, Clement J, Arnold D, Lloyd A (1999) Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development 126(4):671–682. https://doi.org/10.1242/dev.126.4.671
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23(5):1795–1814. https://doi.org/10.1105/tpc.111.083261
Rerie WG, Feldmann KA, Marks MD (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Gene Dev 8(12):1388–1399. https://doi.org/10.1101/gad.8.12.1388
Sallaud C, Giacalone C, Topfer R, Goepfert S, Bakaher N, Rosti S, Tissier A (2012) Characterization of two genes for the biosynthesis of the labdane diterpene Z-abienol in tobacco (Nicotiana tabacum) glandular trichomes. Plant J 72(1):1–17. https://doi.org/10.1111/j.1365-313X.2012.05068.x
Shi P, Fu X, Shen Q, Liu M, Pan Q, Tang Y, Jiang W, Lv Z, Yan T, Ma Y, Chen M, Hao X, Liu P, Li L, Sun X, Tang K (2018) The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol 217(1):261–276. https://doi.org/10.1111/nph.14789
Tan H, Xiao L, Gao S, Li Q, Chen J, Xiao Y, Ji Q, Chen R, Chen W, Zhang L (2015) TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol Plant 8(9):1396–1411. https://doi.org/10.1016/j.molp.2015.04.002
Wang E, Wagner GJ (2003) Elucidation of the functions of genes central to diterpene metabolism in tobacco trichomes using posttranscriptional gene silencing. Planta 216(4):686–691. https://doi.org/10.1007/s00425-002-0904-4
Wu M-L, Cui Y-C, Ge L, Cui L-P, Xu Z-C, Zhang H-Y, Wang Z-J, Zhou D, Wu S, Chen L, Cui H (2020) NbCycB2 represses Nbwo activity via a negative feedback loop in tobacco trichome development. J Exp Bot 71(6):1815–1827. https://doi.org/10.1093/jxb/erz542
Xu J, van Herwijnen ZO, Dräger DB, Sui C, Haring MA, Schuurink RC (2018) SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell 30(12):2988–3005. https://doi.org/10.1105/tpc.18.00571
Yan T, Chen M, Shen Q, Li L, Fu X, Pan Q, Tang Y, Shi P, Lv Z, Jiang W, Ma Y-n, Hao X, Sun X, Tang K (2017) HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol 213(3):1145–1155. https://doi.org/10.1111/nph.14205
Yan T, Li L, Xie L, Chen M, Shen Q, Pan Q, Fu X, Shi P, Tang Y, Huang H, Huang Y, Huang Y, Tang K (2018) A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol 218(2):567–578. https://doi.org/10.1111/nph.15005
Yang C, Li H, Zhang J, Luo Z, Gong P, Zhang C, Li J, Wang T, Zhang Y, Lu Y, Ye Z (2011) A regulatory gene induces trichome formation and embryo lethality in tomato. Proc Natl Acad Sci USA 108(29):11836–11841. https://doi.org/10.1073/pnas.1100532108
Yang C, Ye Z (2013) Trichomes as models for studying plant cell differentiation. Cell Mol Life Sci 70(11):1937–1948. https://doi.org/10.1007/s00018-012-1147-6
Yang C, Gao Y, Gao S, Yu G, Xiong C, Chang J, Li H, Ye Z (2015) Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wov gene in tobacco. BMC Genom 16(1):868. https://doi.org/10.1186/s12864-015-2099-7
Zhang F, Liu X, Zuo K, Zhang J, Sun X, Tang K (2011) Molecular cloning and characterization of a novel Gossypium barbadense L. RAD-Like gene. Plant Mol Biol Rep 29(2):324–333. https://doi.org/10.1007/s11105-010-0234-9
Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135(11):1991–1999. https://doi.org/10.1242/dev.016873
Acknowledgements
We thank Professor Liang Chen (Xiamen University) for helping with vector construction.
Funding
This research was financially supported by the National Science Foundation of Henan province (Grant No. 182300410094), the State Tobacco Monopoly Administration of China [Grant No. 110202101005 (JY-05)], and China Tobacco Henan Industrial Co., Ltd. [Grant No. AW202149].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. HC and ZW designed the study. ZW, XY, YM, and YP performed the experiments. ZW, HZ and HC analyzed the data, ZW and H. C. wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Z., Yan, X., Zhang, H. et al. NtCycB2 negatively regulates tobacco glandular trichome formation, exudate accumulation, and aphid resistance. Plant Mol Biol 108, 65–76 (2022). https://doi.org/10.1007/s11103-021-01222-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01222-z