Abstract
Key message
Genes related to the MAPK cascade, ethylene signaling pathway, Pi starvation response, and NAC TFs were differentially expressed between normal and abortive ovules. Receptor-mediated ethylene signal perception and transmission play an important role in regulating fruit and ovule development.
Abstract
Xanthoceras sorbifolium, a small to medium-sized tree endemic to northern China, is an emerging dedicated oilseed crop designed for applications in advanced biofuel, engine oil, and functional food, as well as for pharmaceutical and cosmetic applications. Despite the importance of Xanthoceras seed oil, low seed productivity has constricted commercial exploitation of the species. The abortion of developing seeds (ovules after fertilization) is a major factor limiting fruit and seed production in the plant. To increase fruit and seed yields, a better understanding of the mechanisms underlying the abortion of fertilized ovules is critical. This study revealed differences in nucellus degeneration, endosperm development, and starch grain content between normally and abnormally developing ovules after fertilization. We constructed 6 RNA-sequencing (RNA-seq) libraries from normally and abnormally developing ovules at the onset of their abortion process. Comparative transcriptome analysis between the normal and abnormal ovules identified 818 differentially expressed genes (DEGs). Among DEGs, many genes involved in mitogen-activated protein kinase (MAPK) cascades, ethylene signaling pathway, and NAC transcription factor genes showed up-regulated expression in abnormal ovules. The RNA-seq data were validated using quantitative reverse-transcription PCR. Using virus-induced gene silencing (VIGS) methods, evaluation of an ethylene receptor gene (XsERS) function indicated that the gene was closely related to early development of fruits and seeds. Based on the data presented here, we propose a model for a MAPK-ethylene signaling-NAC2 gene regulatory cascade that plays an important role in the regulation of the ovule abortion process in X. sorbifolium. The present study is imperative for understanding the mechanisms of ovule abortion after fertilization and identifying the critical genes and gene networks involved in determining the fate of ovule development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Xanthoceras sorbifolium belongs to the monotypic genus Xanthoceras of the family Sapindaceae (Zhou and Liu 2012). It is a diploid species (2n = 2x = 30) endemic to northern China and is a small to medium-sized tree principally cultivated in temperate regions for its seeds. The plant has attracted much interest due to its high seed oil content and unique oil profile with applications in advanced biofuel, engine oil industries, other oil-based industrial products, and as a source for edible oils, as well as pharmaceutical and cosmetic applications (Gu et al. 2019; Wang et al. 2018).
The oil content of Xanthoceras seed ranges from 34 to 38% of seed dry weight. The oil is enriched with high levels of unsaturated fatty acids (85–93%), with predominantly oleic acid (28%) and linoleic acid (46%) present (Gu et al. 2019). The highly unsaturated fatty acid composition makes it well-suited for improving nutritional benefits from foods and may be beneficial to humans by improving cardiovascular health. The seed oil was found to contain high levels of tocopherol (Vitamin E; 106.3 mg/100 g). The antioxidant properties of tocopherols could play a significant role in the therapeutic effects of Xanthoceras seed oil. Another critical health benefit is attributed to the high content of nervonic acid in Xanthoceras seed oil (3.7%). Nervonic acid oils have become important targets for pharmaceutical and nutraceutical applications in the prevention and treatment of neurological disorders and associated diseases (Yang et al. 2018).
Despite multiple uses for and the importance of Xanthoceras seed oil, progress toward large-scale commercial cultivation of the trees has been hampered by low seed productivity (Zhou et al. 2017). Abortion of developing seeds is a major factor limiting fruit and seed production in X. sorbifolium. Hence, it has become crucial to resolve the problems of abortion of fertilized ovules for improved seed production to cope with the rapidly growing market demand for Xanthoceras seed and oil.
Seed development begins with the double fertilization event in flowering plants. The haploid egg cell and diploid central cell of the female gametophyte within the ovule each fuse with one sperm cell from the pollen tube to form the zygote and endosperm cell, respectively (Russell 1993). Divisions of the zygote give rise to the diploid embryo, whereas the fertilized central cell forms the triploid endosperm. The fertilization products are surrounded by the maternal tissues, including the integument and nucellus. Growth and development of the embryo, endosperm, and surrounding maternal tissues must be coordinated to produce the mature seed (Xu et al. 2016).
In response to various environmental stresses, plants have evolved multiple mechanisms to relieve deleterious effects (Sreenivasulu et al. 2007). One of these mechanisms is a reduction in plant reproduction, involved in arresting the ovule and/or pollen development, because of the substantial resources required for reproduction (Sun et al. 2005). Plants adapt to adverse environmental conditions by changing their gene expression profiles after perceiving stress signals, and as a result, cellular, physiological, and biochemical processes are modified (Abdelrahman et al. 2018). Ovule abortion can be triggered by endogenous signals, such as developmental cues and plant hormones, and by external environmental factors, such as nutrient deficit, salinity, drought stress, and extreme temperatures (Savadi 2018).
There are several transcriptome studies related to ovule abortion in some species, such as Arachis hypogaea (Chen et al. 2013), Dimocarpus longan (Liu et al. 2010), Corylus heterophylla (Cheng et al. 2015), and Davidia involucrata (Li et al. 2016). These studies suggest that ovule abortion is an extremely complicated process integrating multiple developmental and environmental signals, which involves extensive reprogramming and modulation of gene expression. Until now, little has been discovered about the mechanisms underlying ovule abortion in X. sorbifolium. To gain insight into these mechanisms, we conducted morphological investigations of the ovules after fertilization and the application of exogenous phosphate (Pi) to adult X. sorbofolium trees. Based on these studies, we made a comparative analysis of Xanthoceras transcriptome profiling of gene expression at the critical stage of ovule development between normally and abnormally developing ovules, using deep RNA sequencing (RNA-seq). Availability of the Xanthoceras reference genome, released in 2019, allowed us to perform transcriptome profiling for the fertilized ovules using a genome-guided assembly approach, unlike the previous studies, in which de novo assembly was used for mapping (Zhou and Zheng 2015; Wang et al. 2018).
Virus-induced gene silencing (VIGS) has been used as an effective tool for functional analysis of genes in X. sorbifolium (Zhou and Cai 2018; Xiong et al. 2020). We previously showed the successful induction of VIGS of the endogenous gene in X. sorbifolium ovules and fruits using tobacco rattle virus virus (TRV) vectors (Zhou and Cai 2018). This study adopted a TRV-based VIGS approach to silence the expression of the candidate gene associated with the abortion of ovules after fertilization.
The approaches used here are helpful to understand the mechanisms of ovule abortion, identifying the genes/gene regulatory networks involved in the determination of ovule development fate, and to select candidate genes to create improved varieties, producing better fruit and seed yields.
Materials and methods
Plant materials
Six-year-old X. sorbifolium trees were cultivated at the experimental farm of the Institute of Botany, Chinese Academy of Sciences (Xiangshan, Beijing, China). Our previous investigations showed that the ovary of X. sorbifolium is tricarpellate and trilocular, with five to eight ovules borne in two rows along the axile placenta of each locule (Zhou and Liu 2012). The reduction of ovule size represents a sign of abortion. Our morphological observations indicated that if there are more than 4 small ovules in an ovarian locule, then the other seemingly normal ovules in the locule arrest development of the embryo sac and begin to abort. The abortion of ovules occurs at different stages between 5 and 20 days after pollination (DAP), depending on the environmental conditions, parent genotypes, pollination times, and floral sites in inflorescence (Zhou et al. 2017). In most female flowers, ovules begin to abort during the period from 5 to 12 DAP. The present study focused on the normally and abnormally developing ovules at the onset of the abortion process at 11 DAP. Fertilized ovules were isolated from the ovaries at 11 DAP. Isolated ovules were used for histological analysis, TUNEL assay, RNA-seq, and real-time quantitative reverse transcription PCR (qRT-PCR) (Fig. 1). All studied samples came from trees of the Xanthoceras genotype 1.
Morphological and histological analysis
Collected ovules were fixed with FAA, dehydrated, infiltrated, and embedded in either Paraplast Plus or Spurr resin. Paraplast Plus sections were cut to 6–10 μm using a steel knife, and stained with safranin and fast green. Resin sections were cut to 1–1.5 μm with a diamond knife, and stained with 0.5% toluidine blue O in 0.1% sodium carbonate (pH 11.1) for general observations and periodic acid–Schiff’s reagent (PAS) for insoluble carbohydrates. Stained sections were observed with a Carl Zeiss microscope.
TUNEL assay
In-situ detection of DNA fragmentation was carried out with a modification of the TUNEL method (Domínguez et al. 2001). Sections were made using the above described Paraplast Plus-embedded materials. Paraplast Plus was removed by treatment with xylol. Sections were then hydrated with a decreasing ethanol series and treated with proteinase K (20 µg ml−1) in PBS (10 mM sodium phosphate buffer, 130 mM NaCl) and rinsed twice in PBS. Endogenous peroxidase activity was then quenched by incubation in 1% (v/v) H2O2 in methanol for 30 min and rinsed twice in PBS. For labeling, sections were incubated for 60 min at 37 °C in the presence of terminal deoxynucleotidyl transferase (Tdt) with the in-situ Cell Death Detection Kit (Boehringer Manheim), according to the manufacturer’s instructions. Controls were performed in which Tdt was omitted.
RNA extraction, library construction and Illumina sequencing
The sampled ovules were immediately frozen with liquid nitrogen and stored at − 80 °C until further use. The total RNA from normal and abnormal ovules was extracted using a Plant Total RNA Isolation Kit (Huayueyang, Beijing, China). A total of 6 RNA preparations (3 biological replicates for each sample) were used to increase the sequencing coverage. The quantity and quality of extracted RNA was evaluated using a Qubit fluorometer (Invitrogen INC. Carlsbad, CA) and bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA), respectively. The mRNA was fragmented using fragmentation buffer. cDNA was then synthesized using the mRNA fragments as templates. The cDNA fragments were purified by Qiaquick PCR Purification Kits (Qiagen, Valencia, USA). The purified cDNA fragments were end repaired, added to poly (A), and ligated to Illumina sequencing adapters. Suitable fragments were selected for the PCR amplification and sequenced by BGISEQ-500 at BGI (Shenzhen, China).
RNA-seq data analysis
Raw RNA-seq reads were filtered using a Perl program to obtain high-quality clean reads by removing low quality sequences (more than 20% bases with quality lower than 15 in one sequence), reads with more than 5% N bases (bases unknown), and reads containing Illumina sequencing adapters. All of the raw data have been deposited in the NCBI Short Read Archive (SRA) under the accession number PRJNA666271. The filtered sequence pairs were aligned to the X. sorbifolium reference genome sequences (Bi et al. 2019), using the alignment tool HISAT2 version 2.1.0 (Kim et al. 2015). The transcript levels of individual transcripts in each sample were normalized as fragments per kilobase of transcript per million mapped reads (FPKM). DEGs were identified using DEseq2 (Wang et al. 2010). Gene Ontology (GO) classification and functional enrichment analysis for DEGs were performed by WEGO software (http://wego.genomics.org.cn/cgi-bin/wego/index.pl). DEGs were mapped to terms in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using BLASTX software. KEGG pathways with the thresholds of q-value ≤ 0.05 were considered to be significantly enriched.
Real-time qRT-PCR analysis
Real-time qRT-PCR was used to estimate the accuracy of RNA-seq data. The same RNA samples used for RNA-seq were used for qRT-PCR. RNA (5 μg) was used for synthesis of cDNA with the PrimeScript RT Master Mix Perfect Real Time Kit (Takara). Real-time qRT-PCR was performed on the StepOnePLUS™ real-time PCR System (Applied Biosystems, USA). Cycling conditions were as follows: 10 min initial denaturation at 95 °C, followed by 40 cycles of 95 °C for 20 s and 60 °C for 30 s. The Actin gene (EVM0013773) was used as an internal reference, and relative expression was calculated using the 2−△△Ct method. Each sample was analyzed under the same conditions using three independent biological replicates and three technical replicate PCR reactions. All of the primers used in this study are listed in Table S1.
VIGS assays
VIGS assays were carried out as described previously (Zhou and Cai 2018). A 443 bp cDNA fragment of an ethylene receptor gene (designated as XsERS) were amplifed using the primers presented in Table S1. The purified PCR products were digested with EcoRI and KpnI and ligated to pTRV2, resulting in the plasmid TRV2-XsERS. The plasmids were sequenced to verify correct insertion of the fragment and were then transformed into Agrobacterium tumefaciens GV3101. The cultures containing pTRV1 and pTRV2/pTRV2-XsERS were mixed in a 1:1 ratio, and then 1 mL of culture was injected into the base of the inflorescence axial 3 days before anthesis in the trees of the genotype 1. Three independent biological replicates were performed for each treatment. Samples were collected 12 days after treatment and used for PCR analysis.
Phosphorus (P) applications
Phosphorus application experiments were carried out with 20 6-year-old trees of 4 Xanthoceras genotypes (1–4; five trees per genotype). Some differences in fruit set were observed among the four genotypes. Phosphorus fertilization doses included no application (control) and 150 g P2O5 per tree. Triple superphosphate (46% P2O5) was used as the source of phosphorus. Applications were realized in the form of a circle (40 cm radius) in front of the root. Plants received P applications at 10 days before anthesis. After P applications, each recipient was immediately irrigated with 50 L of water. Thereafter, the plants were irrigated according to the water demand and to the climatic conditions of the environment. During the experiment, the following cultural practices were performed: manual removal of weeds, superficial scarification of the soil every irrigation and monthly pesticide sprayings for the preventive control of insects and fungal diseases.
Statistical analyses
Statistical analyses were conducted using the SPSS 16.0 software package (SPSS Inc, Chicago, IL, USA). Differences between the means were considered statistically significant at a difference of P < 0.05.
Results
Comparative morphology of normally and abnormally developing ovules after fertilization
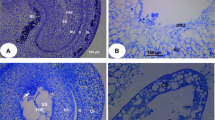
Ovules at 11 DAP consisted of an inner and outer integument, thick nucellus, and large embryo sac (Fig. 2a). Comparative analysis of tissue structures indicated that there were no obvious differences in the outer and inner integuments between the normally and abnormally developing ovules at the onset of the abortion process (Fig. 2b, c). Some cells of the outer integument became impregnated by phenolic compounds. Abundant starch grains were observed in the two integuments, but they were not detected in the nucellus or embryo sac (Fig. 2b, c). The abnormal ovules showed reduced starch content in both integuments (Fig. 2c).
Histological sections of the ovules after fertilization in Xanthoceras sorbifolium. a A portion of a longitudinal resin section of the normal ovules 11 days after pollination (DAP). b A resin section of the normal ovules 11 DAP was stained with periodic acid–Schiff’s reagent (PAS) for insoluble carbohydrates. c A resin section of the abnormal ovules at 11 DAP was stained with PAS. d A Paraplst Plus section of the ovules 8 DAP was observed with fluorescence microscopy, showing bright autofluorescent wall of the postament. e A chalazal portion of a longitudinal resin section of the normal ovules 11 DAP. f A portion of a transverse resin section of the abnormal ovules at 11 DAP. g A micropylar portion of a transverse resin section of the normal ovules at 11 DAP. h A micropylar portion of Paraplast Plus longitudinal section at 17 DAP. i A portion of a transverse resin section of the normal ovules at 18 DAP. j Magnification of micropylar portion in i. k A Paraplast Plus section of a normal ovule 11 DAP was subjected to TUNEL assay. l Localization of DNA fragmentation in an abnormal ovule 11 DAP. CE cellularized endosperm; E embryo; ES embryo sac; FNE free nuclear endosperm; HP hypostase; II inner integument; NU nucellus; OI outer integument; PP postament projection
In both types of ovules, several rows of radially elongated cells (termed postament) were located below the chalazal end of the embryo sac, extending to the hypostase adjacent to the vascular tissue (Fig. 2c, d). The column cells were distinguished from the surrounding nucellus cells by denser cytoplasm and bright autofluorescent walls (Fig. 2d). The morphological features of the postament suggest that its primary function may be involved in the transfer of metabolites from the hypostase to the embryo sac.
A nuclear type of endosperm development was conducted in normally and abnormally developing ovules (Fig. 2a, e, f). The nuclear endosperm became dispersed around the periphery of the embryo sac and surrounded a large central vacuole. The endosperm cytoplasm was prominent in the micropylar end of the embryo sac, whereas in the chalazal end, it formed a thin sheath along the embryo sac boundary. The inner layers of the nucellus degenerated after fertilization, except adjacent to the chalazal vascular bundle, where it formed a postament projection, surviving for a longer period of time (Fig. 2c, d). The degeneration of nucellar cells provided space for the expanding embryo sac. Based on morphological features, four types of nucellus tissue along the radial axis of normal ovules were recognized: (starting from the integument inward) (1) region of dividing cells adjacent to the inner integument; (2) middle zone of expanding cells; (3) autolyzing cells; and (4) cell debris region, proximal to the embryo sac (Fig. 2e). The nucellus was separated from the endosperm by a cavity filled with nucellar lysate (referred to as the “nucellar cavity”). In abnormal ovules at the onset of the abortion process, mitotic activity of the cells in the division region was reduced, and the extent of radial cell expansion in the middle regions was increased (Fig. 2f). The nucellus showed varying degrees of development in different regions of the ovules. The nucellus tissue flanking the large global part of the embryo sac in the chalazal end was predominantly developed compared to the nucellus tissue alongside the slender part of the embryo sac in the micropylar end (Fig. 2e, g).
In normally developing ovules, the free nuclear endosperm was cellularized in the micropylar part of the embryo sac at the early stage of the global embryo, but the endosperm coenocyte persisted in the chalazal part of the embryo sac until it was completely consumed by the developing embryo (Fig. 2h, i, j). In abnormal ovules, endosperm cellularization never occurred before ovule abortion. The endosperm cytoplasm and free nuclei tended to decrease gradually until they disappeared completely in the embryo sac in abnormal ovules. At the same time, the cavity of the embryo sac also decreased in size, resulting in a gradual reduction in size of abnormally developing ovules.
DNA fragmentation in normal and abortive ovules
The internucleosomal fragmentation of DNA generates 3′-OH DNA ends, which may be nick end-labeled with fluorescein-dUTP, mediated by terminal deoxynucleotidyl transferase (TdT), allowing the localization of programmed cell death (PCD) in histological sections. Paraffin sections of normally and abnormally developing ovules at 11 DAP were assayed by the TUNEL technique. The results showed that TUNEL-stained nuclei were located at the nucellar cells of one to three cell layers proximal to the nucellar cavity and some cells of the inner integument in the normal ovules, suggesting that these cells were undergoing PCD (Fig. 2k). More abundant TUNEL-stained nuclei were detected in the cells of the nucellus and inner integument in the abortive ovules (Fig. 2l).
Response of fruit set to phosphorus application in X. sorbifolium
Application of exogenous phosphate to 6-year-old Xanthoceras trees resulted in a significant increase in the number of mature fruits in plants of each genotype, but there were a significant difference among genotypes (Fig. 3). The most obvious effect was observed in the genotype 1 with 300% increase in fruit set. The fruit yield was reduced after 2 years without added Pi, showing Pi deficiency symptoms. All of the ovules developed normally and reached maturity in fruits from the phosphorus application treatment. The increase in fruit number observed in this study may be related to the important effects of phosphorus on seed development. The results suggested that the limited availability of phosphate may affect the development of fruits and ovules and, thus, become a major constraint on seed yield in X. sorbifolium.
Overview of the Xanthoceras ovule transcriptome
To investigate the transcriptional landscape of the ovules at the critical stage of development after fertilization and unravel how the ovule transcriptome is reprogrammed at the onset of the abortion process, 6 cDNA libraries from normally and abnormally developing ovules at 11 DAP were constructed and subjected to Illumina sequencing. We obtained 712.2 million paired-end raw reads (Table 1). With the process of quality control for raw reads, a total of 660.26 million high-quality clean reads were generated, with clean read ratios more than 91%. All of the clean reads were mapped to the X. sorbifolium reference genome, with mapping rates more than 73%. More than 60% of clean reads were uniquely mapped to the reference database (Table 1). All of the samples had a Q20 percentage (proportion of nucleotides with quality value larger than 20 in reads) greater than 97% and GC (guanine and cytosine) greater than 45%, indicating the high quality of RNA-seq.
A total of 15,463 novel transcripts were identified from all samples. Among them, 10,367 were novel isoforms for known coding genes, and 2519 were novel coding genes without any known features. The remaining 2577 were noncoding transcripts. After novel transcript detection, the novel coding transcripts were merged with the reference transcripts to obtain the complete reference. Clean reads were then mapped to the reference using Bowtie2.
Identification of differentially expressed genes (DEGs)
To identify the genes that were differentially expressed between normally and abnormally developing ovules, we compared and analyzed the expression levels of transcripts (Log2 FPKMs) in normal and abnormal ovule samples. We selected the transcripts with fold changes > 1.0 or < − 1.0 and a false discovery rate (FDR) ≤ 0.001 as DEGs. The FPKM method enabled us to identify 818 DEGs (Table S2). Among these DEGs, 324 genes were up-regulated, and 494 genes were down-regulated in normal ovules, compared to abnormal ones. Furthermore, 66 DEGs were only detected in normal ovules, and 49 only in abnormal ones.
Annotation of DEGs and functional category determination
To determine the functional significance of the transcriptional changes and highlight the involvement of the identified DEGs in ovule abortion, DEGs were annotated and categorized into various GO terms. GO classification results indicated that the DEGs were involved in 15 biological processes, 11 cellular components, and 12 molecular functions (Fig. 4). The GO terms related to “cellular process” and “metabolic process” represented the two largest groups in the biological process category; “membrane” and “membrane part” were the most enriched GO terms of the cellular component. Among molecular functions, the terms “binding” and “catalytic activity” were over-represented.
KEGG pathway analysis indicated that 368 DEGs were aligned to the KEGG database and assigned to 110 pathways. The pathway containing the largest number of DEGs was “plant hormone signal transduction” (ko04075), followed by “MAPK signaling pathway” (ko04016) (Fig. 5). Enrichment analysis of DEGs using KEGG showed that the enriched pathways were involved in caffeine metabolism (ko00232), linoleic acid metabolism (ko00591), and flavone and flavonol biosynthesis (ko00944) (Table 2). Among these, the genes encoding cytochrome P450, plant cysteine oxidase, and inorganic pyrophosphatase were up-regulated in abnormal ovules. These annotations provide a valuable resource for studying the biological process and candidate genes involved in Xanthoceras ovule abortion regulation.
DEGs involved in the hormone signal transduction pathway
A total of 36 DEGs were annotated in the hormone signal transduction pathways (ko04075), including ethylene, auxin, abscisic acid, brassinosteroid, and gibberellin signaling pathways. In the ethylene signaling pathway, we identified 12 genes encoding ethylene-responsive transcription factors (AP2/ERF, including ERF098, ERF109; ERF014, and ERF017). Among them, six genes showed up-regulated expression, and the others showed down-regulation in abnormal ovules (Fig. 6). Furthermore, an ethylene receptor gene and 5 ethylene biosynthetic genes (1-aminocyclopropane-1-carboxylate oxidase, ACO, and aminocyclopropane-1-carboxylate synthase, ACS) were also detected, showing uniformly up-regulated expression in abnormal ovules, except a gene coding ACO. In the auxin pathway, we detected 5 genes encoding auxin-responsive proteins, with 3 showing up-regulated expression in normal ovules over abnormal ovules. In the abscisic acid pathway, a gene encoding abscisic acid receptor PYR1 was detected, showing up-regulated expression in abortive ovules. In the brassinosteroid signaling pathway, we found 4 genes encoding protein brassinosteroid insensitive 1, and 3 of them showed down-regulated expression in abnormal ovules. In the gibberellin pathway, we identified a gene encoding gibberellin 20 oxidase 2 (GA20OX2), a key oxidase enzyme in the biosynthesis of gibberellins, which exhibited significantly increased expression in normal ovules. Another gene encoding gibberellin 3-beta-dioxygenase 1, involved in the production of bioactive GA, also showed enhanced expression in normal ovules.
Heatmaps of expression patterns for differentially expressed genes involved in phytohormone signaling pathways, including ethylene (a), auxin (b), brassinosteroid (c), gibberellin (d), and abscisic acid (e). The color bar is the scale for the expression levels of each gene on the basis of FPKM value. AD abnormally developing ovules; ACO 1-aminocyclopropane-1-carboxylate oxidase; ACS aminocyclopropane-1-carboxylate synthase; ND normally developing ovules
DEGs encoding transcription factors
Changes in gene expression that accompany the onset of the ovule abortion process were probably controlled by transcription factors (TFs). We identified 60 DEGs predicted to encode TFs belonging to 17 families (Fig. 7). Of these genes, 41 showed up-regulated expression in the abnormal ovules. The 5 top TFs included AP2-EREBP, NAC, MYB, WRKY, and bHLH. These TFs represent potential crucial regulators of ovule abortion in X. sorbifolium. Any small changes in the abundance of TFs between the normal and abnormal ovules could easily cause a larger number of downstream transcriptome differences.
DEGs related to the phosphate starvation response
Based on the observations that seed development could be affected by exogenously applied Pi in X. sorbifolium, we tried to identify DEGs related to the Pi starvation response in the transcriptome data. Genes encoding purple acid phosphatase, phosphoenol pyruvate carboxylase kinase 1, ubiquitin-conjugating enzyme E2 24 (UBC24)/PHOSPHATE2 (PHO2), MYB62, and WRKY6 transcription factors were detected in the DEG database, which showed significantly up-regulated expression in abnormal ovules. It has been reported that these genes play important roles in the regulation of the Pi starvation responses in Arabidopsis and soybean (Devaiah et al. 2009; Ye et al. 2018; Feder et al. 2020). A gene (EVM0001389) encoding the glucose-6-phosphate/phosphate translocator 1 (GPT1), which has been implicated in the control of pollen and ovule development (Niewiadomski et al. 2005), was exclusively expressed in normal ovules, and no transcripts were detected in abnormal ovules.
Validation of RNA-seq data by qRT-PCR
To validate the quantification of transcript abundance of Xanthoceras genes obtained from the RNA-seq approach, qRT-PCR was used to measure the gene expression of 15 randomly selected DEGs with the same RNA samples used for RNA-seq. The high and significant Pearson correlation coefficient (0.801) of RNA-seq with RT-PCR results suggest high confidence in the expression values obtained from the transcriptome data (Fig. 8).
Silencing of XsERS gene decreased abortion of fruits and fertilized ovules
To validate the role of ethylene signaling pathway in regulation of fruit and ovule development in X. sorbifolium, we conducted an experiment in which we inoculated inflorescences at 3 days before anthesis with pTRV2-XsERS plus pTRV1 plasmids aimed to silence ethylene receptor gene XsERS. The results showed that virus-induced gene silencing of XsERS can be achieved in X. sorbifolium fruits and ovules using tobacco rattle virus vectors. Of the inflorescences inoculated with pTRV2-XsERS plus pTRV1 (N = 60), 41 inflorescences showed higher fruit set rate than the mock-treated inflorescences inoculated with pTRV2-empty (Fig. 9). Mock-treated inflorescences were undistinguishable from untreated inflorescences (N = 20). pTRV2-XsERS inoculated fruits showed normal endosperm development in all the ovules examined (N = 100). RT-PCR analyses confirmed a down-regulation of endogenous XsERS expression levels in the ovules of pTRV2-XsERS inflorescences compared to those of the mock-treated inflorescences (Fig. 10). Expression of XsERS in the ovules of the mock-treated inflorescences was similar to that of untreated inflorescences, suggesting that the viral treatment does not interfere with XsERS expression.
Discussion
This study indicated that abortion of the ovules after fertilization in X. sorbifolium is a complicated process that involves visible changes of microscopic morphology and extensive reprogramming of global gene expression. Comparative morphological investigations between normally and abnormally developing ovules revealed that at the onset of the abortion process, abnormal ovules were characterized by significant modifications, including a decrease in starch grains in the integument cells, reduced mitotic activity of the nucellus cells, progressive degeneration of nuclear endosperm, and arrest of embryo sac growth. Application of exogenous phosphate to Xanthoceras trees affected the development of fruits and ovules. Based on these observations, we studied the complete transcriptome of normal and abnormal ovules using the high-throughput RNA sequencing technology. Comparative transcriptomic studies identified 818 DEGs, at least some of which were probably required for the initiation and progression of the ovule abortion process.
Differences in PCD and ethylene signaling between normal and abnormal ovules are possibly associated with ovule abortion
We showed that the cells of the nucellus were subjected to a degenerative process in normally developing ovules after fertilization. Collapse of the nucellar cells was accompanied by fragmentation of nuclear DNA, considered one of the typical hallmarks of PCD, which occurs in many species such as wheat (Triticum aestivum; Domínguez et al. 2001), barley (Hordeum vulgare; Lin et al. 1998), Ricinus (Greenwood et al. 2005), and Sechium edule (Lombardi et al. 2007). In the normally developing ovules of X. sorbifolium, PCD was localized in the border region of the nucellar tissue adjacent to the expanding embryo sac, whereas in the abnormally developing ovules, the extent of nucellar degeneration increased.
Normal seed development requires fine temporal and spatial regulation of nucellar degeneration, which relies on strict coordination between nucellus elimination and endosperm development (Xu et al. 2016). Fertilization of the central cell is necessary to trigger nucellus elimination in Arabidopsis. The endosperm and nucellus develop antagonistically in rice and Arabidopsis seeds (Xu et al. 2016). Ethylene, having simple structures, small dimensions, and high diffusibility in aqueous and lipid environments, is a key signaling molecule in response to environmental stress and the regulation of many plant processes (Abeles et al. 1992). It might be a good candidate for the role of messenger in the communication between different ovule tissues. In S. edule, the expanding endosperm produces a considerable amount of ethylene, which acts as a signaling molecule to control nucellus PCD (Lombardi et al. 2012). Nucellar degeneration is blocked when ethylene biosynthesis in the endosperm is inhibited. Nucellus PCD is also strongly compromised when ethylene perception in the nucellus is impaired by the addition of specific and competitive inhibitors. In Gingko biloba, ethylene production increased in the ovule at the stage of nucellar PCD preparation, and the ethylene signaling pathway was activated for transcriptional regulation of downstream targets (Li et al. 2019). Ethylene has also been implicated in the regulation of several other PCD-dependent developmental processes, including hypoxia-induced aerenchyma formation (Gunawardena et al. 2001), leaf and petal senescence (Tsanakas et al. 2014), and PCD in the endosperm (Domínguez and Cejudo 2014).
Our transcriptomic analysis detected 18 DEGs related to the ethylene signaling pathway, including 12 ERFs, 5 ethylene biosynthetic genes, and an ethylene receptor gene. Of these genes, 12, including all of the ethylene biosynthetic genes and an ethylene receptor gene, showed enhanced expression levels in the abnormal ovules. Expression of the ethylene receptor gene (EVM0018513) was validated by qRT-PCR analysis. We propose that the ethylene signaling pathway could be involved in the ovule abortion process and cell death in X. sorbifolium. The role of the ethylene signaling pathway in seed abortion has also been reported in other plant species, such as Arabidopsis (Carbonell-Bejerano et al. 2011), Corylus heterophylla (Cheng et al. 2015), maize (Shen et al. 2020), and wheat (Hays et al. 2007). Several lines of evidence support the important role of ethylene in modulating the onset of ovule senescence and fruit set response in Arabidopsis (Sun et al. 2005; Carbonell-Bejerano et al. 2011). Expression of the ethylene biosynthesis gene ACS2 is specifically activated in ovules shortly before their senescence in Arabidopsis (Carbonell-Bejerano et al. 2011). An ACC oxidase gene (TAIR: At1g12010) also showed high expression in the ovules of 2 days post-anthesis unfertilized pistils in Arabidopsis, suggesting activation of ethylene biosynthesis upon the initiation of senescence in unfertilized ovules (Carbonell-Bejerano et al. 2010).
Mitogen-activated protein kinase (MAPK) cascades play important roles in regulating plant growth, development, and responses to stress (Nakagami et al. 2005; Liu et al. 2020). In Arabidopsis transgenic plants, mitogen-activated protein kinase kinase 9 (MKK9) was found to activate MPK3/MPK6, which in turn phosphorylates and positively modulates the stability of ETHYLENE-INSENSITIVE3 (EIN3), a key transcription factor in ethylene signaling (Xu et al. 2008; Yoo et al. 2008). As a consequence, transcription of multiple genes responsible for ethylene biosynthesis, ethylene responses, and camalexin biosynthesis is promoted (Xu et al. 2008). In our DEG analysis database, a gene encoding MKK9 showed up-regulated expression in the abnormal ovules, suggesting that MKK9 possibly plays an important role in ethylene signaling in the abnormal ovules of X. sorbifolium.
Roles of ethylene perception and signal transduction in Xanthoceras fruit development
The ethylene signal is perceived by a small family of receptors that are similar to bacterial two-component histidine kinase (Bleecker et al. 1998). In Arabidopsis, there are five known ethylene receptors (ETR1, ETR2, ERS1, ERS2, and EIN4). Based on sequence analysis and functional studies, the ethylene receptors have been classified into two subfamilies. The subfamily I receptors, ETR1 and ERS1, are the most homologous to histidine kinases. The subfamily II members, ETR2, EIN4 and ERS2, have lost most of the amino acids critical for histidine kinase activity and instead possess serine kinase activity. The ethylene receptors act as negative regulators of the ethylene signaling pathway, following an inverse-agonist model in which ethylene binding to the receptors reduces the activity of the receptors and switches off the downstream signal transmission (Berleth et al. 2019). Expression of ethylene receptors has been detected during fruit development in many important fruit species, such as tomato (Solanum lycopersicum; Tieman and Klee 1999; Chen et al. 2020), peach (Prunus persica; Rasori et al. 2002), and durian (Durio zibethinus; Thongkum et al. 2018), suggesting that receptor-mediated ethylene signal perception and transmission play an important role in regulating fruit development.
To better understand the regulatory role of ethylene signaling responses in Xanthoceras fruit and ovule development, we adopted a VIGS approach to silence the expression of the ethylene receptor gene EVM0018513. As mentioned earlier, the silencing of XsERS gene promoted Xanthoceras fruit development and reduced the rate of fruit and ovule abortion. This study provided direct evidence that the XsERS gene is closely related to development of the fruits and ovules in X. sorbifolium.
Transcriptional regulators are probably involved in X. sorbifolium ovule abortion
Onset and progression of the ovule abortion process, which is accompanied by the massive reprogramming of gene expression, might be tightly regulated by transcription factors. DEG analysis detected 11 NAC TF genes, such as NAC2, NAC72, and NAC13, all of which showed up-regulated expression in abnormal ovules, except one gene encoding NAC domain-containing protein 30. Growing evidence showed that NAC TFs play an important role in the control of senescence (Gregersen and Holm 2007; Kim et al. 2018; Ueda et al. 2020). The best-characterized senescence regulatory NAC TF was ORE1 [also called AtNAC2 and ANAC092 (At5g39610)] that not only plays a key role in the regulation of leaf senescence but also exerts a function in PCD. It was reported that the trifurcate feed-forward pathway, which involves ETHYLENE-INSENSITIVE2 (EIN2/ORE2/ORE3), miRNA164 (miR164), and ORE1, regulates age-dependent leaf senescence and cell death (Li et al. 2013; Kim et al. 2016). EIN3, a well-known key TF in the EIN2-mediated ethylene signaling cascade, has been shown to be involved in the trifurcate feed-forward pathway (Li et al. 2013). EIN3 induces the accumulation of ORE1 transcripts in an age-dependent manner by directly repressing miR164 transcription. Leaf senescence was delayed in mutants lacking the functional ORE1 protein, whereas over-expression of the ORE1 gene promoted senescence (Balazadeh et al. 2010; Kim et al. 2016). ORE1 has also been shown to control lateral root development and to act downstream of the ethylene and auxin signaling pathways in response to salinity stress (He et al. 2005). In the present study, a Xanthoceras homolog to Arabidopsis AtNAC2 was significantly over-expressed in abnormal ovules, suggesting that this gene possibly plays a positive role in the regulation of the ovule abortion process.
Genetic regulation of sucrose transport is likely involved in the ovule abortion process in X. sorbifolium
Following fertilization, massive amounts of sugar have to be imported into rapidly developing fruits and ovules (Hill et al. 2003; Sgromo et al. 2010). Sugars serve as carbon and energy sources and as signaling molecules during seed development (Ruan et al. 2012). Sucrose transport across the plasma membranes of sieve elements and companion cells is essential for the proper partitioning of photoassimilates and, thus, for seed development (Sauer et al. 2004). In Arabidopsis, SUC2 function is essential for loading sucrose into the phloem in leaf minor veins (Srivastava et al. 2008). Mutant plants harboring a T-DNA insertion in the AtSUC2 gene showed reduced sucrose export from leaves. AtSUC8 encodes another functional sucrose transporter and is expressed in the floral tissue of Arabidopsis. It was proposed that AtSUC8, AtSUC1, and possibly also AtSUC9 could generate a signal in pollen tube guidance (Sauer et al. 2004). In our study, we found transcripts of a Xanthoceras homolog of the SUC8 gene, whose expression was up-regulated in abnormal ovules. It is intriguing to speculate that the Xanthoceras SUC8 gene may play an important role in remobilizing stored carbohydrates from the aborting ovules to normally developing ovules or other healthy sink organs.
Another class of plasma membrane sucrose transporters is the SWEET family members (Chen et al. 2010; Geng et al. 2020). They have seven transmembrane domains and are divided into four clades. Clade III SWEETs appear to transport sucrose in a pH-independent manner and are typically involved in cellular efflux processes (Chen et al. 2012). In Arabidopsis, several Clade III SWEET genes, including SWEET11, 12, and 15, are expressed in developing seeds and may serve as sucrose carriers for efflux from the integument into the apoplast, as well as from the endosperm to the embryo, supporting growth and development of the embryo (Dean et al. 2011; Chen et al. 2015). In our study, we detected a Xanthoceras homolog of the SWEET15 gene, showing up-regulated expression in normal ovules. It is likely that the Xanthoceras SWEET15 gene is essential for normal ovule development by mediating sucrose efflux involved in the transfer of sugars from the integuments or nucellus to actively developing endosperm.
Phosphorus deficiency-induced DEGs are probably required for control of the ovule abortion process in X. sorbifolium
Phosphorus is an essential macronutrient for plant growth and development, and it is absorbed by the roots in the form of inorganic phosphate. Because Pi is stable and insoluble, it is not readily available to plants in most soils, and, consequently, P deficiency is a major constraint to crop production (Schachtman et al. 1998). Our studies indicated that limited availability of Pi decreased fruit and seed yield in X. sorbifolium. The application of exogenous Pi to Xanthoceras trees can significantly promote fruit production and alleviate the abortion of ovules after fertilization. These changes appear to be the consequence of altered expression of genes responsible for Pi starvation-induced stress.
It has been reported that the induction of MYB62, an R2R3-type MYB transcription factor, is a specific response to Pi deficiency in Arabidopsis, and its transcript levels decreased with increasing availability of Pi to the plant (Devaiah et al. 2009). The over-expression of MYB62 resulted in altered root architecture, Pi uptake, and acid phosphatase activity, leading to decreased total Pi content in the shoots of Arabidopsis. The expression of several Pi starvation-induced genes was also reduced in MYB62 over-expressing plants. Over-expression of AtMYB62 resulted in a characteristic gibberellic acid (GA)-deficient phenotype (Devaiah et al. 2009). We identified a Xanthoceras homolog of Arabidopsis MYB62, showing significantly up-regulated expression in abnormal ovules after fertilization. Transcript levels of the Xanthoceras MYB62 gene were validated using qRT-PCR. The enhanced expression of MYB62 in the ovules at the onset of the abortion process suggested that these ovules may be under P deficit conditions, and Pi limitation negatively affected Xanthoceras seed development.
Another key regulator for Arabidopsis adaptation to Pi availability is PHO2, which encodes ubiquitin E2 conjugase (UBC24) (Aung et al. 2006). The PHO2/UBC24 gene controls Pi uptake in roots and Pi translocation from roots to shoots by mediating protein degradation of high-affinity Pi transporters and PHOSPHATE1 (PHO1) (Huang et al. 2013). The PHO2/UBC24 functions as a repressor that prevents excessive accumulation of Pi and maintains cellular Pi homeostasis. MicroRNA399 (miR399) directs the cleavage of PHO2 transcripts (Aung et al. 2006). Up-regulated expression of miR399 by Pi deficiency activates Pi uptake and root-to-shoot translocation. Our study identified a Xanthoceras homolog of Arabidopsis PHO2/UBC24 gene, showing enhanced expression in abnormal ovules, which suggests its role in Pi uptake and reallocation to the developing organs coupled with regulation of the ovule abortion process.
Ethylene plays an important regulatory role in plant responses to mineral nutrient availability (Khan et al. 2015). Ethylene biosynthesis varies with the availability of P nutrients (Iqbal et al. 2013). It was shown that P-deficient roots generally produce more ethylene than P-sufficient ones (Borch et al. 1999). Ethylene promotes up-regulation of Pi transporter genes and increases phosphatase activity in P-deficient roots (Lei et al. 2011; García et al. 2015). Ethylene plays an important role in the phosphate starvation response by altering root system architecture (Dinh et al. 2012). In Petunia, ethylene initiates petal senescence and regulates expression of the phosphate transporter gene, PhPT1, which functions in the remobilization of Pi during petal senescence (Chapin and Jones 2009). Based on the gene expression analysis of ovules after fertilization in X. sorbifolium, we predicted that the ethylene signaling pathway is involved in the response of Pi starvation and recycling of P nutrients at the onset and progression of the ovule abortion process.
Hypothetical model for the regulation pattern of ovule abortion in X. sorbifolium
Based on the data presented here, we propose a model for a MAPK-ethylene signaling -NAC2 gene regulatory cascade that might have an important role in the regulation of ovule abortion after fertilization in X. sorbifolium (Fig. 11). According to this putative model, MAPK cascades are activated by nutrient stress, such as Pi starvation, in fertilized ovules. Activation of MAPK cascade proteins (such as MKK9) induces up-regulated expression of multiple genes responsible for ethylene biosynthesis, such as ACO and ACS, and ethylene responses (ERFs). The ethylene signaling cascade promotes transcription of the NAC2 gene that positively regulates ovule abortion and cell death in X. sorbifolium.
Hypothetical model occurring in fertilized ovules of Xanthoceras sorbifolium upon nutrient stress. According to this model, nutrient stress (such as Pi starvation) are recognized through MAPK cascades, ethylene signaling and NAC-dependent pathway. These signaling pathways stimulate transcriptional reprogramming of many genes related to ovule development fate, eventually leading to ovule abortion
Conclusion
In this study, we performed comparative transcriptome analysis of normal and abnormal ovules of X. sorbifolium using next-generation RNA sequencing, based on morphological investigations of ovule development after fertilization. The recent release of the X. sorbifolium reference genome significantly increased both the depth and coverage of RNA-seq data analysis, thus allowing us to widely identify the genes and gene networks involved in controlling ovule development after fertilization. Our studies showed that genes related to the MAPK cascade, ethylene signaling pathway, sucrose transfer, Pi starvation response, and NAC TFs were differentially expressed between normal and abortive ovules, suggesting their crucial roles in regulating the onset and progress of the ovule abortion process. Our data serve as a valuable genetic source that can be used for molecular breeding programs aimed to increase seed production. Considering the multidimensionality of mechanisms involved in ovule abortion, much more sophisticated analyses, including a transcriptome approach integrated into metabolome and proteome approaches, as well as functional characterization of key genes, are required in future studies.
References
Abdelrahman M, Jogaiah S, Burritt DJ, Lam-Son Phan Tran LS (2018) Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ 41:1972–1983
Abeles FB, Morgan PW, Saltveit ME (1992) Ethylene in plant biology, 2nd edn. Academic Press, New York
Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate over accumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141:1000–1011
Balazadeh S et al (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62:250–264
Berleth M et al (2019) Molecular analysis of protein-protein interactions in the ethylene pathway in the different ethylene receptor subfamilies. Front Plant Sci 10:726. https://doi.org/10.3389/fpls.2019.00726
Bi Q et al (2019) Pseudomolecule-level assembly of the Chinese oil tree yellowhorn (Xanthoceras sorbifolium) genome. GigaScience 8:1–11. https://doi.org/10.1093/gigascience/giz070
Bleecker AB, Esch JJ, Hall AE, Rodriguez FI, Binder BM (1998) The ethylene-receptor family from Arabidopsis: structure and function. Phil Trans R Soc Lond B 353:1405–1412
Borch K, Bouma TJ, Lynch JP, Brown KM (1999) Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ 22:425–431
Carbonell-Bejerano P, Urbez C, Carbonell J, Granell T, Perez-Amador MA (2010) A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol 154:163–172
Carbonell-Bejerano P, Urbez C, Granell A, Carbonell J, Perez-Amador MA (2011) Ethylene is involved in pistil fate by modulating the onset of ovule senescence and the GA-mediated fruit set in Arabidopsis. BMC Plant Biol 11:84
Chapin LJ, Jones ML (2009) Ethylene regulates phosphorus remobilization and expression of a phosphate transporter (PhPT1) during Petunia corolla senescence. J Exp Bot 60:2179–2190
Chen LQ et al (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468:527–532
Chen LQ et al (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335:207–211
Chen X et al (2013) Deep sequencing analysis of the transcriptomes of peanut aerial and subterranean young pods identifies candidate genes related to early embryo abortion. Plant Biotechnol J 11:115–127
Chen LQ et al (2015) A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27:607–619
Chen Y et al (2020) Roles of SlETR7, a newly discovered ethylene receptor, in tomato plant and fruit development. Hortic Res 7:17. https://doi.org/10.1038/s41438-020-0239-y
Cheng Y et al (2015) Transcriptome analysis and gene expression profiling of abortive and developing ovules during fruit development in hazelnut. PLoS ONE 10:e0122072. https://doi.org/10.1371/journal.pone.0122072
Dean G et al (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4:1074–1091
Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2:43–58
Dinh PTY, Leung MRS, McManus MT (2012) Regulation of root growth by auxin and ethylene is influenced by phosphate supply in white clover (Trifolium repens L.). Plant Growth Regul 66:179–190
Domínguez F, Cejudo FJ (2014) Programmed cell death (PCD): an essential process of cereal seed development and germination. Front Plant Sci 5:366. https://doi.org/10.3389/fpls.2014.00366
Domínguez F, Moreno J, Cejudo FJ (2001) The nucellus degenerates by a process of programmed cell death during the early stages of wheat grain development. Planta 213:352–360
Feder D, McGeary RP, Mitić N, Lonhienne T, Furtado A (2020) Structural elements that modulate the substrate specificity of plant purple acid phosphatases: avenues for improved phosphorus acquisition in crops. Plant Sci 294:110445. https://doi.org/10.1016/j.plantsci.2020.110445
García MJ, Romera FJ, Lucena C, Alcántara E, Pérez-Vicente R (2015) Ethylene and the regulation of physiological and morphological responses to nutrient deficiencies. Plant Physiol 169:51–60
Geng Y, Wu M, Zhang C (2020) Sugar transporter ZjSWEET2.2 mediates sugar loading in leaves of Ziziphus jujuba Mill. Front Plant Sci 11:1081. https://doi.org/10.3389/fpls.2020.01081
Greenwood JS, Helm M, Gietl C (2005) Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proc Natl Acad Sci USA 102:2238–2243
Gregersen PL, Holm PB (2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol J 5:192–206
Gu LB et al (2019) Comparative study on the extraction of Xanthoceras sorbifolia Bunge (yellow horn) seed oil using subcritical n-butane, supercritical CO2, and the Soxhlet method. LWT Food Sci Technol 111:548–554
Gunawardena A, Pearce DM, Jackson MB, Hawes CR, Evans DE (2001) Characterisation of programmed cell death during aerenchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.). Planta 212:205–214
Hays DB, Do JH, Mason RE, Morgan G, Finlayson SA (2007) Heat stress induced ethylene production in developing wheat grains induces kernel abortion and increased maturation in a susceptible cultivar. Plant Sci 172:1113–1123
He XJ et al (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Hill LM, Morley-Smith ER, Rawsthorne S (2003) Metabolism of sugars in the endosperm of developing seeds of oilseed rape. Plant Physiol 131:228–236
Huang TK et al (2013) Identification of downstream components of ubiquitin conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25:4044–4060
Iqbal N, Trivellini A, Masood A, Ferrante A, Khan NA (2013) Current understanding on ethylene signaling in plants: the influence of nutrient availability. Plant Physiol Biochem 73:128–138
Khan MIR, Trivellini A, Fatma M, Masood A, Francini A, Iqbal N, Ferrante A, Khan NA (2015) Role of ethylene in responses of plants to nitrogen availability. Front Plant Sci 6:927. https://doi.org/10.3389/fpls.2015.00927
Kim D, Langmead B, Salzberg SL (2015) HSAT: a fast spliced aligner with ow memory requirements. Nat Methods 12:257–360
Kim HJ, Nam HG, Lim PO (2016) Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol 33:48–56
Kim J et al (2018) New insights into the regulation of leaf senescence in Arabidopsis. J Exper Bot 69:787–799
Lei M et al (2011) Ethylene signaling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New phytol 189:1084–1095
Li Z, Peng J, Wen X, Guo H (2013) ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25:3311–3328
Li M et al (2016) De novo transcriptome sequencing and gene expression analysis reveal potential mechanisms of seed abortion in dove tree (Davidia involucrata Baill.). BMC Plant Biol 16:82. https://doi.org/10.1186/s12870-016-0772-x
Li D et al (2019) Transcriptomic profiling identifies differentially expressed genes associated with programmed cell death of nucellar cells in Ginkgo biloba L. BMC Plant Biol 19:91. https://doi.org/10.1186/s12870-019-1671-8
Lin IW et al (1998) Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiol 118:1169–1180
Liu H et al (2010) Comparative proteomic analysis of longan (Dimocarpus longan Lour.) seed abortion. Planta 231:847–860
Liu J et al (2020) Involvement of active MKK9-MAPK3/MAPK6 in increasing respiration in salt-treated Arabidopsis callus. Protoplasma 257:965–977
Lombardi L et al (2007) Programmed cell death of the nucellus during Sechium edule Sw. seed development is associated with activation of caspase-like proteases. J Exp Bot 58:2949–2958
Lombardi L, Mariotti L, Picciarelli P, Ceccarelli N, Lorenzi R (2012) Ethylene produced by the endosperm is involved in the regulation of nucellus programmed cell death in Sechium edule Sw. Plant Sci 187:31–38
Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10:339–346
Niewiadomski P et al (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17:760–775
Rasori A, Ruperti B, Bonghi C, Tonutti P, Ramina A (2002) Characterization of two putative ethylene receptor genes expressed during peach fruit development and abscission. J Exp Bot 53:2333–2339
Ruan YL, Patrick JW, Bouzayen M, Osorio S, Fernie AR (2012) Molecular regulation of seed and fruit set. Trends Plant Sci 17:656–665
Russell SD (1993) The egg cell: development and role in fertilization and early embryogenesis. Plant Cell 5:1349–1359
Sauer N et al (2004) AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J 40:120–130
Savadi S (2018) Molecular regulation of seed development and strategies for engineering seed size in crop plants. Plant Growth Regul 84:401–422
Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116:447–453
Sgromo C et al (2010) Isolation and expression analysis of organelle genes involved in the development of olive flowers (Olea europaea L.). Plant Biosyst 144:733–739
Shen S et al (2020) The equilibrium between sugars and ethylene is involved in shading- and drought-induced kernel abortion in maize. Plant Growth Regul 91:101–111
Sreenivasulu N, Sopory SK, Kavi Kishor PB (2007) Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene 388:1–13
Srivastava AC, Ganesan S, Ismail IO, Ayre BG (2008) Functional characterization of the Arabidopsis thaliana AtSUC2 Suc/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol 147:200–211
Sun K, Cui Y, Hauser BA (2005) Environmental stress alters genes expression and induces ovule abortion: reactive oxygen species appear as ovules commit to abort. Planta 222:632–642
Thongkum M et al (2018) The effect of 1-methylcyclopropene (1-MCP) on expression of ethylene receptor genes in durian pulp during ripening. Plant Physiol Biochem 125:232–238
Tieman DM, Klee HJ (1999) Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiol 120:165–172
Tsanakas GF, Manioudaki ME, Economou AE, Kalaitzis P (2014) De novo transcriptome analysis of petal senescence in Gardenia jasminoides Ellis. BMC Genomics 15:554
Ueda H, Ito T, Inoue R, Masuda Y, Nagashima Y, Kozuka T, Kusaba M (2020) Genetic interaction among phytochrome, ethylene and abscisic acid signaling during dark-induced senescence in Arabidopsis thaliana. Front Plant Sci 11:564. https://doi.org/10.3389/fpls.2020.00564
Wang L, Feng Z, Wang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Wang L, Ruan C, Liu L, Du W, Bao A (2018) Comparative RNA-Seq analysis of highand low-oil yellow horn during embryonic development. Int J Mol Sci 19:3071. https://doi.org/10.3390/ijms19103071
Xiong CW, Zhao S, Yu X, Sun Y, Li H, Ruan CJ, Li JB (2020) Yellowhorn drought-induced transcription factor XsWRKY20 acts as a positive regulator in drought stress through ROS homeostasis and ABA signaling pathway. Plant Physiol Biochem 155:187–195
Xu J et al (2008) Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J Biol Chem 283:26996–27006
Xu W et al (2016) Endosperm and nucellus develop antagonistically in Arabidopsis seeds. Plant Cell 28:1343–1360
Yang T, Yu Q, Xu W, Li D, Chen F, Aizhong Liu A (2018) Transcriptome analysis reveals crucial genes involved in the biosynthesis of nervonic acid in woody Malania oleifera oilseeds. BMC Plant Biol 18:247. https://doi.org/10.1186/s12870-018-1463-6
Ye Q, Wang H, Su T, Wu WH, Chen YF (2018) The ubiquitin E3 ligase PRU1 regulates WRKY6 degradation to modulate phosphate homeostasis in response to low Pi stress in Arabidopsis. Plant Cell 30:1062–1076
Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451:789–795
Zhou QY, Liu GS (2012) The embryology of Xanthoceras and its phylogenetic implications. Plant Syst Evol 298:457–468
Zhou QY, Zheng YR (2015) Comparative de novo transcriptome analysis of fertilized ovules in Xanthoceras sorbifolium uncovered a pool of genes expressed specifically or preferentially in the selfed ovule that are potentially involved in late-acting self-incompatibility. PLoS ONE 10:e0140507. https://doi.org/10.1371/journal.pone.0140507
Zhou QY, Zheng YR, Lai LM, Du H (2017) Observations on sexual reproduction in Xanthoceras sorbifolium (Sapindaceae). Acta Bot Occid Sin 37:0014–0022
Zhou QY, Cai Q (2018) The superoxide dismutase genes might be required for appropriate development of the ovule after fertilization in Xanthoceras sorbifolium. Plant Cell Reports 37:727–739
Acknowledgements
We would like to thank Xiuping Xu, Jie Wen, and Fengqin Dong for technical help. This work was supported by National Natural Science Foundation of China (U1903103, 30972344, 31370611 and 31570680) and Beijing Natural Science Foundation (6172028).
Author information
Authors and Affiliations
Contributions
QZ conceived the research; QZ and QC carried out the experiments; QZ analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Q., Cai, Q. Role of ethylene in the regulatory mechanism underlying the abortion of ovules after fertilization in Xanthoceras sorbifolium. Plant Mol Biol 106, 67–84 (2021). https://doi.org/10.1007/s11103-021-01130-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01130-2