Abstract
Acetylation and deacetylation of histones are important for regulating a series of biological processes in plants. Histone deacetylases (HDACs) control the histone deacetylation that plays an important role in plant response to abiotic stress. In our study, we show the evidence that GhHDT4D (a member of the HD2 subfamily of HDACs) is involved in cotton (Gossypium hirsutum) response to drought stress. Overexpression of GhHDT4D in Arabidopsis increased plant tolerance to drought, whereas silencing GhHDT4D in cotton resulted in plant sensitivity to drought. Simultaneously, the H3K9 acetylation level was altered in the GhHDT4D silenced cotton, compared with the controls. Further study revealed that GhHDT4D suppressed the transcription of GhWRKY33, which plays a negative role in cotton defense to drought, by reducing its H3K9 acetylation level. The expressions of the stress-related genes, such as GhDREB2A, GhDREB2C, GhSOS2, GhRD20-1, GhRD20-2 and GhRD29A, were significantly decreased in the GhHDT4D silenced cotton, but increased in the GhWRKY33 silenced cotton. Given these data together, our findings suggested that GhHDT4D may enhance drought tolerance by suppressing the expression of GhWRKY33, thereby activating the downstream drought response genes in cotton.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In plants, modifications of histones, including acetylation, methylation, phosphorylation, ubiquitylation, and adenosine diphosphate ribosylation, can cause dynamic changes of chromatin structure and gene expression (Peterson and Laniel 2004). Acetylation and deacetylation of histones play vital roles in regulating a series of important biological processes in plants (Pfluger and Wagner 2007). The positive charges of histone tails can be neutralized by acetylation, leading to the decreased ability of the histone binding to negatively charged DNA, and making the chromatin structure more loosely to facilitate the entry of transcriptional regulators (Annunziato and Hansen 2001). In contrast, eliminating the acetyl groups by histone deacetylation can lead to an increased affinity of histone binding to DNA, and thence block the availability of transcriptional regulators (Chrun et al. 2018).
The level of histone acetylation is closely related to gene expression and can be used as a marker to understand the transcriptional activity of the related genes. Histone acetylation is modulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Legube and Trouche 2003). HATs catalyze hyperacetylation by adding acetyl groups to the lysine residues of histone, while HDACs bring about deacetylation by eliminating acetyl groups from the lysine residues of histone (Koprinarova et al. 2016).
HDACs are a class of important enzymes widely distributed in plants. They are involved in regulating many biological processes, such as determination of plant cell-type specificity, transition between developmental stages and response to environmental stress (Liu et al. 2014). HDACs can be divided into three subfamilies including RPD3/HDA1 (Reduced Potassium Dependence 3/Histone Deacetylase 1), SIR2 (Silent Information Regulator 2), and HD2 (Histone Deacetylase 2)-related proteins. The SIR2 subfamily has a catalytic domain whose function required nicotine adenine dinucleotide (NAD) as a cofactor, while members of the RPD3/HDA1 subfamily share sequence homology in the HDAC domain, and need Zn2+ cofactor for deacetylase activity (Liu et al. 2014). Besides, the HD2 subfamily is a class of plant-specific HDACs, and have four members in Arabidopsis, including HD2A, HD2B, HD2C and HD2D (Hollender and Liu 2008). It has been indicated that HDACs play vital roles in plant response to environmental stresses. SRT2 acts as a negative regulator that participates in plant basic defense response by suppressing salicylic acid (SA) biosynthesis (Wang et al. 2010). HDA19 forms a protein complex with AtERF7 and AtSin3 to regulate expressions of the abiotic stress-responsive genes, and the hda19 mutant increases the sensitivity to ABA and salt stress (Luo et al. 2012a). Similarly, the T-DNA insertion mutant of HDA6 also shows a highly sensitive phenotype to ABA and salt stress, and the transcription levels of ABA-responsive genes are declined in hda6 mutant (Chen and Wu 2010). The lysine residue 9 of histone H3 (H3K9) is an important acetylation modification site. Lysine 9 acetylation of histone H3 (H3K9ac) is mainly occurred around the transcript start site (TSS) regions, indicating H3K9ac is closely related to the transcriptional activity of genes (Zhou et al. 2010). Previous study showed that the higher expression levels of some water deprivation-related genes were detected in hda19 mutant, and also the higher levels of H3K9 acetylation at promoters of these genes were found in the hda19 mutant, relative to wild-type plants (Chen and Wu 2010). These results illustrate that the H3K9ac level is associated with the expression change of drought-related genes.
Upland cotton (Gossypium hirsutum) is an important crop planted worldwide, and provides natural fiber materials for the textile industry. Additionally, cotton seeds are also utilized as an important source of edible oil, and therefore have important value for the food industry (Gotmare et al. 2004). Drought is an important environmental limiting factor for agricultural production. The water source is very scarce in more than half of the cotton planting area (Pettigrew 2004). Thus, understanding the molecular mechanism and genetic basis in cotton response to drought stress is significant for basic cotton biology, and will contribute to cultivating drought-tolerant cotton cultivars. Although some plant HDACs have been reported to participate in plant response to drought stress (Luo et al. 2012a), the roles of HDACs in cotton response to drought stress remain unclear and need to be explored in details. In this study, we revealed that GhHDT4D participates in cotton response to drought stress. Overexpression of GhHDT4D in Arabidopsis increased drought tolerance of the transgenic plants, whereas silencing GhHDT4D in cotton resulted in the reduced tolerance to drought. Further study revealed that GhHDT4D functions in cotton response to drought stress possibly by modulating the H3K9 acetylation level at the promoter region of GhWRKY33, and thereby suppressing GhWRKY33 expression.
Results
GhHDT4D expression is repressed in cotton by drought
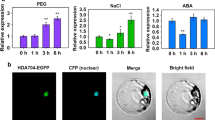
To study the roles of the histone deacetylases in cotton, a total of 33 HDAC genes were identified in upland cotton (Gossypium hirsutum), and the HDAC homologous genes were also identified in diploid A genome species (G. arboreum) and D genome species (G. raimondii) (Table S1). Among them, we found a gene (designated as GhHDT4D, Ghir_D11G035640) may be involved in cotton response to drought stress. The evolutionary analysis revealed that GhHDT4D clusters to the branch with AtHD2A, AtHD2AB, AtHD2C and AtHDT2C (Fig. 1a), and motif analysis indicated that GhHDT4D shares similar motif composition to HD2 subgroup of AtHDACs (Fig. 1b), demonstrating that GhHDT4D belongs to the HD2 subgroup. We further analyzed GhHDT4D expression in cotton based on the cottonFGD database (https://cottonfgd.org/), and found the expression level of GhHDT4D is increased in cotton 1–3 h after PEG treatment, and then declined in cotton tissues under PEG treatment for 6–12 h (Fig. 1c). Furthermore, the 4-week-old cotton seedlings were subjected to water-withholding treatment for 7 days. The leaves of cotton seedlings with normal watering and water-withholding treatment were collected to extract RNA for expression analysis. The results showed that the transcription of GhHDT4D was inhibited in leaves of cotton after drought stress (Fig. 1d), suggesting that GhHDT4D may participate in cotton response to drought stress. We also investigated the expression pattern of HDAC genes under drought stress using the previously published transcriptome data from our laboratory (Chen et al. 2013). Based on transcriptome data, we can find that the expression of GhHDT4D is obviously altered under drought stress (Fig. S1), corresponding to the results above. Besides, the transcriptome data showed that expressions of some other HDAC genes are also altered under drought (Fig. S1), indicating that some other HDAC genes may also be involved in cotton drought response.
Characterization and expression analysis of GhHDT4D. a Phylogenetic analysis of GhHDACs and HD2 subgroup of Arabidopsis. The unrooted phylogenetic tree was constructed using MEGA 6.0 by Neighbor-Joining method and the bootstrap test was performed with 1000 iterations. b Motif structure comparison using MEME Suite (https://meme-suite.org/tools/meme) with a set of parameters as follows: the optimum motif width was set to ≥ 6 and ≤ 50; the maximum number of motifs was set to 15. Different motifs are represented by various colored boxes. Motif 1/3/4/6/8/10/13/14/15 were annotated as the conserved histone deacetylase domain (pfam00850). Motif 2/11/12 were annotated as the nucleoplasmin-like domain (pfam17800). c Expression profile of GhHDT4D in cotton under PEG treatment. d Expression of GhHDT4D in cotton under drought stress. The experiments were repeated three times, and error bars denote the standard deviation calculated from three independent experiments. Asterisks represent Student’s t test in statistical analysis for significant differences: *P < 0.05; **P < 0.01
Ectopic expression of GhHDT4D improves transgenic Arabidopsis drought tolerance
To explore the function of GhHDT4D in response to drought, the coding sequence of GhHDT4D under the control of CaMV35S promoter was ectopically expressed in Arabidopsis Col-0 plants. The expression levels of GhHDT4D in the transgenic Arabidopsis lines were detected by quantitative RT-PCR (Fig. 2a), and the transgenic line L1, L7 and L15 with higher expression levels of GhHDT4D were selected for further analysis. To observe the phenotype of the transgenic Arabidopsis, the four-week-old GhHDT4D transgenic plants and wild type were subjected to water-withholding treatment for 15 days. As shown in Fig. 2b, the growth status of the transgenic lines was better than that of wild type. Besides, the contents of MDA, proline, chlorophyll, and activities of SOD (Superoxide dismutase) and POD (Peroxidase) were measured in leaves of the transgenic plants and wild type under drought stress and normal growth condition. The experimental results showed that no significant difference was found between wild type and GhHDT4D transgenic lines under normal growth condition. However, the contents of proline and chlorophyll, and activities of SOD and POD in leaves of the GhHDT4D transgenic lines were higher, but MDA content in the transgenic plants were lower than those in wild type under drought treatment (Fig. 2c–g). Reducing the stomatal opening is an important way for plant to reduce water loss when facing drought stress. So we observed the stomatal aperture of wild type and the transgenic plants under normal condition and drought treatment. As shown in Fig. 2h, i, there was no obvious difference in stomatal aperture opening between wild type and the transgenic plants under normal growth condition. However, the stomata apertures of wild type plants opened wider than those of the transgenic plants under drought stress, suggesting that the transgenic lines had lower rate of water loss. The above results indicated that the transgenic plants enhanced their drought tolerance owing to the ectopic expression of GhHDT4D in Arabidopsis.
Phenotypic analysis of GhHDT4D overexpression transgenic Arabidopsis under drought stress. a Quantitative RT-PCR analysis of GhHDT4D expression in the transgenic lines and wild type. b Phenotypic analysis of 4-week-old wild type and GhHDT4D transgenic Arabidopsis grown in soil under drought stress by withholding water for 15 days. c–g Several physiological indexes measured in the transgenic plants and wild type under normal growing condition (control) and drought treatments. c Proline content. d MDA content. e Chlorophyll content. f SOD activity. g POD activity. h Images of stoma in wild type and transgenic lines under normal growing conditions (control) and drought treatment. i Comparative stomatal aperture measurements (ratio of width to length). The experiments were repeated three times, and error bars denote the standard deviation calculated from three independent experiments. Asterisks represent Student’s t-test in statistical analysis for significant differences: *P < 0.05; **P < 0.01. WT, wild type; L1, L7, L15, three GhHDT4D overexpression transgenic lines
GhHDT4D positively regulated drought tolerance in cotton
To further explore the role of GhHDT4D in cotton response to drought, we conducted a virus induced gene silencing (VIGS) experiment. The plants infected with TRV2:GhCLA appeared the albino phenotype, indicating the success of the VIGS experiment (Fig. 3a). Then the expression level of GhHDT4D was detected in TRV2:00 and TRV2:GhHDT4D plants to confirm the expression of GhHDT4D was effectively suppressed. Quantitative RT-PCR analysis revealed that the expression levels of GhHDT4D were significantly reduced in TRV2:GhHDT4D plants compared with the TRV2:00 plants (Fig. 3b). After four weeks of growth, the TRV2:00 and TRV2:GhHDT4D plants were subjected to drought treatment. After water-withholding treatment for 10 days, we observed that the TRV2:GhHDT4D plants were more seriously wilted than those TRV2:00 plants (Fig. 3c), indicating that silencing GhHDT4D impared cotton tolerance to drought. These results suggested that GhHDT4D may have an important effect on cotton response to drought.
Phenotypic analysis of GhHDT4D VIGS cotton plants under drought stress. a Albino phenotypes of positive control cotton plants. b Quantitative RT-PCR analysis of GhHDT4D expression in mock and GhHDT4D VIGS cotton plants. Experiments were repeated three times, and error bars denote the standard deviation calculated from three independent experiments. Asterisks represent Student’s t test in statistical analysis for significant differences: *P < 0.05; **P < 0.01. c Phenotypic analysis of mock and GhHDT4D VIGS cotton plants under drought stress by withholding water for 10 days (bottom)
GhHDT4D modulates the H3K9 acetylation level of GhWRKY33 promoter in cotton
H3K9 is an important epigenetic modification site that is positively correlated with transcriptional activity of stress-responsive genes in plants (Kim et al. 2015). To investigate whether silencing GhHDT4D can affect H3K9 acetylation level in cotton, we detected the H3K9ac level in the TRV2:00 and TRV2:GhHDT4D plants. As shown in Fig. 4a and b, H3K9ac level in TRV2:GhHDT4D plants was higher than that in TRV2:00 plants, suggesting that GhHDT4D could regulate H3K9 acetylation level in cotton. A previous report showed that overexpression of GhWRKY33 resulted in the transgenic Arabidopsis sensitivity to drought stress (Wang et al. 2019). Here, we found that the transcription level of GhWRKY33 is increased in TRV2:GhHDT4D plants compared with TRV2:00 plants (Fig. 4c). Therefore, the H3K9 acetylation level of GhWRKY33 promoter was detected by the Chip-qPCR assay using the anti-H3K9ac antibody. As shown in Fig. 4d, the H3K9 acetylation level of GhWRKY33 promoter was increased in TRV2:GhHDT4D plants relative to the controls. To determine whether GhWRKY33 is the direct target gene of GhHDT4D, the GhHDT4D-GFP fusion construct was introduced into cotton hypocotyls by Agrobacterium-mediated DNA transfer. After three months, the transformed callus cells were used for the Chip-qPCR assay. As shown in Fig. 4e, GhHDT4D-GFP fusion proteins were expressed successfully in the callus cells. Chip-qPCR assay, using anti-GFP antibody, showed that GhHDT4D can directly bind to the promoter of GhWRKY33 (Fig. 4f). The above results suggested that GhHDT4D negatively regulates GhWRKY33 expression by decreasing the H3K9 acetylation level of GhWRKY33 promoter for cotton response to drought stress.
Silencing of GhHDT4D changes the expression and H3K9 acetylation level of GhWRKY33 in cotton. a and b Comparison of H3K9 acetylation levels between mock and silenced GhHDT4D cotton plants. a The expression analysis of GhWRKY33 in mock and silenced GhHDT4D cotton plants. b Immunoblot assay for histone acetylation levels in mock and silenced GhHDT4D cotton plants, probed with anti-H3K9ac and antiH3 antibodies. The intensities of blotting signals were quantified using Image J software. c Quantitative RT-PCR analysis of GhHDT4D expression in mock and silenced GhHDT4D cotton plants. Experiments were repeated three times, and error bars denote the standard deviation calculated from three independent experiments. Asterisks represent Student’s t test in statistical analysis for significant differences: *P < 0.05; **P < 0.01. d Chip-qPCR analysis of H3K9 acetylation level of GhWRKY33. The 1000 bp promoter sequence of GhWRKY33 was divided into four segments (P1–P4) for Chip-qPCR. Experiments were repeated three times, and error bars denote the standard deviation calculated from three independent experiments. Asterisks represent Student’s t test in statistical analysis for significant differences: *P < 0.05; **P < 0.01. e Western blot analysis of expression of GhHDT4D-GFP fusion protein in transgenic cotton callus cells. f ChIP-PCR assay of GhHDT4D binding to GhWRKY33 promoter. The transgenic callus cells overexpressing GhHDT4D-GFP were used for the ChIP assay with anti-GFP antibody, and the precipitated DNA was quantified by semiquantitative RT-PCR analysis
GhWRKY33 negatively regulates cotton drought tolerance
To further understand the role of GhWRKY33 in cotton response to drought, similarly, we silenced GhWKY33 in cotton via VIGS technique. As shown in Fig. 5a, the expressions of GhWRKY33 and GhHDT4D were effectively repressed in the target gene-silenced plants. Then, the four-week-old TRV2:00, TRV2:GhWRKY33 and TRV2:GhHDT4D plants were subjected to drought treatment. After 15 days of water-withholding treatment, the degree of leaf wilting in the TRV2:GhHDT4D plants was more serious compared with that in TRV2:00 and TRV2:GhWRKY33 plants, whereas the TRV2:GhWRKY33 plants displayed the best growth status (Fig. 5b). Additionally, we measured several physiological parameters (including contents of MDA, proline and chlorophyll and activities of SOD and POD) in leaves of plants under both drought stress and normal growth condition. As shown in Fig. 5c–g, no significant difference was observed in these parameters between the target gene-silenced plants and TRV2:00 controls under normal growth condition. After drought treatment, however, contents of chlorophyll and proline, and activities of SOD and POD were higher, but the MDA content was lower in leaves of the TRV2:GhWRKY33 plants, compared with the TRV2:00 controls. Conversely, the TRV2:GhHDT4D plants showed the opposite phenotype. Besides, the stomatal aperture was observed in TRV2:00, TRV2:GhWRKY33 and TRV2:GhHDT4D plants. The experimental results revealed that there was no significant difference in stomatal aperture opening among the target gene-silenced plants and TRV2:00 controls under normal growth condition. However, the opening of stomatal apertures in the TRV2:GhWRKY33 plants was smaller than that in the TRV2:00 plants, whereas the opening of stomatal apertures in TRV2:GhHDT4D plants was bigger than that in the TRV2:00 plants under drought stress (Fig. 5h, i). As the opening size of stomatal apertures was closely related to the rate of water loss in plants, our results indicated that silencing GhWRKY33 resulted in the enhanced drought tolerance of cotton owing to the decreased water loss in the plants.
Phenotypic analysis of the silenced GhWRKY33 and GhHDT4D cotton plants under drought stress. a Quantitative RT-PCR analysis of GhWRKY33 and GhHDT4D expressions in mock and silenced GhWRKY33 and GhHDT4D cotton plants. b Phenotypic analysis of mock and silenced GhWRKY33 and GhHDT4D cotton plants under drought stress by withholding water for 15 days. c–e Several physiological indexes measured in the target gene-silenced cotton plants and mock controls under normal growing condition and drought treatments. c Proline content. d MDA content. e Chlorophyll content. f SOD activity. g POD activity. h Images of stoma in silenced GhWRKY33 and GhHDT4D cotton plants and mock controls under normal growing condition and drought treatment. i Comparative stomatal aperture measurements (ratio of width to length). The experiments were repeated three times, and error bars denote the standard deviation calculated from three independent experiments. Asterisks represent Student’s t test in statistical analysis for significant differences: *P < 0.05; **P < 0.01
GhHDT4D affects expressions of the drought stress-related genes in cotton
It was reported that DREB2C, DREB2A, RD20, SOS2 and RD29A play critical roles in plant response to drought stress (Kasuga et al. 1999). To elucidate the regulatory mechanism of GhHDT4D-GhWRKY33-mediated cotton response to drought stress, we analyzed the expression levels of these drought-related genes in cotton. The experimental results showed that the transcription levels of DREB2C, DREB2A, RD20, SOS2 and RD29A were all increased in the TRV2:GhWRKY33 plants, but decreased in the TRV2:GhHDT4D plants compared with those in the TRV2:00 plants (Fig. 6). Therefore, we supposed that GhWRKY33 may negatively regulate the expressions of these drought-related genes in cotton. Possibly, GhHDT4D plays a positive role in cotton response to drought by negatively regulating GhWRKY33 to affect expressions of the drought-related genes.
Quantitative RT-PCR analysis of expression of the drought stress-related genes in the silenced GhWRKY33 and GhHDT4D cotton plants. Transcript levels of GhDREB2A, GhDREB2C, GhSOS2, GhRD20-1, GhRD20-1 and GhRD29A in mock and silenced GhWRKY33 and GhHDT4D plants were determined by quantitative RT-PCR using GhUBI1 as a quantification control. Mean values and standard errors (bars) are shown from three independent experiments. Asterisks represent Student’s t test in statistical analysis for significant differences: *P < 0.05; **P < 0.01
GhHDT4D interacts with GhHDA19
Previous study reported that HD2 proteins could interact with RPD3-type histone deacetylase for forming a corresponding dimer (Luo et al. 2012a, b, c). Thus, the yeast two-hybrid assay was conducted to search potential proteins interacting with GhHDT4D. Results showed that GhHDT4D can interact with GhHDA19 in yeast (Fig. 7a). To further determine the interaction between GhHDT4D and GhHDA19, the coding sequences of GhHDT4D and GhHDA19 were inserted into JW771 and JW772 vectors, respectively, to perform the bimolecular fluorescence complementation (BiFC) assay. As shown in Fig. 7b, GhHDT4D could interact with GhHDA19 in tobacco leaves. The above results showed that GhHDT4D can interact with GhHDA19, an RPD3-type histone deacetylase, suggesting that GhHDT4D may form a dimer with GhHDA19 for its function in cotton.
GhHDT4D interacts with GhHDA19 in vitro and in vivo. a Yeast two-hybrid assay of GhHDT4D interacted with GhHDA19. BD, GAL4 DNA binding domain; AD, GAL4 activation domain. Left panel: The yeast cells grown on 2-dropout medium. Right panel: The yeast cells grown on selective 4-dropout medium. b Bimolecular fluorescence complementation (BiFC) assay of GhHDT4D interacted with GhHDA19. cLUC, the carboxy terminus of luciferase; nLUC, the amino terminus of luciferase
Discussion
GhHDT4D is involved in cotton response to drought stress
The HD2 genes are a class of plant-specific genes that have been reported to be involved in plant response to abiotic stress (Zhou et al. 2004; Luo et al.2017). In Arabidopsis, the transcription of HDT1 (HDT2A), HDT2 (HDT2B), HDT3 (HD2C) and HDT4 (HD2D) is suppressed under ABA treatment and high salt stress (Luo et al. 2012a; Zhou et al. 2004). In rice, the transcription level of HDT701 is reduced dramatically 1 h after ABA, NaCl and PEG treatments, but recovered after ABA and PEG exposure for 3 h. Similarly, expression of HDT702 in rice is reduced 1 h after ABA treatment, but dramatically enhanced after ABA, NaCl, and PEG exposure for 3 h, indicating that the expressions of HDT701 and HDT702 could be regulated by abiotic stresses (Zhao et al. 2015). In this study, we found that the expression of GhHDT4D is repressed in cotton under drought treatment, suggesting that GhHDT4D may be involved in cotton drought response. Moreover, previous study revealed that overexpression of AtHD2D in Arabidopsis enhanced plant tolerance to drought and salt stresses (Han et al. 2016; Farhi 2015). Overexpression of AtHD2C also increased plant resistance to drought and salt stresses by regulating the expressions of some ABA-responsive genes (Sridha and Wu 2006). Similarly, overexpression of HDT701 in rice enhanced plant salt and osmotic stress resistance. On the contrary, hdt701 mutant seedlings displayed the increased sensitivity to both salt and osmotic stresses with the decreased expression levels of stress-inducible genes (Zhao et al. 2015; Wai and An 2018). Likewise, we found that overexpression of GhHDT4D strengthened the drought tolerance of the transgenic Arabidopsis, while virus-induced gene silencing of GhHDT4D remarkably reduced cotton drought resistance, suggesting that GhHDT4D may play an important role in cotton response to drought stress.
GhHDT4D regulates the H3K9ac level in cotton
Histone deacetylases regulate the histone acetylation levels on DNA for influencing the transcription activities of genes (Ma et al. 2013; Liu et al. 2014). It has been demonstrated that lysine residue 9 of histone H3 (H3K9) is a critical site for the acetylation/deacetylation (Zhou et al. 2010). This epigenetic modification site is positively correlated with transcriptional activity of stress-responsive genes in plants (Kim et al. 2015; Zhou et al. 2010). In Arabidopsis, the HDA9 mutation increases the expressions of 47 stress-related genes that are accompanied by higher H3K9ac in their promoters (Zheng et al. 2016). The hda19 mutant has higher levels of H3K9ac in the PR1 and PR2 promoter regions compared with wild type (Choi et al. 2012). Overexpression of HDC1 increased the transcript levels and H3K9K14ac levels of the salt stress-regulated genes (Perrella et al. 2013). In hd2c mutant, the expression and H3K9K14ac levels of ABI1 and ABI2 are higher than those in wild type plants (Luo et al. 2012a, b, c). In srt1-1/srt2-1 double mutant, the H3K9ac level was significantly increased compared with that in wild type. SRT1 and SRT2 suppress expressions of ethylene-repressed genes by regulating H3K9ac levels of them (Zhang et al. 2018). In Brachypodium, overexpression of BdHD1 leads to a decrease of H3K9ac level. BdHD1 acts as a global regulator to link the H3K9ac level and gene expression to respond to drought stress (Song et al. 2019). In this study, we found that the H3K9ac level in the TRV2:GhHDT4D plants was higher than that in TRV2:00 plants, indicating that GhHDT4D may regulate the H3K9ac level for responding to drought stress in cotton.
GhHDT4D plays a positive role in cotton response to drought by suppressing GhWRKY33 expression
It is known that the histone deacetylases can repress the expressions of the target genes by decreasing the histone acetylation levels of these genes (Chen et al. 2015; Liu et al. 2014). Therefore, we speculated that the histone acetylation levels and expression levels of putative target genes of GhHDT4D should be increased in the GhHDT4D silenced cotton. Furthermore, a study indicated that histone deacetylase 701 enhances rice abiotic stress resistance at the seedling developmental stage by suppressing expression of OsWRKY45 (Wai and An 2018). Additionally, WRKY transcription factors play essential roles in plant responsed to drought stress (Chen et al. 2012). In Arabidopsis, three WRKY genes, WRKY46, WRKY54 and WRKY70, participate in drought stress responses as negative regulators, and the triple mutant wrky46/wrky54/wrky70 exhibits the enhanced drought stress tolerance (Chen et al. 2017). Overexpression of GhWRKY33 increased plant sensitivity to drought (Wang et al. 2019). In this study, silencing GhWRKY33 increased cotton tolerance to drought, suggesting that GhWRKY33 plays a negative role in cotton response to drought stress. So we detected the H3K9 acetylation level and expression of GhWRKY33 in TRV2:00 and TRV2:GhHDT4D plants. As expected, both acetylation level and expression level of GhWRKY33 were increased in the GhHDT4D silenced plants compared with TRV2:00 controls. Furthermore, Chip-PCR analysis showed that GhHDT4D could directly bind to the promoter region of GhWRKY33. These results suggested that GhHDT4D suppresses the expression of GhWRKY33 by modulating its H3K9ac level.
As WRKY transcription factors can bind to W-box cis-elements existed in promoter regions of the target genes to regulate their transcription activities (Chen et al. 2012), to elucidate the regulatory pathway governed by GhHDT4D depended on GhWRKY33, we analyzed the expressions of some drought-related genes, including DREB2A, DREB2C, RD20, SOS2 and RD29A, with W-box motifs on their promoters, in TRV2:00, TRV2:GhHDT4D and TRV2:GhWRKY33 plants. These genes have been reported to contribute to drought tolerance in different plants (Nakashima et al. 2000; Sakuma et al. 2006; Xiu et al. 2016; Je et al. 2014; Zhao et al. 2013; Aubert et al. 2010; Liu et al. 2000; Xiao et al. 2009). In this study, the transcriptional levels of DREB2A, DREB2C, RD20, SOS2 and RD29A were decreased in the TRV2:GhHDT4D plants, but increased in the TRV2:GhWRKY33 plants compared with those in the TRV2:00 plants. The above results indicate that the increased sensitivity of GhHDT4D silenced cotton to drought stress is due to the reduced expression of these drought-related genes. It also means that increased expression of drought stress-related genes contributes to increase the drought tolerance of GhWRKY33 silenced cotton. GhHDT4D can enhance expressions of the drought stress-related genes, but GhWRKY33 can suppress their expressions. Also, because GhHDT4D can repress the expression of GhWRKY33, we infer that GhHDT4D may enhance the expression of these drought-related genes by suppressing the transcription activity of GhWRKY33. In brief, our data suggest that GhHDT4D plays a positive role in cotton response to drought stress. GhHDT4D might enhance the osmotic stress tolerance of cotton via suppressing the expression of GhWRKY33, thereby releasing the inhibition of the downstream drought-related genes by GhWRKY33.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana (Columbia ecotype) seeds were surface-sterilized with 10% NaClO for 5 min, followed by washing three times with sterile water. Then the seeds were plated on Murashige and Skoog (MS) medium. After placed at 4 °C for 72 h in darkness, the seeds were transferred to the growth incubator for germination and development (22 °C, 16 h light/8 h dark). Seven days later, the seedlings were transplanted into soil and grew in the growth chamber (22 °C, 16 h light/8 h dark).
Cotton (Gossypium hirsutum cv. Coker312) seeds germinated and grew under controlled condition (25 °C, 16 h light/8 h dark) in culture room. Tissues were harvested from these seedlings for further study.
Analyses of phylogenetic relationship and motifs of GhHDACs
The protein sequences of GhHDACs were searched in cotton (Gossypium hirsutum L., cultivar TM-1 (AD1)) genome database (https://cottonfgd.org/). To identify GhHDACs, the Arabidopsis HDAC proteins were employed as queries to search cotton database. The MEGA 6.0 was used to construct the unrooted phylogenetic tree. The protein sequences were subjected to the Motif Elicitation (MEME) online program (https://meme.sdsc.edu/meme/intro.html) to predict conserved motif (Bailey et al. 2015). The InterProScan (https://www.ebi.ac.uk/Tools/InterPro-Scan/) was used to annotate the identified motifs (De Castro et al. 2006).
Transcriptome data for analyzing gene expressions in cotton
The transcriptomic data of cotton under drought stress used in this study was obtained from our laboratory (Chen et al. 2013). The fragments per kilobase per million reads (FPKM) value of genes is discarded if all three samples are less than one (Zhu et al. 2018). Then, the remaining expression data were used for generation of heatmap using TBtools (Chen et al. 2018).
Arabidopsis transformation and phenotypic analysis
The coding sequence of GhHDT4D was cloned into PK2GW7.0 vector to generate 35S:GhHDT4D construct. The construct was introduced into Arabidopsis through Agrobacterium-mediate transformation. Seeds of GhHDT4D homozygous lines (T3 generation) were selected by kanamycin resistance and were used for further study. For drought treatment, the four-week-old seedlings were subjected to water-deficit treatment for 15 days. The phenotypes of the plants were observed and photographed after drought treatments. Each of the experiments was performed at three independent biological replicates.
Test of virus induced gene silencing (VIGS) in cotton
The VIGS experiment was carried out by the method as described previously (Gao et al 2016). The fragments (400 bp) of GhHDT4D and GhWRKY33 genes were inserted into TRV2 vector to generate TRV2:GhHDT4D and TRV2:GhWRKY33 constructs, respectively. The constructs were transferred into Agrobacterium tumefaciens strain GV3101. The transformed agrobacteria solution containing the TRV1 and TRV2:00 or TRV2:GhHDT4D or TRV2:GhWRKY33 was mixed and injected into cotyledons of cotton seedlings. After detecting the expression of GhHDT4D and GhWRKY33 by quantitative RT-PCR analysis, the four-week-old cotton seedlings were subjected to water-deficit treatment. Each of the experiments was performed at three independent biological replicates. Nine individuals were used in each treatment and control, respectively.
Determination of drought stress-related physiological parameters
Malondialdehyde (MDA) content was determined from 0.1 g cotton leaf tissues by using MDA Quantification Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). For quantification of proline content, 0.1 g samples of cotton leaves were prepared and followed the procedure as described by the manufacturer of Proline Quantification Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The measurement of peroxidase (POD) and superoxide dismutase (SOD) enzyme activities in the stressed plants and controls was performed as described by the manufacturer of Peroxidase (POD) assay kit and Superoxide Dismutase (SOD) assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Stomatal aperture was observed in leaves of cotton under normal condition and drought treatment by microscopy, and the ratio of stomatal length to width was measured (n > 50 stoma per sample).
Yeast two-hybrid assay
The coding sequence of GhHDA19 was inserted into pGADT7 vector and transferred into yeast strain AH109, while the coding sequence of GhHDT4D was inserted into pGBKT7 vector and transferred into yeast strain Y187, using the high-efficiency lithium acetate transformation procedure following the manufacturer’s instructions (Clontech). Then AH109 and Y187 yeast haploid strains were mated. The mated yeast diploid strains were identified on yeast double drop-out (DDO) medium lacking Leu and Trp, and were incubated on DDO medium at 30 °C for 3–4 days. The positive interactions were identified on yeast quadruple dropouts (QDO) medium lacking Leu, Trp, His and Ade.
Bimolecular fluorescence complementation (BiFC) assay
The bimolecular fluorescence complementation experiment was carried out as described by Chen et al (2008). The coding sequences of GhHDT4D and GhHDA19 were cloned into JW771 and JW772 vectors, respectively. In the constructs, GhHDT4D was fused to the amino terminus of luciferase (nLUC) to generate nLUC-GhHDT4D, and GhHDA19 was fused to the carboxy terminus of LUC (cLUC) to generate GhHDA19-cLUC, respectively, using cLUC and nLUC as negative controls. The vectors were transferred into Agrobacterium tumefaciens stain GV3101. Resuspending Agrobacterium cells to OD1.0 with infiltration buffer (10 mM MgCl2, 10 mM MES (2-(N-morpholino) ethanesulfonic acid) pH 5.7, 150 mM acetosyringone). Then, the Agrobacterium cell solution was injected into tobacco (Nicotiana benthamiana) leaves. After 72 h of co-cultivation, the tobacco leaves were used for detecting luciferase activity using chemiluminescence image analysis system (Tanon 4600SF).
Western blot analysis
Total proteins were extracted from leaves of cotton seedlings or callus cells by the method described previously (Yao et al. 2006). The protein samples were loading onto SDS-PAGE gels. After separation by electrophoresis, proteins were transferred to a nitrocellulose membrane (Amersham) using a Trans-Blot Semi-DryElectrophoretic Transfer Cell (Bio-Rad). The primary antibodies anti-H3K9ac (Millipore), anti-H3 (Millipore) and anti-GFP (ABclonal) were incubated overnight at 4 degrees Celsius. The membrane was washed three times for 15 min each time by using TBST solution. Secondary rabbit antibody (CWBIO) was incubated with the membrane for 1.5 h. Then The membrane was washed three times for 15 min each time by using TBST solution. Proteins were detected using the eECL Western Blot Kit (CWBIO) and developed using chemiluminescence imaging system(LI-COR).
Chromatin immunoprecipitation (ChIP) assay
ChIP experiment was conducted as described previously (Liu et al. 2019). In brief, 3 g of leaves of cotton seedlings or callus cells were harvested and then fixed in 1% formaldehyde solution for 20 min in a vacuum. The cross-linking reactions were stopped by adding 0.125 M glycine. The samples were ground in liquid nitrogen and chromatin was extracted with extraction buffers as described in the protocol. Sonication was applied to shear chromatin into pieces with an average length of 500 bp. The chromatin fragments were immunoprecipitated with the specific antibody anti-H3K9ac and anti-GFP (Millipore and ABclonal). The DNA fragments were purified and used as templates for ChIP-PCR analysis. All the primers used in the experiments are listed in Table S2.
References
Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Galaud J (2010) RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 51:1975–1987
Annunziato AT, Hansen JC (2001) Role of histone acetylation in the assembly and modulation of chromatin structures. Gene expression. J Liver Res 9(1–2):37–61
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:W39–W49
Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146:368–376
Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, Yin Y (2017) Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29:1425–1439
Chen LT, Wu K (2010) Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav 5:1318–1320
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys A 1819:120–128
Chen C, Chen H, He Y, Xia R (2018) TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. https://doi.org/10.1101/289660
Chen Y, Liu ZH, Feng L, Zheng Y, Li DD, Li XB (2013) Genome-wide functional analysis of cotton (Gossypium hirsutum) in response to drought. PLoS ONE 8(11):120–128
Chen HP, Zhao YT, Zhao TC (2015) Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog. 20:35–47
Chrun ES, Modolo F, Daniel FI (2018) HDAC1 (histone deacetylase 1). Atlas of genetics and cytogenetics in oncology and haematology.
Choi SM, Song HR, Han SK, Han M, Kim CY, Park JJ, Noh B (2012) HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J 71:135–146
De Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Hulo N (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34(2):W362–W365
Farhi JA (2015) HD2D is a regulator of abscisic acid responses in Arabidopsis. Electronic thesis and dissertation repository 3467.
Gao W, Long L, Xu L, Lindsey K, Zhang X, Zhu L (2016) Suppression of the homeobox gene HDTF1 enhances resistance to Verticillium dahliae and Botrytis cinerea in cotton. J Integr Plant Biol 58:503–513
Gotmare V, Singh P, Mayee CD, Deshpande V, Bhagat C (2004) Genetic variability for seed oil content and seed index in some wild species and perennial races of cotton. Plant Breed 123:207–208
Han Z, Yu H, Zhao Z, Hunter D, Luo X, Duan J, Tian L (2016) AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front Plant Sci 7:310
Hollender C, Liu Z (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50:875–885
Je J, Chen H, Song C, Lim CO (2014) Arabidopsis DREB2C modulates ABA biosynthesis during germination. Biochem Biophys Res Commun 452:91–98
Kim JM, Sasaki T, Ueda M, Sako K, Seki M (2015) Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Frontiers Plant Sci 6:114
Koprinarova M, Schnekenburger M, Diederich M (2016) Role of histone acetylation in cell cycle regulation. Curr Top Med Chem 16(7):732–744
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287
Legube G, Trouche D (2003) Regulating histone acetyltransferases and deacetylases. EMBO Rep 4:944–947
Liu X, Yang S, Zhao M, Luo M, Yu CW, Chen CY, Wu K (2014) Transcriptional repression by histone deacetylases in plants. Mol Plant 7:764–772
Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97:3730–3734
Liu ZH, Chen Y, Wang NN, Chen YH, Wei N, Lu R, Li XB (2019) A basic helix—loop—helix (bHLH) protein (GhFP1) promotes fiber elongation of cotton (Gossypium hirsutum) via modulating BR biosynthesis and signaling. New Phytol. https://doi.org/10.1111/nph.16301
Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K (2012a) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta 1819:129–136
Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y, Wu K (2012b) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63:3297–3306
Luo M, Wang YY, Liu X, Yang S, Wu K (2012c) HD2 proteins interact with RPD3-type histone deacetylases. Plant Signal Behav 7:608–610
Luo M, Cheng K, Xu Y, Yang S, Wu K (2017) Plant responses to abiotic stress regulated by histone deacetylases. Frontiers Plant Sci 8:2147
Ma X, Lv S, Zhang C, Yang C (2013) Histone deacetylases and their functions in plants. Plant Cell Rep 32(4):465–478
Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration-and high-salinity-responsive gene expression. Plant Mol Biol 42:657–665
Perrella G, Lopez-Vernaza MA, Carr C, Sani E, Gosselé V, Verduyn C (2013) Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25:3491–3505
Peterson CL, Laniel MA (2004) Histones and histone modifications. Curr Biol 14:R546–R551
Pettigrew WT (2004) Physiological consequences of moisture deficit stress in cotton. Crop Sci 44:1265–1272
Pfluger J, Wagner D (2007) Histone modifications and dynamic regulation of genome accessibility in plants. Curr Opin Plant Biol 10:645–652
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309
Song J, Henry HA, Tian L (2019) Brachypodium histone deacetylase BdHD1 positively regulates ABA and drought stress responses. Plant Sci 283:355–365
Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46(1):124–133
Wai AH, An G (2018) Histone Deacetylase 701 enhances abiotic stress resistance in rice at the seedling stage by suppressing expression of OsWRKY45. 2nd Myanmar-Korea conference on useful plants, Dagon University.
Wang C, Gao F, Wu J, Dai J, Wei C, Li Y (2010) Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol 51(8):1291–1299
Wang NN, Xu SW, Sun YL, Liu D, Zhou L, Li Y, Li XB (2019) The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci Rep 9:724
Xiao BZ, Chen X, Xiang CB, Tang N, Zhang QF, Xiong LZ (2009) Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol Plant 2:73–83
Xiu Y, Iqbal A, Zhu C, Wu G, Chang Y, Li N, Wang H (2016) Improvement and transcriptome analysis of root architecture by overexpression of Fraxinus pennsylvanica DREB2A transcription factor in Robinia pseudoacacia L. ‘Idaho’. Plant Biotechnol J 14:1456–1469
Yao Y, Yang YW, Liu JY (2006) An efficient protein preparation for proteomic analysis of developing cotton fibers by 2-DE. Electrophoresis 27:4559–4569
Zhang F, Wang L, Ko EE, Shao K, Qiao H (2018) Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30(1):153–166
Zhao J, Zhang J, Zhang W, Wu K, Zheng F, Tian L, Duan J (2015) Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front Plant Sci 5:764
Zhao K, Shen X, Yuan H, Liu Y, Liao X, Wang Q, Li T (2013) Isolation and characterization of dehydration-responsive element-binding factor 2C (MsDREB2C) from Malus sieversii Roem. Plant Cell Physiol 54:1415–1430
Zheng Y, Ding Y, Sun X, Xie S, Wang D, Liu X, Zhou DX (2016) Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J Exp Bot 67:1703–1713
Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek-Green M, Wu K (2004) Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J 38(5):715–724
Zhou DX, Hu Y (2010) Regulatory function of histone modifications in controlling rice gene expression and plant growth. Rice 3(2):103–111
Zhou J, Wang X, He K, Charron JBF, Elling AA, Deng XW (2010) Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol 72:585–595
Zhu G, Li W, Zhang F, Guo W (2018) RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. BMC Genom 19(1):73
Acknowledgements
This work was financially supported by National Key R&D Program of China (Grant No. 2016YFD0100505), and National Natural Sciences Foundation of China (Grant No. 31871667).
Author information
Authors and Affiliations
Contributions
XBL and JBZ conceived and designed the research; JBZ, SPH, JWL, XPW and DDL performed the experiments; JBZ and XBL analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, JB., He, SP., Luo, JW. et al. A histone deacetylase, GhHDT4D, is positively involved in cotton response to drought stress. Plant Mol Biol 104, 67–79 (2020). https://doi.org/10.1007/s11103-020-01024-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01024-9