Abstract
Histone deacetylation catalyzed by histone deacetylases is an important type of histone modification. Histone deacetylases affect various processes of plant development and involve in responding to hormones and biotic and abiotic stresses. Here, we report a tomato PRD3/HDA1 histone deacetylase gene, SlHDA5, which is expressed ubiquitously in different tissues and development stages. Expression profiles in hormone treatments showed that SlHDA5 was induced by abscisic acid (ABA) and methyl jasmonate (MeJA). Seedlings growth of SlHDA5-RNAi lines were more inhibited on the medium containing salt compared with wild type (WT). Under salt stress, chlorophyll in mature leaves degraded earlier in transgenic leaves than that in WT, and transgenic plants displayed wilting earlier and more severe than WT. After drought treatment, transgenic plants wilted and dehydrated earlier than WT, which was confirmed by lower water and chlorophyll content, and higher malondialdehyde (MDA) content in transgenic plants manifesting that the tolerance of transgenic plants to drought receded. Under the treatment of ABA, root length of transgenic seedlings was more strongly repressed by contrast with WT, suggesting repression of SlHDA5 increased seedling sensibility to ABA. Our study indicated that silencing of SlHDA5 resulted in decreasing tolerance to salt, drought, and ABA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotic cells, the basic unit of chromatin is nucleosome, which is an octamer assembled by two copies of H3, H4, H2A, and H2B and wrapped by 145–147 bp DNA (Luger et al. 1997). In the process of genetic information transmission, DNA sequences and epigenetic markers control the activation of genes together (Berger 2007). The known epigenetic events contain DNA methylation and histone modifications. Until now, the histone post-translation modifications which have been recognized are as follows: acetylation, methylation (lysines and arginines), phosphorylation, ubiquitylation, sumoylation, ADP-ribosylation, deimination, carbonylation, glycosylation, and proline isomerization (Fuchs et al. 2006; Kouzarides 2007a).
Among these histone modifications, research on acetylation is the earliest. Histone modifications often occur in the N-terminal tails. For example, histone acetylation and deacetylation appear at the N-terminal lysines 5, 8, 12, 16, 20 of H4, 9, 14, 18, 23, 36, 56 of H3, 5 of H2A and 12, and 15 of H2B (Kouzarides 2007b; Loidl 2004; Lusser et al. 2001). It is worth noting that the histone acetylation is a dynamic process which is catalyzed by two kinds of enzymes known as histone acetyltransferases (HATs) and histone deacetylases (HDACs). Previous researches showed that HDACs widely exist in yeast, human, and plants (Taunton et al. 1996), and it was firstly isolated from plants (Sendra et al. 1988). Subsequently, more and more HDACs had been characterized and functionally studied. Based on the homology with yeast, HDACs in plants were classified into three groups: reduced potassium dependence 3/histone deacetylase 1 (RPD3/HDA1) superfamily, silent information regulator 2 (SIR2) family, and histone deacetylase 2 (HD2) family (Pandey et al. 2002; Yang and Seto 2007). In the past few years, HDACs in different species have been isolated, such as pea (Sendra et al. 1988), potato (Lagace et al. 2003), barley (Demetriou et al. 2009), Arabidopsis (Tian and Chen 2001), maize (Pipal et al. 2003), rice (Jang et al. 2003), tobacco (Bourque et al. 2011), tomato (Zhao et al. 2015a), and so on. So far, there are 18 HDACs that have been identified from Arabidopsis genome, and among these genes, 12 genes belong to RPD3/HDA1 superfamily, 4 genes belong to HD2 family, and 2 genes belong to SIR2 family (Alinsug et al. 2009; Pandey et al. 2002). However, in tomato, there are only 14 HDACs, and RPD3/HDA1 superfamily, HD2 family, and SIR2 family contain nine, three, and two members, respectively (Zhao et al. 2015a).
Histone acetylation and deacetylation determine the activation and silence of eukaryotic genes. For example, acetylation of H3 Lys9 (H3K9) is the marker for active genes while deacetylation of H3K9 and H3K14 is the marker for silenced genes (Chen and Tian 2007; Earley et al. 2006). Furthermore, HDACs integrate histone modification and DNA methylation to regulate gene silencing (Liu et al. 2012a). Researches showed that in the process of plant development, HDACs play vital roles in various events such as the leaf morphology construction, growth of hypocotyl and root, flowering time, and fruit ripening (Wang et al. 2014). In Arabidopsis, RPD3/HDA1 superfamily member HDA6 and its homologous HDA19 are the most studied histone deacetylase. HDA6 and HDA19 redundantly function in modulating the germination process and embryonic properties after germination by repressing embryo-specific gene function (Chen et al. 2010; Chen and Wu 2010; Tanaka et al. 2008). Additionally, HDA6 mediates heterochromatin silencing, transposable element silencing by interacting with DNA methyltransferase MET1 and histone demethylase FLD (Liu et al., 2012a; Liu et al., 2012b; To TK et al., 2011). Moreover, HDA6 interacts with FLC (FLOWERING LOCUS C) to regulate flowering time in Arabidopsis (Yu et al. 2011). Other Arabidopsis HAD genes have also been proved participating in various developmental processes, such as gametophyte, embryo, and root epidermis cell development (Cigliano et al. 2013; Liu et al. 2013; Luo, et al. 2015).
Besides the important role of HDACs in plant development, they also take part in responding to hormones and biotic and abiotic stresses. HDA6 is involved in jasmonate response, and the expression of the jasmonate responsive genes is down-regulated when HDA6 is repressed (Wu et al. 2008). Experiments on hda19-1, a mutant of HDA19, axe1-5, a mutant of HDA6, and HDA6 interfering plants showed that respective deletion of HDA6 and HDA19 increases the hypersensitivity to ABA and salt stress in Arabidopsis and plant deficiency in HDA6 and HDA19 display decreased expression of ABA and abiotic stress-responsive genes as well (Chen et al. 2010; Chen and Wu 2010). Nevertheless, HDA19 interacts with WRKY38 and WRKY62 and abolishes their activation to fine-tune plant basal defense responses (Kim et al. 2008a). In rice, overexpression of HDA705 decreases ABA and salt stress resistance during seed germination and enhances osmotic stress resistance during the seedling stage which indicating its role in regulating seed germination and the response to abiotic stresses in rice (Zhao et al. 2016).
In tomato, SlHDA1, SlHDA3, and SlHDA4 have been proved interacting with MADS-box proteins TAG1 (TOMATO AGAMOUS1) and TM29 (TOMATO MADS BOX 29), which are involved in reproductive development, suggesting that SlHDAC genes may contribute to plant reproductive development (Zhao et al. 2015a). Although HDACs in tomato have been identified and classified, the functional process and molecular mechanism are not very clear. Here, we focused on a tomato RPD3/HDA1 superfamily member SlHDA5 whose homolog, HDA2, was expressed primarily in embryos and dry seeds in Arabidopsis (Hollender and Liu 2008; Schmid et al. 2005). Previous report revealed that SlHDA5 is localized in nucleus and accumulated to a high level in flowers and fruit of 10 dpa, but decreased as fruit development and ripening (Zhao et al. 2015a). Furthermore, SlHDA5 was induced by abiotic stress, such as high temperature, dehydration, and salt (Guo et al. 2017). To further explore the roles of SlHDA5 in drought and salt stress response, we generated tomato plants silencing SlHDA5 by RNA interference, and the transgenic plants showed reduced tolerance to salt, drought, and ABA. These phenotypes were further confirmed by analysis of physiological and biochemical features.
Materials and Methods
Plant Materials and Growth Conditions
Solanum lycopersicon Mill. cv. Ailsa Craig++ (AC++) was used as wild type. All tomato seedlings used for hormone and treatments and tolerance assay were grown under greenhouse condition: 16/8 h day/night cycle, 25/18 °C day/night temperature, 250 μmol m−2 s−1 light intensity, and 80% humidity.
Hormone Treatments
Seedlings about 35 days old that are consistent in growth status were chosen for treatment. The whole tomato plants were sprayed with 100 μM ABA, 50 μM MeJA, 50 μM auxin (IAA), and 50 μM salicylic acid (SA) (all plant hormones are manufactured by Sigma) solution respectively while seedlings for control were sprayed with distilled water (Fujita et al. 2004). Seedlings were enclosed in plastic immediately after spraying and collected leaves at 0, 1, 2, 4, 8, 12, and 24 h for further analysis. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C for RNA extraction.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted using RNA trizol (Takara) according to the manufacturer’s instructions. The first strand cDNA was synthesized by M-MLV reverse transcriptase (Promega) using 1-μg RNA as template and Oligo (dT)18 as primer. Quantitative real-time PCR was carried out using CFX96™ Real-Time System (Bio-Rad), and the reaction system was 10 μL (5 μL 2 × SYBR Premix, 0.5 μL primers, 1 μL of cDNA, and 3.5 μL distilled water). NTC (no template control) and NRT (no reverse transcription control) were performed. Each sample was repeated for three times and standard curves were run at the same time. SlCAC gene (primer sequence: SlCAC-F: CCTCCGTTGTGATGTAACTGG, SlCAC-R: ATTGGTGGAAAGTAACATCATCG) and SlEF1α (primer sequence: SlEF1α-F: TACTGGTGGTTTTGAAGCTG, SlEF1α-R: AACTTCCTTCACGATTTCATCATA) were used as internal standard (Exposito-Rodriguez et al. 2008). The primers qSlHDA5-F (AGTGCCAAAGTTATTGCTGATTCC) and qSlHDA5-R (TTCGCCTCTGCTTTTCCCA) were used to detect the transcript level of SlHDA5 in tissues and treated materials.
Construction of SlHDA5 RNA Interference Vector and Plant Transformation
The SlHDA5-RNAi vector was constructed using pBIN19 vector. A 401-bp specific fragment of SlHDA5 was amplified with specific primers SlHDA5-F (CGGGGTACCATCGATAGCATGTCTTTGCATAGCTACTTAA) and SlHDA5-R (CCGCTCGAGGGATCCGAGGTACGACGAGAACTTGATTG). After purifying, amplified products were digested with Cla I/BamH I and Kpn I/Xho I and then linked into pHANNIBAL plasmid. The double-stranded RNA expression unit, containing the cauliflower mosaic virus 35S promoter (the sense-orientated SlHDA5 fragment (PDK intron) and the antisense-orientated SlHDA5 fragment (OCS terminator)), was digested with Sac I and Xba I. Then, the unit was linked in pBIN19 and transferred into Agrobacterium LBA4404 strain. The final vector carried SlHDA5-RNAi unit was transferred into wild-type tomato by Agrobacterium-mediated plant transformation method (Chen et al. 2004). The positive transgenic tomato plants were selected for kanamycin and detected by PCR with NPTII-F (CTCAGAAGAACTCGTCAAGAAGG) and NPTII-R (GACTGGGCACAACAGACAATC) primers.
Salt and Drought Treatment of Transgenic Tomato
Three experiments were conducted to research the effect of SlHDA5 on tomato salinity tolerance. Experiment 1: transgenic lines and WT seeds were sterilized and placed in culture flask with sterilized water in it. Then, the culture flask was put in constant temperature shaker which was 100 revolutions per minute and 28 °C. The germinant seeds were sowed on prepared culture flasks containing MS medium with 0 and 100 mM NaCl. A week later, picture was taken and the length of shoot and root was measured. Experiment 2: leaves of similar size, age, and position were detached from transgenic and WT plants and dipped in 300 mM NaCl for 4 days. Pictures were taken to record the phenotypes and the chlorophyll content was measured. Experiment 3: 35-day-old transgenic and WT plants were irrigated with 200 mL 400 mM NaCl solution every 3 days. Pictures were taken to record the phenotypes.
For drought treatment, 35-day-old T1 transgenic and WT plants with the similar growth status were selected and watered daily. Once drought treatment began, plants were withholding water until 30 days. All the plants were kept in a greenhouse under condition described above. Pictures were taken to record the phenotypes. Leaves were sampled at 0, 25, and 30 days after the onset of drought treatment to measure relative water content (RWC), total chlorophyll, and MDA contents.
Quantitation of RWC, Total Chlorophyll, and MDA Contents
Tomato leaves were detached from the plants and weighted (fresh weight, FW), then placed in culture dishes with water filled for 24 h. The water was removed from leaf surface using absorbent paper and weight to obtain turgid weight (TW). The leaves were placed in 50-mL centrifuge tubes and dried at 60 °C for 24 h, and then dry weight (DW) was weighted. The RWC was calculated using the following formula: RWC (%) = (FW−DW)/(TW−DW) × 100%.
For total chlorophyll measurement, leaves with the same weight were excised from control and treated plants. Samples were grinded with liquid nitrogen and extracted with 3 mL 80% aqueous acetone (v/v). The extract was kept in dark place for a night and centrifuged at 4000×g for 5 min. Then, the supernatant was diluted by 80% aqueous acetone and absorbance was recorded at 645 and 663 nm. The content of chlorophyll was calculated using the following formula: Chl = (20.21 × A645 + 8.02 × A663) (Pei et al. 1997).
To measure the content of MDA, leaves of the same weight were detached respectively from control and transgenic plants. Then, the samples were grinded with liquid nitrogen and added 5 mL trichloroacetic acid (TCA), mixed, and centrifuged at 4000×g for 10 min. A 2-mL supernatant was removed to a new 5-mL centrifuge tube, and 2 mL distilled water was set as control; then, we diluted the extraction with 2 mL thiobarbituric acid (TBA). The mixture was incubated in boiling water for 10–15 min and then immediately cooled in ice. The absorbance in 450, 532, and 600 nm was recorded. MDA content was calculated using the following formula: MDA contents (nmol g−1 fresh weight) = [6.45 × (A532–A600)–0.56 × A450]/fresh weight (Lim et al. 2012).
Assay for ABA Sensitivity of Transgenic Tomato Seedlings
We obtained germinating transgenic and WT seeds as described above and sown them respectively on MS medium with 0, 5, and 10 μM ABA. After a week, the phenotype was observed and root length was measured.
Results
SlHDA5 Gene Was Induced by ABA and MeJA
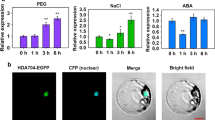
So far, researches have proved that HDACs respond to hormones and participate in hormone-induced development processes in other plants (Liu et al. 2014; Luo et al. 2012; Zhao et al. 2015b). To investigate the response of SlHDA5 gene to hormones, the expression patterns of SlHDA5 under various treatments were studied. As shown in Fig. 1a, the expression of SlHDA5 only significantly increased at 12 h while no obvious difference was observed at other time compared with control under IAA treatment. When suffered with exogenous ABA, the expression of SlHDA5 had no obvious change at 1–4 h, but increased clearly from 8 h and then peaked at 12 h which was about sixfold by contrast with control (Fig. 1b). However, for the treatments of exogenous SA, the expression of SlHDA5 was slight up-regulated at 4, 8, and 12 h while at other time points, the expression remained no distinct change (Fig. 1c). When suffered with exogenous MeJA, SlHDA5 gene was always up-regulated at 2–12 h and the peak expression about fourfold compared with control appeared at 2 h (Fig. 1d).
Detection of SlHDA5 under treatment of hormones by qPCR. Expression profile of SlHDA5 in WT leaf treating with IAA (a), ABA (b), SA (c), and MeJA (d). Seedlings about 35-day-old were treated by hormones. Each sample was repeated for three times. The asterisks indicate significant differences between the treated and contrast seedlings (P < 0.05)
Repression of SlHDA5 Increased the Sensibility to Salt
Existing research indicated that SlHDA5 was induced by salt stress both in root and leaf (Guo et al. 2017). We selected the T1 generation of two SlHDA5-RNAi transgenic tomato lines with better silencing efficiency (Fig. 2), RNAi-17 and RNAi-21 for NaCl treatment. When the seedlings were growing on MS medium, the difference between WT and transgenic seedlings could barely be distinguished. When growing on MS medium with 50 mM NaCl, both WT and transgenic seedlings were inhibited, and the hypocotyl and root of transgenic seedlings were significant shorter than those of WT. When the salt concentration increased to 100 mM, seedlings were inhibited more significantly, and the difference of hypocotyl and root length between WT and RNAi seedlings was also more obvious (Fig. 3a, c, d). As shown in Fig. 3b, after soaking in 300 mM NaCl solution for 4 days, the leaves from transgenic plants became more transparent compared with WT, and the chlorophyll degradation was severer. The result of total chlorophyll content measurement was consistent with the phenotype we observed (Fig. 3e). In addition, we treated the 6-week-old plants of WT and transgenic lines with 400 mM NaCl. After a week, the leaves of transgenic plants turned wilted, and the lower leaves were yellow obviously while WT was a little wilted. At about 2 weeks after treatment, leaves of transgenic plants turned severe wilted and yellow, and the lower leaves fell off while leaves of WT just showed wilted and a little yellow (Fig. 3f). In conclusion, the SlHDA5-RNAi transgenic plants showed reduced salt tolerance at the seedling and whole plant stages.
Phenotype of WT and SlHDA5-RNAi seedlings, leaves, and plants under salt treatment. a WT and transgenic seedlings treating with 0, 50, and 100 mM NaCl. Scale bars = 1 cm. Hypocotyl length (c) and root length (d) of seedlings. b Leaves soaking with 300 mM NaCl for 4 days and chlorophyll content (e). f WT and transgenic plants treated with 400 mM NaCl for 7 and 14 days. Scale bars = 10 cm. Data are the mean from three independent replicates with three biological repeats. Asterisks indicate significant difference from WT (P < 0.05)
Silencing of SlHDA5 Gene Reduced the Tolerance to Drought
Previous researches also demonstrated that the expression SlHDA5 was induced by dehydration stress (Guo et al. 2017), so we speculated that the tolerance of SlHDA5-RNAi transgenic plants to drought may be altered. To confirm this, the drought tolerance test was carried out. Figure 4a displayed that the lower leaves of transgenic plants turned yellow and a bit of wilted at 25 days after drought stress while the WT plants had no evident change. After 30 days, the transgenic plants were entirely yellow, wilted, and collapsed. Nevertheless, the WT plants were only a little wilted in lower leaves and the upper leaves were still green. At the same time, we sampled the leaves of transgenic and WT plants at 0, 25, and 30 days to measure RWC, contents of total chlorophyll, and MDA. Figure 4b revealed that in WT tomato leaves, RWC reduced about 15% at 25 days and 23% at 30 days, but in transgenic plants, RWC reduced by 30% at 25 days and 50% at 30 days. At 25 and 30 days after drought stress, the total chlorophyll contents of WT leaves decreased by 50 and 58%, but the chlorophyll contents of two transgenic lines’ leaves were lessened respectively by 55 and 77% at 25 days and 73 and 91% at 30 days (Fig. 4c). Figure 4d displayed that after 25 and 30 days since onset of drought stress, the contents of MDA in WT plants increased by 0.75- and 1.5-fold while in two transgenic lines, the uplift amounts were respectively 1.4-fold, 2.8- and 1.4-fold, and 3.4-fold. We concluded that silencing of SlHDA5 reduced the RWC and total chlorophyll contents, increased the MDA content, and accelerated the drying of transgenic plants under drought stress. Overall, silencing SlHDA5 led to increasing sensibility to drought in tomato.
Tomato Seedlings Lacking of SlHDA5 Were More Sensitive to ABA
Detection of SlHDA5 gene expression in hormone treatment implied that SlHDA5 was induced by ABA (Fig. 1b). So we designed experiment to verify whether SlHDA5-RNAi transgenic lines had difference in response to ABA with WT plants. As shown in Fig. 5a, seedlings of transgenic and WT had no obvious difference grew on MS medium, but after adding 5 μM ABA, seedlings were inhibited and the root length of transgenic seedlings was short than that of WT. When growing in the condition of 10 μM ABA, the difference of root length between transgenic and WT seedlings was more pronounced (Fig. 5b). In summary, compared with WT, the SlHDA5-RNAi seedlings were more sensitive to ABA.
Discussion
Various stresses in the natural environment affect the agricultural economical characters and crop production. To obtain the stress-tolerance crop varieties is always one of the main breeding goals. Recent decades, the research of epigenetics was more detailed. Increasing enzymes modulating the reactions of DNA and histone modification were identified affecting the tolerance to biotic and abiotic stresses, which provides a method for resistance breeding. In Arabidopsis HDA6 mutant and HDA6 RNA interference plants, the salt-stress signaling pathways were inhibited in a 2-week-old plant treating with NaCl solution (Chen et al. 2010). hda19-1, a mutant of HDA19, showed lower seed germination than wild type under the treatment of 200 mM NaCl (Chen and Wu 2010).
But in tomato, the roles of HDA genes play in controlling tolerance to abiotic stresses have not been studied. Former work on the expression of SlHDA5 under the treatment of various abiotic stresses showed that expression of SlHDA5 was induced distinctly under the treatments of salt and dehydration, indicating that SlHDA5 may play a role in responding to salt and drought (Guo et al. 2017). Experiment on post-germination seeds indicated that the elongation of transgenic seedling hypocotyl and root was more inhibited by salt than that of WT. Chlorophyll of SlHDA5-RNAi detached leaf degraded faster than that of WT. When treated with NaCl solution, 6-week-old transgenic plants turned wilted and yellow earlier than WT. These results mean that the photosynthetic capacity of transgenic plants decreased faster than that of WT with the existence of salt stress. Taken these results together, we concluded that repression of SlHDA5 impaired the tolerance to salt stress in multiple development stage in tomato.
The transcriptional responsiveness of drought stress-up-regulated genes was found to be correlated with changes in histone modification (To TK and Kim, 2014). Under strong drought conditions, the histone modifications H3K4me3 and H3K9ac on drought stress-up-regulated genes, such as RD20 and RD29A, were more highly enriched than under moderate drought conditions, and the nucleosome loss in the same region of RD29A under strong drought conditions was more than that under moderate drought (Kim et al. 2012; Kim et al. 2008b). Overexpression of Arabidopsis HD2-type HDAC, HD2C, plants showed enhanced tolerance to drought (Sridha and Wu 2006). In this work, we examined the tolerance of RNAi plants to drought, and the results indicated that when treated with drought, RNAi plants showed desiccation symptoms such as leaf rolling and wilting earlier than WT. MDA is a decomposition product of poly-unsaturated fatty acid hydroperoxides in osmotic stress (Heath and Packer 1968). The content of MDA manifests the damage of membrane. RWC is also an index to evaluate the damage caused by osmotic stress. In our work, SlHDA5-RNAi plant had lower chlorophyll content, RWC, and higher MDA content. These physiological indices are consistent with the morphology change. Overall, we deduced that SlHDA5 was a positive regulator in responding to the osmotic stress caused by drought and salt.
The Arabidopsis HDA6 mutant, axe1-5, and HDA6 RNA interference plants displayed down-regulated expression of ABA-responsive genes when treated with ABA (Chen et al. 2010). The seed germination of hda19-1 was lower than that of wild type under 2 μM ABA, and the ABA synthesis genes were decreasing, suggesting the increasing sensitivity to ABA with deletion of HDA19 (Chen and Wu 2010). Members of histone deacetylases HD2 family have also been proved to regulate ABA responses. When HD2C was over-expressed, the transgenic plants showed higher germination rate and longer root length by contrast with WT under the treatment of ABA (Sridha and Wu 2006). In this study, the expression profile showed that SlHDA5 was induced by ABA. To sum up, the reported histone deacetylase genes are almost positively related to ABA sensitivity. Experiment of SlHDA5-RNAi seedlings revealed that the development of transgenic seedlings was repressed, which root was shorter than that of WT with the treatment of ABA, suggesting that repression of SlHDA5 improved seedling sensitivity in tomato. These results were consistent with the former research that histone deacetylases positively influence the stress to salt, drought, and ABA.
In addition, we displayed that SlHDA5 was also remarkably induced by MeJA, which plays a crucial role in the signaling pathways involved in responding to biotic stress such as wounding and pathogen attack (Benedetti et al. 1998). Researches demonstrated that overexpression of HDA19 enhanced the resistant to pathogen Alternaria brassicicola in Arabidopsis and up-regulated the expression of PATHOGENSIS-RELATED genes, Basic Chitinase and β-1, and 3-glucanase which are regulated by jasmonic acid (Zhou et al. 2005). Therefore, we speculated that SlHDA5 may involve in other biotic and abiotic stresses, such as pathogenic bacteria.
Histone deacetylase was multifunctional in plant development and resisting adverse effects from environment. Some molecular mechanisms of histone deacetylase taking part in development process and stress-responding have been clarified in Arabidopsis. However, in other species, research on histone deacetylases is little. Here, we revealed the molecular characters of a tomato histone deacetylase gene, SlHDA5. Expression profiles under the treatment of hormones were also investigated. Besides, we obtained SlHDA5-RNAi transgenic plants and seeds. Further experiments showed that the tolerance of transgenic tomato to drought and salt stress were decreased, and the sensitivity of seedlings to ABA was increased. These results provide significant basis for breeding. But the molecular mechanisms of processes were not very clear. To elucidate the mechanism, it remains to be identified the genes that are regulated by SlHDA5 via histone deacetylation.
References
Alinsug MV, CW Y, Wu K (2009) Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol 9(1):37. https://doi.org/10.1186/1471-2229-9-37
Benedetti CE, Costa CL, Turcinelli SR, Arruda P (1998) Differential expression of a novel gene in response to coronatine, methyl jasmonate, and wounding in the Coi1 mutant of Arabidopsis. Plant Physiol 116(3):1037–1042. https://doi.org/10.1104/pp.116.3.1037
Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447(7143):407–412. https://doi.org/10.1038/nature05915
Bourque S, Dutartre A, Hammoudi V, Blanc S, Dahan J, Jeandroz S, Pichereaux C, Rossignol M, Wendehenne D (2011) Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol 192(1):127–139. https://doi.org/10.1111/j.1469-8137.2011.03788.x
Chen ZJ, Tian L (2007) Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta Gene Struct. Expr. 1769(5-6):295–307. https://doi.org/10.1016/j.bbaexp.2007.04.007
Chen LT, Wu K (2010) Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav 5(10):1318–1320. https://doi.org/10.4161/psb.5.10.13168
Chen GP, Hackett R, Walker D, Taylor A, Lin ZF, Grierson D (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136(1):2641–2651. https://doi.org/10.1104/pp.104.041608
Chen LT, Luo M, Wang YY, KQ W (2010) Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot 61(12):3345–3353. https://doi.org/10.1093/jxb/erq154
Cigliano RA, Cremona G, Paparo R, Termolino P, Perrella G, Gutzat R, Consiglio MF, Conicella C (2013) Histone deacetylase AtHDA7 is required for female gametophyte and embryo development in Arabidopsis. Plant Physiol 163(1):431–440. https://doi.org/10.1104/pp.113.221713
Demetriou K, Kapazoglou A, Tondelli A, Francia E, Stanca MA, Bladenopoulos K, Tsaftaris AS (2009) Epigenetic chromatin modifiers in barley: I. Cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment. Physiol. Plant. 136(3):358–368. https://doi.org/10.1111/j.1399-3054.2009.01236.x
Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS (2006) Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 20(10):1283–1293. https://doi.org/10.1101/gad.1417706
Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8(1):131. https://doi.org/10.1186/1471-2229-8-131
Fuchs J, Demidov D, Houben A, Schubert I (2006) Chromosomal histone modification patterns—from conservation to diversity. Trends Plant Sci 11(4):199–208. https://doi.org/10.1016/j.tplants.2006.02.008
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LSP, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39(6):863–876. https://doi.org/10.1111/j.1365-313X.2004.02171.x
Guo J-E, Hu Z, Guo X, Zhang L, Yu X, Zhou S, Chen G (2017) Molecular characterization of nine tissue-specific or stress-responsive genes of histone deacetylase in tomato (Solanum lycopersicum). J Plant Growth Regul 36(3):566–577. https://doi.org/10.1007/s00344-016-9660-8
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts .I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125(1):189–198. https://doi.org/10.1016/0003-9861(68)90654-1
Hollender C, Liu Z (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50(7):875–885. https://doi.org/10.1111/j.1744-7909.2008.00704.x
Jang IC, Pahk YM, Song SI, Kwon HJ, Nahm BH, Kim JK (2003) Structure and expression of the rice class-I type histone deacetylase genes OsHDAC1-3: OsHDAC1 overexpression in transgenic plants leads to increased growth rate and altered architecture. Plant J 33(3):531–541. https://doi.org/10.1046/j.1365-313X.2003.01650.x
Kim KC, Lai ZB, Fan BF, Chen ZX (2008a) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20(9):2357–2371. https://doi.org/10.1105/tpc.107.055566
Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, Toyoda T, Kimura H, Shinozaki K, Seki M (2008b) Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant & Cell Physiology 49(10):1580–1588. https://doi.org/10.1093/pcp/pcn133
Kim JM, To TK, Ishida J, Matsui A, Kimura H, Seki M (2012) Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant & Cell Physiology 53(5):847–856. https://doi.org/10.1093/pcp/pcs053
Kouzarides T (2007a) Chromatin modifications and their function. Cell 128(4):693–705. https://doi.org/10.1016/j.cell.2007.02.005
Kouzarides T (2007b) SnapShot: histone-modifying enzymes. Cell 131(4):822–822.e1. https://doi.org/10.1016/j.cell.2007.11.005
Lagace M, Chantha SC, Major G, Matton DP (2003) Fertilization induces strong accumulation of a histone deacetylase (HD2) and of other chromatin-remodeling proteins in restricted areas of the ovules. Plant Mol Biol 53(6):759–769. https://doi.org/10.1023/B:PLAN.0000023665.36676.89
Lim MY, Pulla RK, Park JM, Harn CH, Jeong BR (2012) Over-expression of l-gulono-gamma-lactone oxidase (GLOase) gene leads to ascorbate accumulation with enhanced abiotic stress tolerance in tomato. In Vitro Cell. Dev. Biol. Plant 48:453–461
Liu XC, CW Y, Duan J, Luo M, Wang KC, Tian G, Cui YH, KQ W (2012a) HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol 158(1):119–129. https://doi.org/10.1104/pp.111.184275
Liu X, Luo M, Wu K (2012b) Epigenetic interplay of histone modifications and DNA methylation mediated by HDA6. Plant Signal Behav 7(6):633–635. https://doi.org/10.4161/psb.19994
Liu C, Li LC, Chen WQ, Chen X, ZH X, Bai SN (2013) HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell 25(1):257–269. https://doi.org/10.1105/tpc.112.107045
Liu XC, Yang SG, Zhao ML, Luo M, CW Y, Chen CY, Tai R, KQ W (2014) Transcriptional repression by histone deacetylases in plants. Mol Plant 7(5):764–772. https://doi.org/10.1093/mp/ssu033
Loidl P (2004) A plant dialect of the histone language. Trends Plant Sci 9(2):84–90. https://doi.org/10.1016/j.tplants.2003.12.007
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 389(6648):251–260. https://doi.org/10.1038/38444
Luo M, Wang YY, Liu XC, Yang SG, Lu Q, Cui YH, KQ W (2012) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63(8):3297–3306. https://doi.org/10.1093/jxb/ers059
Luo M, Tai R, CW Y, Yang SG, Chen CY, Lin WD, Schmidt W, KQ W (2015) Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J 82(6):925–936. https://doi.org/10.1111/tpj.12868
Lusser A, Kolle D, Loidl P (2001) Histone acetylation: lessons from the plant kingdom. Trends Plant Sci 6(2):59–65. https://doi.org/10.1016/S1360-1385(00)01839-2
Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30(23):5036–5055. https://doi.org/10.1093/nar/gkf660
Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9(3):409–423. https://doi.org/10.1105/tpc.9.3.409
Pipal A, Goralik-Schramel M, Lusser A, Lanzanova C, Sarg B, Loidl A, Lindner H, Rossi V, Loidl P (2003) Regulation and processing of maize histone deacetylase Hda1 by limited proteolysis. Plant Cell 15(8):1904–1917. https://doi.org/10.1105/tpc.013995
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37(5):501–506. https://doi.org/10.1038/ng1543
Sendra R, Rodrigo I, Salvador ML, Franco L (1988) Characterization of pea histone deacetylases. Plant Mol Biol 11(6):857–866. https://doi.org/10.1007/BF00019525
Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46(1):124–133. https://doi.org/10.1111/j.1365-313X.2006.02678.x
Tanaka M, Kikuchi A, Kamada H (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146(1):149–161. https://doi.org/10.1104/pp.107.111674
Taunton J, Hassig CA, Schreiber SL (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272(5260):408–411. https://doi.org/10.1126/science.272.5260.408
Tian L, Chen ZJ (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development (vol 98, pg 200, 2001). Proc Natl Acad Sci U S A 98:7647–7647
To TK, Kim JM (2014) Epigenetic regulation of gene responsiveness in Arabidopsis. Front Plant Sci 4. https://doi.org/10.3389/fpls.2013.00548
To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, Kimura H, Yokoyama S, Shinozaki K, Seki M (2011) Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 7(4):e1002055. https://doi.org/10.1371/journal.pgen.1002055
Wang Z, Cao H, Chen FY, Liu YX (2014) The roles of histone acetylation in seed performance and plant development. Plant Physiol. Biochem. 84:125–133. https://doi.org/10.1016/j.plaphy.2014.09.010
Wu K, Zhang L, Zhou C, CW Y, Chaikam V (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59(2):225–234. https://doi.org/10.1093/jxb/erm300
Yang XJ, Seto E (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26(37):5310–5318. https://doi.org/10.1038/sj.onc.1210599
Yu CW, Liu XC, Luo M, Chen CY, Lin XD, Tian G, Lu Q, Cui YH, KQ W (2011) HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol 156:173–184
Zhao LM, JX L, Zhang JX, PY W, Yang SG, KQ W (2015a) Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front Plant Sci 5. https://doi.org/10.3389/fpls.2014.00760
Zhao JH, Zhang JX, Zhang W, KL W, Zheng F, Tian LN, Liu XC, Duan J (2015b) Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front Plant Sci 5. https://doi.org/10.3389/fpls.2014.00764
Zhao JH, Li MZ, Gu DC, Liu XC, Zhang JX, Wu KL, Zhang XH, da Silva JAT, Duan J (2016) Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 470(2):439–444. https://doi.org/10.1016/j.bbrc.2016.01.016
Zhou CH, Zhang L, Duan J, Miki B, KQ W (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17(4):1196–1204. https://doi.org/10.1105/tpc.104.028514
Funding
This work was supported by National Natural Science Foundation of China (no. 31572129) and the Natural Science Foundation of Chongqing of China (cstc2015jcyjA80026).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yu, X., Gao, Q., Chen, G. et al. SlHDA5, a Tomato Histone Deacetylase Gene, Is Involved in Responding to Salt, Drought, and ABA. Plant Mol Biol Rep 36, 36–44 (2018). https://doi.org/10.1007/s11105-017-1057-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-017-1057-8