Abstract
Key message
Expression of synthesized cecropin B genes in the citrus phloem, where Candidatus Liberibacter asiaticus resides, significantly decreased host susceptibility to Huanglongbing.
Abstract
Huanglongbing (HLB), associated with Candidatus Liberibacter asiaticus bacteria, is the most destructive disease of citrus worldwide. All of the commercial sweet orange cultivars lack resistance to this disease. The cationic lytic peptide cecropin B, isolated from the Chinese tasar moth (Antheraea pernyi), has been shown to effectively eliminate bacteria. In this study, we demonstrated that transgenic citrus (Citrus sinensis Osbeck) expressing the cecropin B gene specifically in the phloem had a decreased susceptibility to HLB. Three plant codon-optimized synthetic cecropin B genes, which were designed to secrete the cecropin B peptide into three specific sites, the extracellular space, the cytoplasm, and the endoplasmic reticulum, were constructed. Under the control of the selected phloem-specific promoter GRP1.8, these constructs were transferred into the citrus genome. All of the cecropin B genes were efficiently expressed in the phloem of transgenic plants. Over more than a year of evaluation, the transgenic lines exhibited reduced disease severity. Bacterial populations in transgenic lines were significantly lower than in the controls. Two lines, in which bacterial populations were significantly lower than in others, showed no visible symptoms. Thus, we demonstrated the potential application of the phloem-specific expression of an antimicrobial peptide gene to protect citrus plants from HLB.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Huanglongbing (HLB), also known as citrus greening, is currently the most devastating disease of citrus worldwide. The disease is associated with the phloem-limited bacterium Candidatus Liberibacter asiaticus (Calas) (Duan et al. 2009). This disease is spread by the Asian citrus psyllid (ACP) vector, Diaphorina citri (Bové 2006). This disease was first reported in China in the early twentieth century, and has now rapidly spread throughout the citrus-growing areas (Bové 2014). Calas has a wide host range and can infect most rutaceous species and some solanaceous species (Duan et al. 2009; Wang and Trivedi 2013). HLB-diseased citrus plants often show yellow shoots, asymmetric blotchy mottling of older leaves, and inverted fruit coloring. At advanced disease stages, twig dieback, tree decline, and tree mortality occur, causing tremendous losses to the citrus industry. HLB management mainly includes quarantine measures to prevent infected plant materials from entering HLB-free orchards, destruction of infected trees, and controlling psyllid populations as much as possible (Bové 2006). However, disease management is not only very difficult, but also expensive. No efficient cure is currently available for infected plants.
The replacement of susceptible citrus cultivars with those showing field resistance is a potential way to control HLB. However, citrus improvement using conventional breeding is difficult and time consuming because of numerous issues, such as male/female sterility, a long juvenile period, a high degree of heterozygosity, and polyembryony (Donmez et al. 2013; Gong and Liu 2013). Genetic engineering, which has been widely employed to produce disease-resistant materials without greatly altering the existing genetic background, is still the fastest method for improving existing citrus cultivars. Recently, Dutt et al. (2015) showed that the expression of an Arabidopsis NPR1 gene in transgenic citrus exhibits enhanced resistance against HLB.
Different genetic strategies have been used to construct disease-resistant plants, including the expression of antimicrobial genes from plant and non-plant organisms, as well as the use of host disease-response pathway components (Gurr and Rushton 2005b). Because no resistant cultivars or active resistance genes have been found in citrus, it is difficult to generate, through a molecular breeding program, resistant materials using only citrus genes. Moreover, the expression of antibacterial genes from plant sources has resulted in only modest levels of protection against pathogens because plant pathogens have already evolved tolerances to these plant-derived proteins (Coca et al. 2006). In contrast, the expression of genes encoding antimicrobial peptides (AMPs) from animal, fungal, or bacterial species in transgenic plants has conferred higher levels of protection and a broad spectrum of resistance against pathogens (Holásková et al. 2015).
AMPs are important components of the innate immune defense system against microbial pathogens in nearly all living organisms, including insects, mammals, and plants (Holásková et al. 2015; Salas et al. 2015). They are active against a broad range of pathogenic organisms and, when compared with traditional antibiotics, they kill bacteria rapidly. The modes of AMP action may involve interactions between the peptides and the microbial membrane, followed by pore formation, which can lead to one or more of the following: bacterial membrane disruption, cytoplasmic leakage, and interference with intracellular macromolecule synthesis (Melo et al. 2009; Nawrocki et al. 2014; Straus and Hancock 2006). Cecropins, which are natural lytic peptides found in Antheraea pernyi, Hyalophora cecropia, and Bombyx mori, possess antibacterial activities (Jaynes et al. 1993; Sharma et al. 2000). Even at low concentrations (0.1–5 µM), cecropins show antibacterial activities against a number of Gram-negative and some Gram-positive bacteria, but do not affect eukaryotic cells (Chen et al. 1997; Holásková et al. 2015; Mills and Hammerschlag 1993). This makes them potentially useful for engineering bacterial resistance in plants. Moreover, in the cecropin family, cecropin B shows the strongest activity against Gram-negative bacteria and, therefore, has been used to increase plant resistance against bacterial diseases (He et al. 2011; Jan et al. 2010; Jaynes et al. 1993; Sharma et al. 2000).
In this study, a phloem-specific promoter, GRP1.8, and three synthesized cecropin B genes (CB, PRlaCB, and PRlaCBer) were assessed to determine whether they could be used to engineer resistance to citrus HLB. The three genes were designed to enable the accumulation of the cecropin B peptide in the cytoplasm and apoplast, as well as in the lumen of the endoplasmic reticulum (ER). Under the control of the GRP1.8 promoter, the expression of the cecropin B gene in the phloem tissues, where Calas resides, significantly decreased the host’s susceptibility to HLB.
Materials and methods
Plant and growth conditions
Tarocco blood orange (Citrus sinensis Osbeck) was chosen for the transformation experiments. Citrus seeds were collected from the National Citrus Germplasm Repository, Chongqing, China. Seed regeneration was performed as described by Zou et al. (2008). The basal medium for plant culturing was Murashige and Skoog (MS) medium (Murashige and Skoog 1962) supplemented with 30 g l−1 sucrose and solidified with 2 g l−1 Gelrite (Promega, Fitchburg, WI, USA). The pH was adjusted to 5.8 before autoclaving.
Plasmid construction
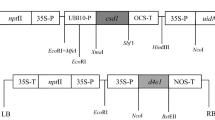
Three synthesized cecropin B genes were constructed for plant transformation (Supplementary Fig. 1a). First, the synthesized CB gene, without a signal peptide, was designed to retain cecropin B in the cytoplasm. Second, the synthesized PR1aCB gene was designed to secret the cecropin B peptide into the apoplastic space. Finally, the synthesized PR1aCBer gene was designed to retain cecropin B in the lumen of the ER. In the PR1aCB and PR1aCBer constructs, the signal sequence from a secreted plant protein, the tobacco PR1a protein (Cornelissen et al. 1987), was fused to the N terminus of the cecropin B sequence. In the PR1aCBer construct, an ER retention signal sequence was fused to the C terminus of the mature cecropin B. All of the genes were fully synthesized by Invitrogen. The sequence AACAAUGGC (underlined translation start codon) was inserted between positions −4 and 5 for efficient translation (Lutcke et al. 1987). BamHI and SalI sites were introduced immediately upstream and downstream of these gene sequences.
The gene fragments were digested by BamHI/SalI and inserted into a BamHI/SalI-digested pGN vector (Zou et al. 2014a) to generate an intermediate vector. In the pGN vector, the gus::npt-II fusion gene (Datla et al. 1991), under the control of the CaMV 35S promoter, was used as the selectable marker and reporter for the genetic transformation of citrus mediated by Agrobacterium. Finally, a selected phloem-specific promoter (Supplementary Table 1), GRP1.8, from the French bean (Phaseolus vulgaris) was amplified with flanking HindIII and BamHI restriction sites (Supplementary Table 2) and inserted upstream of the synthesized cecropin B genes in the intermediate vector to yield the pGC, pGA, and pGE plant expression vectors (Supplementary Fig. 1b). In these vectors, the expression levels of the synthesized cecropin B genes were controlled by the GRP1.8 promoter. All of the constructs were verified by restriction analyses and DNA sequencing. Each plasmid was introduced independently into A. tumefaciens EHA 105 by electroporation using the Gene PulserTM Xcell system (Bio-Rad, Hercules, CA, USA). Transformants were selected on YEB (Vervliet et al. 1975) solid medium supplemented with 50 mg/L kanamycin, and further confirmed by restriction enzyme and PCR analyses.
Plant transformation
The Agrobacterium-mediated transformation of Tarocco blood orange epicotyl explants was performed as previously described (Zou et al. 2014b). Because the gus::npt-II fusion reporter gene was present in these constructs (Supplementary Fig. 1b), kanamycin-resistant shoots were first analyzed by β-glucuronidase (GUS) histochemical staining to identify transformants (Jefferson et al. 1987; Zou et al. 2008). GUS-positive shoot tips were grafted on Troyer citrange [Poncirus trifoliata (L.) Raf. × C. sinensis (L.) Osbeck] seedlings in vitro. The recovered shoots were further grafted onto Troyer citrange seedlings in the greenhouse. As a control, the pGN vector (Zou et al. 2014a) without cecropin B gene was also introduced into Tarocco blood orange.
Molecular confirmation of the transgenic plants
The integration of foreign genes in transgenic lines was confirmed by PCR analysis. Total genomic DNA was prepared using the Plant DNeasy Prep Kit (Qiagen, Beijing, China). The primers g-f/c-r were used to amplify GRP1.8::CB and GRP1.8::PRlaCB cassettes in pGC and pGA-containing transgenic lines, while g-f/e-r primers were used to amplify the GRP1.8::PRlaCBer cassette in pGE-containing transgenic lines (Supplementary Fig. 1b and Supplementary Table 2). The predicted fragments were 755, 1009, and 1025 bp long for pGC-, pGA-, and pGE-containing transgenic lines, respectively. PCR reactions were carried out as follows: 94 °C for 5 min and 30 cycles of 94 °C for 30 s, 60 °C for 1 min, and 72 °C for 8 min, followed by a final extension at 72 °C for 10 min.
Southern blot analysis was also applied to confirm gene integration and the copy number of transgenes in the citrus genome. In total, 35 µg of genomic DNA from each transgenic and control plants was digested with HindIII, subjected to electrophoresis in a 0.8% (w/v) agarose gel, transferred onto a nylon membrane (Hybond-N+; Amersham Biosciences, Buckinghamshire, UK) and cross-linked to the membrane with ultraviolet irradiation. The digoxigenin-labeled PCR product of the gus fragment (Supplementary Fig. 1b) was used as the probe. DNA labeling, hybridization, and immunological detection were carried out according to the manufacturer’s instructions (DIG High Prime DNA Labeling and Detection Starter Kit II; Roche, Basel, Switzerland).
Quantitative reverse transcription-polymerase chain reaction (qPCR)
RNA extraction was constructed according to the instructions of the EASYspin Plant RNA Extraction kit (Aidlab, Beijing, China). cDNA was synthesized from 1 µg total RNA with an iScript cDNA Synthesis kit (Bio-Rad). The primers qCB-f/qCB-r (Supplementary Table 2) were used to detect the synthesized cecropin B gene in transgenic plants. Transcripts of the citrus actin gene (GenBank Accession No. GU911361.1), as an internal control, were detected with the primers CtAct-f/CtAct-r (Supplementary Table 2). cDNAs were amplified in 10-µL reaction mixtures using 2 × iQ™ SYBR Green Supermix (Bio-Rad). PCR reactions were carried out as follows: a pretreatment at 95 °C for 5 min followed by 40 amplification cycles of 95 °C for 20 s and 60 °C for 1 min. Experiments were repeated three times. Using the WT control as a reference, transgene expression levels were calculated as described by Zou et al. (2014b).
Western blot analysis
Citrus protein extraction was performed following the instructions of the Plant Protein Extraction kit (KeyGEN BioTECH, Jiangsui, China). Briefly, 40 µg total proteins per sample were separated by 18% (wt/vol) SDS–PAGE and electrotransferred onto a polyvinylidene difluoride nylon membrane (Millipore Corp., Burlington, MA, USA). Immunodetection was performed using 0.5 µg/ml polyclonal rabbit anti-Cecropin B antibody (Abcam, Shanghai, China) as the primary antibody and a goat anti-rabbit HRP-conjugated secondary antibody (Abgent, San Diego, CA, USA), which had been diluted to 1:2500. Hybrid signals were detected according to the Opti-4CN Substrate kit manual (Bio-Rad), and the images were recorded on X-ray film.
Microscopic observations
WT and transgenic tissues were prepared for light microscopic observations. Sample preparations of paraffin-embedded sections were performed as previously described (Aritua et al. 2013). The 20-μm cross-sections were made using a KD-1508A microtome (KEDI Instrumental Equipment Co. Ltd, Zhejiang, China). The samples were observed using a BX51 system microscope equipped with a DP70 digital camera (Olympus, Tokyo, Japan).
Immunohistochemical localization of cecropin B
The cecropin B protein immunolocalization procedure was performed essentially as described previously (Hou and Huang 2005). Briefly, 1 µg/ml polyclonal rabbit anti-Cecropin B antibody (Abcam) was used to blot cecropin B proteins in citrus tissue. After the first and second immunoreactions, the slides were rinsed three times in RSR solution (10 mM PBS, 0.1% Tween-20, 0.8% BSA, 0.8% NaCl) and once in PBS (pH 7.4). The slides were incubated with 100 µL of western blotting 3,3′,4,4′-diaminobenzidine staining kit (BOSTER, Wuhan, China) for approximately 1 h in the dark at room temperature. The samples were photographed with a light microscope system.
HLB resistance analysis
Citrus samples containing Calas were harvested from naturally infected sweet orange (Citrus sinensis Osbeck) in a citrus orchard in Guilin, Guangxi Province, China. The scions were reproduced and maintained in Tarocco blood sweet orange by grafting in a greenhouse with restricted access. Using the primers Cla16s-f/Cla16s-r (Supplementary Table 2), the presence of the Calas pathogen in the plants was confirmed by PCR.
Transgenic and control lines were propagated by grafting on Troyer citrange rootstock in the greenhouse, and all of the plant growth was directed by maintaining only a single stem. After 1 year, three well-grown plants per line were selected to evaluate resistance to HLB. The inoculation was performed by graft transmission from axillary buds containing the Calas pathogen to transgenic stems. Each tested plant was inoculated with three infected buds. The inoculated and non-inoculated plants, which acted as controls, were maintained in a greenhouse at the Citrus Research Institute in Chongqing, China. The Calas pathogen levels in plants was quantified every 3 months using the qPCR method.
The Calas bacterial populations were tested according to the protocol of Tatineni et al. (2008) with some modifications. Three leaves per plant were selected randomly to be tested. Their midrib tissues were pooled and DNA was isolated from the pooled tissues. Using the 2 × iQ™ SYBR Green Supermix (Bio-Rad), the Calas 16S and citrus 18S genes were amplified using qCla16s-f/ qCla16s-r and Ct18s-f /Ct18s-r primers, respectively (Supplementary Table 2). The bacterial populations per µg citrus DNA were calculated using the formula: Calas cells µg−1 citrus \(\text{DNA}=\left[ {{{10}^{\left( -0.2718 \times \text{C}{{\text{t}}_{\text{16S}}} +10.624\right)}}}/{{{10}^{\left( -0.2749 \times \text{C}{{\text{t}}_{\text{18S}}}+4.0531 \right)}}} \right]\times {{10}^{3}}\left( 12.7 < \text{C}{{\text{t}}_{\text{16S}}} <31.3\ \text{and}\ 8.4 <\text{C}{{\text{t}}_{\text{18S}}} <26.5 \right)\) (Supplementary Fig. 2). The data analysis was performed using the SPSS v22.0 statistical package (IBM Corp., Armonk, NY, USA). Data are presented as the means ± standard deviations. Significant differences were subjected to a Tukey’s test (P < 0.05).
Results
Production of transgenic citrus plants
Based on the amino acid sequence of the cecropin B from the Chinese tasar moth (A. pernyi) (Jaynes et al. 1993), three codon-optimized cecropin B genes, CB, PR1aCB, and PR1aCBer, which were designed to locate cecropin B protein in the cytoplasm, apoplastic space, and ER, respectively, were synthesized (Supplementary Fig. 1). Moreover, to express these genes specifically in citrus phloem, four phloem-associated promoters were analyzed using the gene encoding GUS as a reporter in Tarocco blood orange (Supplementary Table 1). Histochemical staining showed that the GRP1.8 promoter from the French bean had phloem-specific expression characteristics in Tarocco blood orange and was the most efficient promoter compared with the others tested (Supplementary Fig. 3). Thus, this promoter was selected to direct the expression of the synthesized cecropin B genes in citrus, and the corresponding vectors, pGC, pGA, and pGE, were constructed (Supplementary Fig. 1).
These constructs were introduced into the genome of Tarocco blood orange by an Agrobacterium-mediated transformation. Transgenic plants were screened based on GUS activity using histochemical staining. Transformation efficiencies of ~10% were observed. GUS-positive shoots were grafted on Troyer citrange seedlings in vitro, and the recovered shoots were further grafted onto 2-year-old Troyer citrange rootstocks in the greenhouse. In total, 17 pGC, 22 pGA, and 32 pGE independent lines, containing the GRP::CB, GRP::PR1aCB, and GRP::PR1aCBer transgenes, respectively, were recovered in this study.
Molecular confirmation of transgenic plants
The presence of the cecropin B gene in GUS-positive plants was confirmed by PCR (Fig. 1a). Specific fragments, confirming the integration of GRP1.8::CB, GRP1.8::PR1aCB, and GRP1.8::PR1aCBer cassettes into the citrus genome, were detected in corresponding transformants. These specific fragments were not detected in WT control plants.
Molecular confirmation of transgenic citrus plants. a PCR analysis of transgenic plants. The primers g-f/c-r, g-f/a-r, and g-f/e-r were used to confirm the integration of GRP1.8::CB (gc), GRP1.8::PRlaCB (ga), and GRP1.8::PRlaCB (ge) cassettes into the citrus genome, respectively. The predicted fragments were 755, 1009, and 1025 bp long, respectively. a representative 10 lines per vector are shown. The amplified fragments were further confirmed by DNA sequencing. Lane M, DNA molecular size marker; lane WT, template from wild type; other lanes, templates from transgenic lines. b–d Southern blot analysis of transgenic plants. Total genomic DNA of transgenic plants was digested with HindIII, electrophoresed, and probed with a digoxigenin-labeled amplified fragment of the gus gene
Southern blotting was also used to verify stable transgene integration and copy number in transgenic plants. One to five integration events were detected in the analyzed lines. No hybridizing bands were detected in WT control plants, or in the pGC9 and pGC15 lines. Representative Southern blot results are shown in Fig. 1b–d.
According to PCR and Southern blot data, 15 pGC-, 22 pGA-, and 32 pGE-containing transgenic lines were used to investigate the cecropin B transcript levels in their veins by qPCR. Representative results are shown in Fig. 2. High cecropin B expression levels were detected in all of these transgenic lines although variable levels of transgene expression were detected. No transgenic transcript was detected in the WT controls.
Real-time PCR analysis of the expression levels of synthetic cecropin B in (a) pGA-, (b) pGC-, and (c) pGE-containing transgenic citrus plants. Total RNA from veins were extracted for PCR analysis. The expression levels of citrus actin were used as the internal control. Relative expression levels were calculated compared with the WT control. WT wild type
Evaluation of transgenic citrus’ resistance to HLB
The resistance levels of the 69 independent transgenic lines were evaluated in the greenhouse. To evaluate the resistance of transgenic plants to pathogens, control and propagated transgenic plants were inoculated by grafting them with axillary buds containing the Calas pathogen. First, three replicated clones per line were inoculated by graft infection. After 3 months, the presence and population levels of the Calas pathogen in the leaf tissues of transgenic lines were determined by qPCR (Table 1). In our study, threshold values above 31.3 were taken to indicate that no bacteria were detected in the citrus plant (Supplementary Fig. 2). As shown in Supplementary Table 3, 22 out of 69 transgenic lines had bacterial population levels significantly lower than those in both the WT and pGN controls. After another 3 months, the 22 lines showing enhanced resistance were tested further by qPCR. Eight (pGA5, pGA6, pGA12, pGA21, pGC8, pGC16, pGE3, and pGE6) of these lines still had significantly lower bacterial cell levels compared with control plants (Table 2). After 1 year of evaluation, the bacterial population levels in the eight transgenic lines were still significantly lower than those of the WT and pGN controls (Table 2). Our test showed that no significant difference in Calas populations was detected between the WT and pGN controls. In the line pGA6 and pGC8, the bacterial population levels were significantly lower than in the other lines (Table 2). These data showed that transformation with the cecropin B gene could significantly inhibit Calas reproduction in citrus.
After 6 months of infection, the majority of the plants tested, including control plants, began to develop symptoms in new leaves and new flush, and in another 6 months, the control plants showed severe symptoms. However, the line pGA6 and pGC8 showed no visual symptoms for the duration of the experiment (Fig. 3a–c). Moreover, during 2 years of greenhouse evaluation, there were still no visible symptoms in the pGA6 and pGC8 lines (Fig. 3d, e), and their bacterial population levels were still significantly lower than that in the control (Fig. 3f).
Evaluation of HLB resistance in transgenic citrus lines growing in a greenhouse. a HLB symptoms in transgenic lines pGA6 and pGC8 and a wild type (WT) control 6 months after infection. Red arrows indicate the sites where the infected scions were grafted. b pGA6 and c wild type, close-ups of flush and leaves. d, e HLB symptoms in transgenic line pGA6 and a wild type (WT) control 18 and 24 months after infection, respectively. f Quantification analysis of Calas populations in the transgenic line pGA6 and pGC8. The bacterial populations (Calas cells µg−1 of citrus DNA) 18 and 24 months after infection were investigated using qPCR. Standard errors were calculated from three plants per line. Different letters on top of the bars indicate significant differences from the WT control based on a Tukey’s test (P < 0.05)
Light microscopy analyses showed differences between the midribs from transgenic and WT control plants 12 months after infection (Fig. 4). The number of cell layers in the phloem of enhanced resistant transgenic plants was far fewer than in the control. In transgenic plants, the phloem cell walls were markedly thinner than in the control. These results indicate that the overexpression of cecropin B could significantly inhibit the abnormal hyperplasia of the infected phloem cells.
Microscopic analyses of midribs of transgenic (pGA6) and wild type (WT) plants 12 months after graft infection. The slides were stained with 0.5% Fast green FCF. Light microscopy of the midrib cross-sections showing the phloem, xylem, and parenchymal cells. Pa parenchyma, Ph phloem, Xy xylem, F phloem fibers. Bar 50 µm
Expression characteristics of cecropin B in transgenic plants
The expression characteristics of cecropin B under control of the phloem-specific promoter GRP1.8 were investigated by in situ hybridization using a cecropin B polyclonal antibody. First, transgene expression in the veins of these lines was analyzed by western blotting (Supplementary Fig. 4). Positive signals, indicating the presence of the mature cecropin B peptide, were detected in all of the transgenic lines with enhanced disease resistance. Cecropin B was not detected in the WT control. The in situ hybridization analysis showed that the cecropin B protein was detected specifically in the phloem of the midribs (Fig. 5). The data were in agreement with the results of GUS staining for GRP1.8::GUS citrus (Supplementary Fig. 3). Moreover, the in situ hybridization analysis revealed high levels of cecropin B expression in companion cells and some weak expression in sieve elements.
Localization of the cecropin B protein in transgenic plants by in situ hybridization. Midrib sections from transgenic and nontransgenic lines were hybridized using the polyclonal rabbit anti-cecropin B antibody. The data from the line pGC8, pGA6, and pGE3 are presented. WT wild type, Pa parenchyma, Ph phloem, Xy xylem. Bar 50 µm
Discussion
HLB is one of the oldest citrus diseases, being recognized over a century ago. However, this disease was largely ignored until its recent outbreak affected the global citrus industry (da Graca et al. 2016). Calas causes HLB and is capable of infecting all known commercial citrus varieties and several close relatives, resulting in dramatic economic losses (Wang and Trivedi 2013). Because no resistant cultivars have been identified, the introduction of selected genes from other species into citrus is a promising approach to improve the resistance of citrus cultivars against HLB disease. In this study, three synthesized cecropin B genes were introduced into Tarocco blood orange (C. sinensis Osbeck) to produce transgenic plants resistant to HLB. The presented data showed that the overexpression of cecropin B in the phloem significantly decreased the host’s susceptibility to HLB.
Successfully producing new plant cultivars with enhanced pathogen resistance through the expression of transgenic antibacterial genes is dependent on the nature of the recipient plant, the specific pathogen, and the gene source. During the early stages of plant responses to pathogen infection, AMPs, together with antimicrobial metabolites and stress-related proteins, act as components of a non-specific basal defense mechanism (Bent and Mackey 2007; Holásková et al. 2015). Insect cecropin peptides, including cecropin B, have attracted attention from plant biotechnologists because of their high toxicity against many important plant pathogens. In the present study, cecropin B from the Chinese oak silkworm (A. pernyi) was chosen for the genetic improvement of citrus because both the inducible and purified antibacterial peptides from Escherichia coli-immunized Chinese oak silkworms showed in vitro killing capabilities against HLB-associated pathogens (Zhang et al. 1995). Data presented here indicated that the overexpression of cecropin B in citrus resulted in an enhanced resistance to HLB disease, implying that cecropin B has in vivo capabilities against HLB-associated pathogens. To our knowledge, the use of cecropin B is the first successful engineering of resistance to the HLB-associated pathogen, Calas.
The expression of cecropin B in plants has produced variable and contradictory results regarding disease resistance. The transgenic expression of cecropin B in tobacco plants did not confer resistance to bacterial infections (Florack et al. 1995; Hightower et al. 1994), while transgenic rice and tomato plants expressing the gene showed enhanced resistance to bacterial diseases (Jan et al. 2010; Sharma et al. 2000). The failure to develop pathogen resistance was due to the degradation of the peptide by host proteases present in the intracellular space (Mills et al. 1994). Secreting cecropin B into the host apoplastic space was considered to efficiently protect the peptide from potential cecropin-degrading proteolytic activities (Coca et al. 2006; Florack et al. 1995; Jan et al. 2010). According to the modes of AMP action (Band and Weiss 2015; Shaw et al. 2006), the direct interactions between antibacterial peptides and the microbial membrane are key steps for the killing functions of antibacterial peptides. Moreover, phytopathogenic bacteria normally multiply in the intercellular space before attacking plant cells (Alfano and Collmer 1996). Thus, secreting antibacterial peptides to the host apoplastic space facilitates the interaction between peptides and pathogens, thereby enhancing the efficacy of the antibacterial activity (Boscariol et al. 2006; Coca et al. 2006; Jan et al. 2010). Interestingly, Coca et al. (2006) showed that targeting cecropin A from the giant silk moth Hyalophora cecropia into the ER was a useful method for protecting rice plants against the rice blast fungus Magnaporthe grisea. Thus, the strategy behind directing the subcellular localization of AMP peptides should be based on the targeted pathogen’s colonization sites in the plant. Theoretically, retaining antibacterial peptides in the citrus cytoplasm should be a preferred strategy for cecropin B to effectively battle Calas because the pathogen multiplies in the host cells (Fu et al. 2014; Hilf et al. 2013; Zhang et al. 2011). However, in some studies, it was demonstrated that cecropin B peptides are highly susceptible to degradation by plant cytoplasmic proteases and that this susceptibility varies from one plant species to another (Cui et al. 2008; Mills et al. 1994). Thus, there is no guarantee that a peptide that was effective in one host against one pathogen will be effective in a different host against a different pathogen. In citrus, there are no reports on the susceptibility of the cecropin B peptide to endogenous plant proteases or its antibacterial activity against Calas. In this study, three synthetic cecropin B genes, which were each designed to deliver the cecropin B peptide to a single site, the intercellular space, cytoplasm, or ER, were used to investigate cecropin B-associated resistance against HLB. Our results showed that 18% (4 out of 22) pGA lines demonstrated enhanced resistance, while 13% (2 out of 15) pGC, and 6% (2 out of 32) pGE transgenic lines had enhanced resistance during the 1-year evaluation period, indicating that delivering the cecropin B peptide into the intercellular space more efficiently killed the HLB pathogen. In situ hybridization (Fig. 5) showed that delivering cecropin B into the intercellular space could allow the peptides into the host cytoplasm where Calas existed. Passive diffusion into the cytoplasm may allow cecropin B peptides to kill cytoplasmic pathogens.
Calas transmission into citrus was usually performed by grafting infected citrus tissues (bud, bark, or leaf tissue) or by exposure to the ACP vector because the pathogen cannot be cultured in vitro (Dutt et al. 2015; Shokrollah et al. 2009). Here, a graft infection method was selected to evaluate disease resistance to the Calas pathogen. To assure the successful transmission of the Calas pathogen to each plant tested, the following three procedures were performed carefully:first, the presence of the Calas pathogen in the buds used for the transmission were confirmed by PCR before grafting; second, three buds were grafted onto each transgenic plant; and finally, plants were decapitated 30 days after graft inoculation to promote grafted buds sprouting. In this way, our study showed that most of the grafted buds sprouted two to three new leaves 3 months after grafting. The Calas pathogen was detected by PCR in these leaves from at least one grafted bud per plant.
To investigate the bacterial populations in citrus plants, two logarithmic standard curves were developed for calculating Calas pathogen cells per µg citrus genome as described in the methods (Supplementary Fig. 2) and Tatineni et al. (2008). Using pCalas16S as a template, the plasmid concentration corresponding to 1.3 × 101 Calas cells µg−1 of citrus DNA gave a consistent fluorescent signal, with an average Ct value of 31.3, while the lower concentrations did not amplify consistently. Hence, this concentration was defined as the detection limit for the conditions used in this study. It was assumed that no bacteria were detected in citrus plants when the threshold value was higher than 31.3. In our study, none of the transgenic plants tested was free of bacteria, indicating that transformations with synthesized cecropin B genes can confer plant tolerance to the HLB disease. Trivedi et al. (2009) showed that there was a minimal Calas concentration required for HLB symptoms in sweet orange trees (C. sinensis). Symptomatic leaves exhibited 9.17 × 105 to 6.60 × 106 bacteria cells per µg citrus genome, while asymptomatic leaves had a concentration lower than 4.81 × 105 bacteria cells per µg citrus genome (Trivedi et al. 2009). Our 1-year evaluation showed that the pGA6 and pGC8 transgenic lines, which displayed no visible symptoms, had less than 9.29 × 103 to 2.93 × 104 and 4.89 × 103 to 5.38 × 104 bacteria cells per µg citrus genome, respectively. Thus, it is possible to enhance citrus resistance by decreasing the bacterial concentration to less than that required for HLB symptoms. This might provide an effective strategy for controlling HLB damage in the field.
The expression of AMPs in transgenic plants has usually been driven by strong and constitutive promoters, including the CaMV 35S promoter, its derivatives, and ubiquitin promoters from various sources (Holásková et al. 2015). This regulatory strategy results in constitutive transgene expression throughout the plant at a high level, which might have a deleterious effect on plant growth and yield, and may even cause plant death (Company et al. 2014; Gurr and Rushton 2005a; Nadal et al. 2012). Calas lives in the citrus phloem tissue; thus, the phloem-specific expression of trait genes is a desirable regulatory pattern for HLB-resistant modifications (Dutt et al. 2012, 2015; Miyata et al. 2012). In this study, the GRP1.8 promoter from French bean was selected to direct the production of the synthesized Cecropin B gene in transgenic plants. GUS histochemical staining and in situ hybridization showed that the promoter can efficiently overexpress target genes in the citrus phloem.
The Calas pathogen colonized sieve elements (Tatineni et al. 2008; Zhang et al. 2011), although some bacterial cells were also detected in companion cells (Fu et al. 2014). However, the hybridization showed that the cecropin B protein content was greater in companion cells compared with in sieve elements. Thus, the transgenic lines showed tolerance to the pathogen, and it is possible that the increasing accumulation of the cecropin B protein in sieve elements further enhanced the resistance of transgenic plants to HLB. In general, mature sieve elements do not produce proteins because they have no nucleus. Most of the phloem proteins are produced in companion cells and then transferred into mature sieve elements (Sjolund 1997). Thus, we could use this similar targeting mechanism (the signal peptide of some phloem proteins) to deliver antibacterial proteins or mRNAs into the sieve elements (Weise et al. 2000; Xoconostle-Cázares et al. 1999) to increase the chance of an interaction between the peptides and bacterial cells that would efficiently kill the pathogen. Additionally, in our experiment, the rootstocks used were wild type plants and the root systems of most of the transgenic lines were damaged by HLB infection (data not shown), which could accelerate the disease progression in the canopy of scions. Johnson et al. (2014) indicated that early root infection, prior to the development of visible foliar symptoms, plays a central role in HLB disease development and spread. In various phytoplasma-related diseases, including phloem-limited bacterial diseases, resistant rootstocks can reduce bacterial replication and enhance the disease resistance of scions (Albrecht and Bowman 2012; Bertaccini and Duduk 2010). Based on these results, the expression of cecropin B in rootstocks should further enhance the resistance of the transgenic plants.
In summary, this study showed that the overexpression of the synthesized cecropin B gene in the phloem can significantly enhance resistance to HLB disease. This strategy provides an effective and promising approach for engineering citrus resistance against bacterial diseases. Further efforts are in progress to increase the concentration of cecropin B in the sieve elements. Additionally, we are transforming the synthesized cecropin B into rootstock varieties and evaluating the resistance of the transgenic scion and transgenic rootstock combinations against HLB. Finally, the resistance of these transgenic lines to HLB will be further investigated in fields exposed to free-flying Calas-positive ACPs.
References
Albrecht U, Bowman KD (2012) Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci 185:118–130. doi:10.1016/j.plantsci.2011.09.008
Alfano JR, Collmer A (1996) Bacterial pathogens in plants: life up against the wall. Plant Cell 8:10. doi:10.1105/tpc.8.10.1683
Aritua V, Achor D, Gmitter FG, Albrigo G, Wang N (2013) Transcriptional and microscopic analyses of citrus stem and root responses to Candidatus Liberibacter asiaticus infection. PloS one 8:e73742. doi:10.1371/journal.pone.0073742
Band VI, Weiss DS (2015) Mechanisms of antimicrobial peptide resistance in gram-negative bacteria. Antibiotics 4:18–41. doi:10.3390/antibiotics4010018
Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45:399–436. doi:10.1146/annurev.phyto.45.062806.094427
Bertaccini A, Duduk B (2010) Phytoplasma and phytoplasma diseases: a review of recent research. Phytopathol Mediterr 48:355–378. doi:10.1146/annurev.micro.54.1.221
Boscariol RL et al (2006) Attacin A gene from Tricloplusia ni reduces susceptibility to Xanthomonas axonopodis pv. citri in transgenic Citrus sinensis ‘Hamlin’. J Am Soc Hortic Sci 131:530–536
Bové JM (2006) Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol 88:7–37. doi:10.1371/journal.pone.0111032
Bové JM (2014) Huanglongbing or yellow shoot, a disease of Gondwanan origin: will it destroy citrus worldwide? Phytoparasitica 42:579–583. doi:10.1007/s12600-014-0415-4
Chen HM, Wang W, Smith D, Chan SC (1997) Effects of the anti-bacterial peptide cecropin B and its analogs, cecropins B-1 and B-2, on liposomes, bacteria, and cancer cells. Bba Gen Subj 1336:171–179. doi:10.1016/S0304-4165(97)00024-X
Coca M, Penas G, Gomez J, Campo S, Bortolotti C, Messeguer J, Segundo BS (2006) Enhanced resistance to the rice blast fungus Magnaporthe grisea conferred by expression of a cecropin A gene in transgenic rice. Planta 223:392–406. doi:10.1007/s00425-005-0069-z
Company N et al (2014) The production of recombinant cationic α-helical antimicrobial peptides in plant cells induces the formation of protein bodies derived from the endoplasmic reticulum. Plant Biotechnol J 12:81–92. doi:10.1111/pbi.12119
Cornelissen BJ, Horowitz J, van Kan JA, Goldberg RB, Bol JF (1987) Structure of tobacco genes encoding pathogenesis-related proteins from the PR-1 group. Nucleic Acids Res 15:6799–6811. doi:10.1093/nar/15.17.6799
Cui L et al (2008) Histone acetyltransferase inhibitor anacardic acid causes changes in global gene expression during in vitro Plasmodium falciparum development. Eukaryot Cell 7:1200–1210. doi:10.1128/EC.00063-08
da Graca JV, Douhan GW, Halbert SE, Keremane ML, Lee RF, Vidalakis G, Zhao H (2016) Huanglongbing: an overview of a complex pathosystem ravaging the world’s citrus. J Integr Plant Biol 58:373–387. doi:10.1111/jipb.12437
Datla RS, Hammerlindl JK, Pelcher LE, Crosby WL, Selvaraj G (1991) A bifunctional fusion between beta-glucuronidase and neomycin phosphotransferase: a broad-spectrum marker enzyme for plants. Gene 101:239–246
Donmez D, Simsek O, Izgu T, Kacar YA, Mendi YY (2013) Genetic Transformation in citrus. The Scientific World J 2013:1–8. doi:10.1155/2013/491207
Duan Y et al (2009) Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol Plant Microbe Interact 22:1011–1020. doi:10.1094/MPMI-22-8-1011
Dutt M, Ananthakrishnan G, Jaromin MK, Brlansky RH, Grosser JW (2012) Evaluation of four phloem-specific promoters in vegetative tissues of transgenic citrus plants. Tree Physiol 32:83–93. doi:10.1093/treephys/tpr130
Dutt M, Barthe G, Irey M, Grosser J (2015) Transgenic citrus expressing an Arabidopsis NPR1 Gene exhibit enhanced resistance against Huanglongbing (HLB; citrus greening). PloS one 10:e0137134. doi:10.1371/journal.pone.0137134
Florack D, Allefs S, Bollen R, Bosch D, Visser B, Stiekema W (1995) Expression of giant silkmoth cecropin B genes in tobacco. Transgenic Res 4:132–141. doi:10.1007/BF01969415
Fu SM, Hartung J, Zhou CY, Su HN, Tan J, Li ZA (2014) Ultrastructural changes and putative phage particles observed in sweet orange leaves infected with ‘Candidatus Liberibacter asiaticus’. Plant Dis 99:320–324. doi:10.1094/PDIS-01-14-0106-RE
Gong X, Liu J (2013) Genetic transformation and genes for resistance to abiotic and biotic stresses in Citrus and its related genera. Plant Cell Tissue Org Cult 113(2):137–147. doi:10.1007/s11240-012-0267-x
Gurr SJ, Rushton PJ (2005a) Engineering plants with increased disease resistance: how are we going to express it? Trends Biotechnol 23:283–290. doi:10.1016/j.tibtech.2005.04.009
Gurr SJ, Rushton PJ (2005b) Engineering plants with increased disease resistance: what are we going to express? Trends Biotechnol 23:275–282. doi:10.1016/j.tibtech.2005.04.007
Shokrollah H, Abdullah TL, Sijam K, Abdullah NK, Abdullah AP (2009) Differential reaction of citrus species in Malaysia to huanglongbing (HLB) disease using grafting method. Am J Agric Bio Sci 4:32–38
He Y, Chen S, Peng A, Zou X, Xu L, Lei T (2011) Production and evaluation of transgenic sweet orange (Citrus sinensis Osbeck) containing bivalent antibacterial peptide genes (Shiva A and Cecropin B) via a novel Agrobacterium-mediated transformation of mature axillary buds. Sci Hortic 128:99–107. doi:10.1016/j.scienta.2011.01.002
Hightower R, Baden C, Penzes E, Dunsmuir P (1994) The expression of cecropin peptide in transgenic tobacco does not confer resistance to Pseudomonas syringae pv tabaci. Plant Cell Rep 13:295–299. doi:10.1007/BF00233324
Hilf ME, Sims KR, Folimonova SY, Achor DS (2013) Visualization of ‘Candidatus Liberibacter asiaticus’ cells in the vascular bundle of citrus seed coats with fluorescence in situ hybridization and transmission electron microscopy. Phytopathology 103:545–554. doi:10.1094/PHYTO-09-12-0226-R
Holásková E, Galuszka P, Frébort I, Öz MT (2015) Antimicrobial peptide production and plant-based expression systems for medical and agricultural biotechnology. Biotechnol Adv 33:1005–1023. doi:10.1016/j.biotechadv.2015.03.007
Hou ZX, Huang WD (2005) Immunohistochemical localization of IAA and ABP1 in strawberry shoot apexes during floral induction. Planta 222:678–687. doi:10.1007/s00425-005-0014-1
Jan PS, Huang HY, Chen HM (2010) Expression of a synthesized gene encoding cationic peptide cecropin B in transgenic tomato plants protects against bacterial diseases. Appl Environ Microbiol 76:769–775. doi:10.1128/AEM.00698-09
Jaynes JM, Nagpala P, Destefanobeltran L, Huang JH, Kim JH, Denny T, Cetiner S (1993) Expression of a cecropin-B lytic peptide analog in transgenic tobacco confers enhanced resistance to bacterial wilt caused by Pseudomonas Solanacearum. Plant Sci 89:43–53. doi:Doi:10.1016/0168-9452(93)90169-Z
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Johnson EG, Wu J, Bright DB, Graham JH (2014) Association of ‘Candidatus Liberibacter asiaticus’ root infection, but not phloem plugging with root loss on huanglongbing-affected trees prior to appearance of foliar symptoms. Plant Pathol 63:290–298. doi:10.1111/ppa.12109
Lutcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA (1987) Selection of AUG initiation codons differs in plants and animals. EMBO J 6:43–48
Melo MN, Ferre R, Castanho MA (2009) Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat Rev Microbiol 7:245–250. doi:10.1038/nrmicro2095
Mills D, Hammerschlag FA (1993) Effect of cecropin-B on peach pathogens, protoplasts, and cells. Plant Sci 93:143–150. doi:Doi:10.1016/0168-9452(93)90043-Y
Mills D, Hammerschlag FA, Nordeen RO, Owens LD (1994) Evidence for the breakdown of cecropin B by proteinases in the intercellular fluid of peach leaves. Plant Sci 104:17–22. doi:10.1016/0168-9452(94)90186-4
Miyata LY, Harakava R, Stipp LC, Mendes BM, Appezzato-da-Gloria B, de Assis Alves Mourao Filho F (2012) GUS expression in sweet oranges (Citrus sinensis L. Osbeck) driven by three different phloem-specific promoters. Plant Cell Rep 31:2005–2013. doi:10.1007/s00299-012-1312-2
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nadal A et al (2012) Constitutive expression of transgenes encoding derivatives of the synthetic antimicrobial peptide BP100: impact on rice host plant fitness. BMC Plant Biol 12:159–181. doi:10.1186/1471-2229-12-159
Nawrocki KL, Crispell EK, McBride SM (2014) Antimicrobial peptide resistance mechanisms of Gram-positive bacteria. Antibiotics 3:461–492. doi:10.3390/antibiotics3040461
Salas CE, Badillo-Corona JA, Ramirez-Sotelo G, Oliver-Salvador C (2015) Biologically active and antimicrobial peptides from plants. Biomed Res Int 2015:102–129. doi:10.1155/2015/102129
Sharma A, Sharma R, Imamura M, Yamakawa M, Machii H (2000) Transgenic expression of cecropin B, an antibacterial peptide from Bombyx mori, confers enhanced resistance to bacterial leaf blight in rice. FEBS Lett 484:7–11. doi:10.1016/S0014-5793(00)02106-2
Shaw JE, Alattia JR, Verity JE, Prive GG, Yip CM (2006) Mechanisms of antimicrobial peptide action: studies of indolicidin assembly at model membrane interfaces by in situ atomic force microscopy. J Struct Biol 154:42–58. doi:10.1016/j.jsb.2005.11.016
Sjolund RD (1997) The phloem sieve element: a river runs through It. Plant Cell 9:1137–1146. doi:10.1105/tpc.9.7.1137
Straus SK, Hancock RE (2006) Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta 1758:1215–1223. doi:10.1016/j.bbamem.2006.02.009
Tatineni S, Sagaram US, Gowda S, Robertson CJ, Dawson WO, Iwanami T, Wang N (2008) In planta distribution of ‘Candidatus Liberibacter asiaticus’ as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology 98:592–599. doi:10.1094/PHYTO-98-5-0592
Trivedi P et al (2009) Quantification of viable Candidatus Liberibacter asiaticus in hosts using quantitative PCR with the aid of ethidium monoazide (EMA). Eur J Plant Pathol 124:553–563. doi:10.1007/s10658-009-9439-x
Vervliet G, Holsters M, Teuchy H, Van Montagu M, Schell J (1975) Characterization of different Plaque-forming and defective temperate phages in agrobacterium strains. J Gen Virol 26(1):33–48
Wang N, Trivedi P (2013) Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 103:652–665. doi:10.1094/PHYTO-12-12-0331-RVW
Weise A, Barker L, Kuhn C, Lalonde S, Buschmann H, Frommer WB, Ward JM (2000) A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12:1345–1355 doi:10.1105/tpc.12.8.1345
Xoconostle-Cázares B et al (1999) Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283:94–98. doi:10.1126/science.283.5398.94
Zhang Q, Zhang J, Huang Z, Tan S, Guo Z (1995) The bactericidal effect of antibacterial peptide from Chinese Oak silkworm on the pathogen of bacterial ulcer and yellow shoot disease in citrus. Acta Sericol Sin 21:77–81
Zhang S et al (2011) ‘Ca. Liberibacter asiaticus’ carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Mol Plant Microbe Interact 24:458–468. doi:10.1094/MPMI-11-10-0256
Zou X, Li D, Luo X, Luo K, Pei Y (2008) An improved procedure for Agrobacterium-mediated transformation of trifoliate orange (Poncirus trifoliata L. Raf.) via indirect organogenesis. In Vitro Cell Dev Biol Plant 44:169–177. doi:10.1007/s11627-008-9106-5
Zou X et al (2014a) Secreted expression of cecropin B gene enhances resistance to Xanthomonas axonopodis pv. citri in transgenic citrus sinensis‘Tarocco rsquo. Acta Hortic Sin 41:417–428
Zou X et al (2014b) Activation of three pathogen-inducible promoters in transgenic citrus (Citrus sinensis Osbeck) after Xanthomonas axonopodis pv. citri infection and wounding. Plant Cell Tissue Org Cult 117:85–98. doi:10.1007/s11240-013-0423-y
Acknowledgements
This work was supported by grants from the National Natural Sciences Foundation of China (31272150, to XZ), the Earmarked Fund for China Agriculture Research System (CARS-27, to SC), and the Fundamental Research Funds for the Central Universities (XDJK2012B023, to XZ).
Author contributions
XZ designed the experiments, constructed the vectors, and wrote the manuscript. LX and AP performed the citrus genetic transformations. XJ evaluated the resistance to HLB. TL and YH performed molecular analyses. LY performed the Southern blot analysis. SC analyzed the data. All of the authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zou, X., Jiang, X., Xu, L. et al. Transgenic citrus expressing synthesized cecropin B genes in the phloem exhibits decreased susceptibility to Huanglongbing. Plant Mol Biol 93, 341–353 (2017). https://doi.org/10.1007/s11103-016-0565-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0565-5