Abstract
Although the main genes in rice involved in the biosynthesis of secondary wall components have been characterized, the molecular mechanism underlying coordinated regulation of genes expression is not clear. In this study, we reported a new rice variety, cef1, showed the culm easily fragile (CEF) without other concomitant phenotypes. The CEF1 gene encodes a MYB family transcription factor OsMYB103L, was cloned based on map-based approach. Bioinformatics analyses indicated that CEF1 belongs to the R2R3-MYB subfamily and highly similar to Arabidopsis AtMYB103. Expression pattern analysis indicated that CEF1 is mainly expressed in internodes and panicles. Biochemical assays demonstrated that OsMYB103L is a nuclear protein and shows high transcriptional activation activity at C-terminus. OsMYB103L mediates cellulose biosynthesis and secondary walls formation mainly through directly binding the CESA4, CESA7, CESA9 and BC1 promoters and regulating their expression. OsMYB103L may also function as a master switch to regulate the expression of several downstream TFs, which involved in secondary cell wall biosynthesis. Furthermore, OsMYB103L physically interacts with SLENDER RICE1 (SLR1), a DELLA repressor of GA signaling, and involved in GA-mediated regulation of cellulose synthesis pathway. Our findings revealed that OsMYB103L plays an important role in GA-regulating secondary cell wall synthesis, and the manipulation of this gene provide a new strategy to help the straw decay in soil.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cell wall in plant is unique. It plays critical role in plant growth and development, resistance to pathogen invasion, quality of plant-based foods and the properties of plant fibers and fuels (Keegstra 2010). However, it also brings many challenges for the plant breeders, for instance, the straw decomposition is very slow when directly decay in the fields. This is mainly due to the high cellulose content in the cell wall, which was negatively correlated with the decomposition rate (Gunnarsson and Marstorp 2002; Rahn et al. 2003). In Asia, a large number of rice straws were processed by stubble burning, which caused severe environment pollution. Therefore, it is urgent to understand the cell wall biosynthesis. The plant cell walls are usually divided into two categories: primary cell walls that surround growing cells or cells retain growing capability; and secondary cell walls that are thickened structures containing lignin and surrounding specific cells such as vessel elements or fiber cells (Keegstra 2010). The secondary walls are typically composed of cellulose, xylan and lignin.
In the last decade, many studies have been done for secondary walls biosynthesis mainly focused on identification and functional characterization of genes involved in the biosynthesis of the secondary cell wall components. In rice, many studies have been performed to characterize the genes controlling culm mechanical strength by analysis the brittle culm mutants. The BC7/BC11, BC6/BC13 and OsCESA7 are the cellulose synthase catalytic subunit genes, involves in cellulose synthesis (Kotake et al. 2011; Song et al. 2013; Tanaka 2003; Yan et al. 2007; Zhang et al. 2009). The BC1 gene encodes a COBRA-like protein which controls cellulose deposition of the secondary cell wall (Li 2003). The BC3, encoding a classical dynamic OsDRP2B essential which might function in vesicle trafficking of CESA (Hirano et al. 2010; Xiong et al. 2010). The rice BC10 gene encodes a Golgi-located type II membrane protein that functions as a glycosyltransferase and mainly expressed in the developing sclerenchyma and vascular bundle cells (Zhou et al. 2009). The rice gene BC15 encodes a membrane-associated chitinase-like protein that is a Golgi-localized type II membrane protein and lacks classical chitinase activity. It mediates cellulose biosynthesis and cell wall remodeling (Wu et al. 2012). Another rice gene BC14, which encodes a Golgi nucleotide sugar transporter that provides the glucosyl substrate for the formation of polysaccharide and thereby regulates the synthesis of cellulose (Zhang et al. 2011). The rice BC12 gene encodes a dual-targeting kinesin-4 protein, controls cell-cycle progression and cellulose microfibril deposition (Li et al. 2011; Zhang et al. 2010b). Other genes involved in xylan and lignin biosynthesis have also been identified, the CslF6 (Vega-Sanchez et al. 2012) and CslD4 (Li et al. 2009a) are encode cellulose synthase-like protein, which is a glycosyltransferase family believed to be involved in the biosynthesis of cell-wall polymers. The FC1 (Li et al. 2009b) and OsCAD (Hirano et al. 2012) are involved in lignin biosynthesis, loss-of-function of these genes can reduce the lignin content. Although many genes involved in secondary cell wall biosynthesis have been reported, how these genes coordinately expressed and what controls the developmental program of secondary wall biosynthesis are less well-understood.
Recently the transcriptional switch mechanism for secondary cell wall biosynthesis have been discovered in dicot species, including Arabidopsis (Ko et al. 2014; Kubo et al. 2005; Ohman et al. 2013; Zhang et al. 2007; Zhong et al. 2006, 2007b; Zhong and Ye 2012), and Poplar (McCarthy et al. 2010). Most of the transcription factors (TFs) regulating the secondary walls biosynthesis belong to two large families: NAC and MYB. A group of the NAC family TFs function as the top master regulators to mediate secondary walls formation in fibers and/or vessels (Zhong et al. 2008). Some members of the MYB TFs family act as the second and/or the third level master regulators to control the secondary walls relevant genes expression (Zhao and Dixon 2011). Many of the MYB TFs involved in secondary walls formation can be up-regulated by SND1 (Zhong et al. 2008). The MYB46 and MYB83, expressed in both fibers and vessels, are direct targets of SND1 and its close homologs VND6, VND7, NST1 and NST2 (Zhong et al. 2008). Some members of MYB TFs including MYB20, MYB42, MYB52, MYB54, MYB103, and MYB63, are also up-regulated by MYB46 and MYB83 (McCarthy et al. 2009; Zhong et al. 2007a). All of these NACs and MYBs collectively regulate the downstream genes expression. A few studies have been investigating the transcription switches to secondary walls formation in monocot species. To date, few TFs have been reported to be associated with secondary cell wall formation in rice (Hirano et al. 2013; Yang et al. 2014; Zhang et al. 2010c; Zhong et al. 2011). The OsMYB103L is an important TF regulates cellulose biosynthesis. Overexpression of OsMYB103L results in leaf rolling, increased cellulose content, and upregulated the expression of genes encoding cellulose synthase (CESA). Moreover, the RNA interference (RNAi)-knockdown of OsMYB103L leads to a weakened mechanical strength and decreased cellulose content in leaves (Yang et al. 2014). The OsMYB61 can directly binding the CESAs promoter and regulate these genes expression. Overexpression of OsMYB61 also leads leaf rolling and increased cellulose content (Huang et al. 2015). Hence finding more transcriptional switches in rice may provide valuable approach for genetically modifying grass crops for biofuel production.
In this report, we isolated a culm easily fragile (cef1) rice plant from Wandao60 (M1148), and developed a pair of near isogenic lines (NIL), NIL-CEF1 and NIL-cef1. The NIL-cef1 showed normal phenotype except the culm breaks easily compare to NIL-CEF1. We further cloned the gene responsible for the corresponding phenotype, CEF1. Molecular characterization of CEF1 suggested it encodes an OsMYB103L transcription factor that regulates secondary cell wall components biosynthesis.
Materials and methods
Plant materials and growth conditions
The culm easily fragile (cef1) rice (Oryza sativa L.) was isolated from a Wandao60 (M1148), an Anhui late japonica rice variety. An F2 mapping population was generated from the cross between cef1 and a polymorphic indica cultivar, NanJing11. We also developed a pair of near isogenic lines (NIL), NIL-cef1 and NIL-CEF1, from the progenies of the cross between Wandao60cef1 and Xiushui110. All of rice plants used in this study were grown in the experimental fields at the Institute of Technical Biology and Agriculture Engineering, Hefei Institute of Physical Science, Chinese Academy of Sciences (Hefei, China) and Sanya (Hainan province, China) in the natural growing season.
Measurement of breaking force and microscopy
Breaking force of the second internodes and flag leaves of NIL-CEF1 and NIL-cef1 were determined with a digital force/length tester (5848 microtester, Instron; http://www.instron.com) by cutting the tested samples into segments with equal length and width. We used the Newton (N) as the unit of mechanical force. For transmission electron microscopy, the second internodes of NIL-cef1 and NIL-CEF1 plants were cut with a razor and immediately placed in 70 % ethanol (V/V), 5 % acetic acid (V/V), and 3.7 % formaldehyde (V/V) for at least 2 h. Samples were dried to the critical point, sputter-coated with gold, and observed with a scanning electron microscope (S570; Hitachi, Tokyo, Japan).
Phenotypic evaluation
Several agronomic traits, including plant height, number of tillers per plant, 1000-grain weight, panicle length, setting rate and number of grains per panicle were chosen for production evaluation. The plant height was measured from the ground surface to the tallest panicle. The number of tillers per plant was the number of effective tillers with corresponding panicle has 10 or more grains. The 1000-grain weight was calculated based on 100 grains and the converted to 1000 grain weight. The panicle length was the distance from length top to the first internode top. The number of grains per panicle was the total number of grains per panicle. The setting rate was scored as the number of full grains per panicle divided by the number of grains per panicle. All the traits were analyzed at mature stage. The data from fifty plants in each NIL in 2013 and 2014 was used for analysis. The independent sample t test program SPSS 10.0 for Windows was used to compare the mean values.
Cell wall composition analysis
The second internodes of NIL-CEF1, NIL-cef1 and complementary transgenic plants (NIL-cef1 pCEF1::CEF1) at mature stage were used to prepare alcohol-insoluble residues (AIRs) of the cell walls. De-starched AIRs were produced as previously described (Xiong et al. 2010). The AIRs were hydrolyzed in 67 % v/v H2SO4 for 1 h at room temperature, and then in 2M H2SO4 at 121 °C for 1 h. The alditol acetate derivatives were prepared from AIRs and then determined by GC–MS. The crystalline cellulose was measured with a modified Updegraff method, as previously described (Updegraff 1969).
Map-based cloning
The cef1 locus was mapped and cloned using 2307 mutant F2 plants, which were selected from the population of Wandao60cef1 and Nanjing11. Molecular markers distributed throughout the whole rice genome were used for cef1 locus rough mapping. Molecular markers for fine mapping were developed to narrow down the mutated locus to a 102-kb region on chromosome 8. The corresponding DNA fragments in this mapping region were amplified from mutants and wild type plants using LA-Taq (TaKaRa, http://www.takara-bio.com) and sequenced using an Applied Biosystems 3730 sequencer (ABI, http://www.appliedbiosystems.com). For complementation of cef1 mutant, the full-length CEF1 coding sequence, a 2.5-kb fragment of upstream sequence from ATG and 0.2-kb fragment of downstream sequence from TAA were inserted into binary vector pCAMBIA2300 between the EcoRI and BamHI sites to generate the construct pCEF1::CEF1, which was introduced into the NIL-cef1 by the Agrobacterium-mediated transformation procedure as described previously (Raineri et al. 1990).
Bioinformatics analysis of OsCEF1
Domain prediction for OsCEF1 was performed using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1). A search for R2R3-MYB-related family members in rice and Arabidopsis was performed using the NCBI BLAST server (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A phylogenetic tree of R2R3-MYB-related proteins in Arabidopsis and rice was generated using MAGE4 (Tamura et al. 2007). The number of bootstrap replicates was 1000.
Transactivation analysis in yeast cells and rice protoplasts
A yeast strain (AH109) containing the HIS3 and ADE2 reporter genes was used to analyst transactivation of OsMYB103L and truncation forms as described previously (Wang et al. 2012). The full length coding sequence of OsMYB103L and sequences encoding N-terminal and C-terminal truncation products were amplified and cloned into pGBKT7 and fused with the GAL4 DNA-binding domain to construct the necessary serial vectors, which were then transformed into the AH109 yeast strain. The empty pGBKT7 (BD) and fusing the GAL4 vectors were used as negative and positive controls, respectively. The transactivation activity of these proteins were evaluated according to the growth on SD/-Trp (synthetic defined, Tryptophan dropout) and SD/-Trp, His, Ade (synthetic defined, Tryptophan, Histidine, Adenine dropout).
The transactivation activity was also examined in the rice protoplast system. To construct the effecter vectors, the full length coding sequence of OsMYB103L and sequences encoding N-terminal and C-terminal truncation products were amplified and fused with GAL4 binding domain (GAL4BD). The empty GAL4BD and fused with VP16 were used as negative and positive controls, respectively. For the reporter construct, the pUC19 containing the firefly luciferase (LUC) reporter gene driven by the minimal TATA box of the 35S promoter plus five GAL4 binding elements was used. A pTRL plasmid that contained a CaMV (Cauliflower mosaic virus) 35S promoter and Renilla LUC, was used as an internal control. The pTRL, effector and reporter were simultaneously transformed into the rice protoplast system, then kept in dark for 16 h. The LUC activity was measured as described previously (Ohta et al. 2000).
Subcellular localization of OsMYB103L
To observe the OsMYB103L subcellular localization, GFP fused to the C-terminus of OsMYB103L and inserted into the pCAMBIA2300 between the EcoRI and BamHI sites. Rice protoplasts expressing GFP positive OsMYB103L were observed using confocal laser scanning microscope (Leica TCS SP5). The cell nucleus was stained by DAPI dye.
Expression analysis
Total RNA was extracted from various rice tissues using TRIzol reagent (Invitrogen), as described previously (Wadsworth et al. 1988). The complementary DNA (cDNA) was synthesized from total RNA using a reverse transcriptional kit (TransGen, http://www.transgen.com.cn/). Quantitative RT-PCR was performed using relevant primers and qRT-PCR kit (TransGen, http://www.transgen.com.cn/) on a quantitative 7500 PCR system (ABI). All assays were repeated at least three times, the Actin gene was used as an internal control.
Yeast one-hybrid assay
The MYB103L encoding sequence fused with activation domain (B42AD) to construct effector (pB42AD-MYB103L) (Fig. 7a), The 2 kb fragment of upstream sequence from start codon of BC1, CESA4, CESA7, CESA9 were amplified and inserted into pLacZi to construct the reporters (pLacZi-BC1, pLacZi-CESA4, pLacZi-CESA7, pLacZi-CESA9) (Fig. 7a). The effectors and reporters were cotransformed into the yeast strain EGY48. The transformants were grown on synthetic dropout plates without tryptophan or uracil containing 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside for color development. The empty pB42AD and pLacZi were used as negative control.
Yeast two-hybrid assay
Yeast two-hybrid assays were performed as described previously (Liu et al. 2013b). The full length cDNA for SLR1 was amplified and cloned into pGBKT7 vector (Clontech), and the full length OsMYB103L cDNA sequence was inserted into the pGADT7 vector (Clontech). The vectors were transformed into yeast strain AH109. The resulting transformants were grown on a synthetic dropout (SD) medium (Difco Yeast Nitrogen Base without amino acid; BD) lacking tryptophan and leucine and also spotted on an SD medium lacking tryptophan, leucine, histidine and adenine.
BiFC assay
The BiFC assay was performed as described previously (Bracha-Drori et al. 2004). The full length CDS sequence of OsMYB103L was amplified and inserted into the pSY735 vector, the full length of SLR1 CDS sequence was amplified and cloned into the pSY736 vector. Arabidopsis protoplasts were prepared, transfected and visualized as described elsewhere (Sun et al. 2014).
Coimmunoprecipitation assay
Coimmunoprecipitation assay was performed as described elsewhere (Liu et al. 2013b). The SLR1-Flag and MYB103L-HA or BC1-HA simultaneously expressing in WT Arabidopsis protoplasts. Anti-Flag (Tiangen) immunoprecipitation was carried out as described previously (Zhang et al. 2010a), and the amount of MYB103L-HA or BC-HA in the immune precipitates was analyzed by anti-HA immunoblot.
Results
Identification and map-based cloning of CEF1
Wandao60cef1 showed the culm easily fragile, we found the Wandao60cef1 internodes were smashed by rice combine harvester and evenly distribute into the field (Fig. 1b, d, e), compare with other varieties (Fig. 1a, c, e). To find out the genetic basis of the cef1 phenotype and isolate the causative gene, we crossed the Wandao60cef1 with an indica cultivar, Nanjing11 to generate genetic analysis and mapping population. All of the F1 plants showed the normal phenotype. Within the F2 population, 256 and 84 individual plants showed normal and brittle culm phenotype, respectively. This results was consistent with the Mendelian segregation ratio of 3:1 (χ2 = 0.004 < χ 20.05 = 3.84) (Table S1). The result suggested that the culm easily fragile phenotype is controlled by a single recessive gene. We try to isolate the CEF1 by map-based cloning method. After screening 108 pleomorphic simple sequence repeat (SSR) markers that distribute evenly among the 12 rice chromosomes, the cef1 locus was mapped between markers N1-1 and HSR4 on chromosome 8 (Fig. 2a), and further narrowed to an approximate 102-kb region between markers N1-4 and N3-9 (Fig. 2b), which contains 15 putative open-reading frames (ORFs) (Fig. 2c). Then we sequenced all of the candidate genes and a 13-bp deletion was found in LOC_Os08g05520 (Fig. 2d). The mutated site was confirmed by amplifying the DNA fragment covering the deletion site (Fig. 2e). This deletion occurred in the first exon of the ORF, resulting a frame-shift and premature translational product of 56 amino acids (Fig. 2d). Therefore, cef1 is very likely to be a loss-of-function mutation. To further confirm that LOC_Os08g05520 corresponds to the cef1 locus, a 3.6-kb DNA fragment containing the 2.5-kb putative promoter and the coding region was cloned vector pCAMBIA2300 to generate the plasmid pCEF1::CEF1 (Fig. 2f), which was introduced into the NIL-cef1 plants by Agrobacterium tumefaciens-mediated transformation. The transgenic NIL-cef1 pCEF1::CEF1 plants complete rescued the culm easily fragile and the cell wall composition phenotypes (Fig. 2g; Table 1). Therefore the LOC_Os08g05520 was the CEF1 gene responsible for the mutant phenotypes described above.
Map-based cloning of the CEF1 gene. a Rough mapping of CEF1 gene. The gene was mapped to the region between markers N1-1 and HSR4 on chromosome 8. b Fine mapping of CEF1 gene. The CEF1 locus was narrowed to an approximately 102 kb region between markers N1-4 and N3-9. Vertical lines represent the positions of molecular markers and the number of recombinants. c 15 predicted ORFs within the fine mapping region. d Genomic structure of CEF1, boxes indicate exons and the mutation site is in the first exon (13-bp deletion). e The amplified DNA fragments that cover the mutation site to confirm the genotypes of NIL-CEF1 and NIL-cef1. f Constructs for complementation and biochemical assays. g Folding the internodes of rice plants to show the rescued mechanical property in the complemented plants
Phenotypic characterization of NIL-CEF1 and NIL-cef1
We also developed a pair of near isogenic lines, NIL-cef1, which carries an approximately 1.2-Mb segment including the Wandao60 cef1 allele in the Xiushui110 background, and its matching line, NIL-CEF1, carries the homologous segment from Xiushui110. Compare with the NIL-CEF1, the plant height of NIL-cef1 slightly decreased (Fig. 3a, h). The internodes of the NIL-cef1 plants were easily broken and the forces applied to break NIL-cef1 internodes were reduced to approximately 35 % compared with those of the NIL-CEF1 (Fig. 3b, c). Apart from the plant height and the break force, other traits, viz. the number of tillers per plant (Fig. 3i), 1000-grain weight (Fig. 3j), panicle length (Fig. 3k), seed setting rate (Fig. 3m) and number of grains per panicle (Fig. 3l) showed no significant difference in the two NILs.
Comparison of the phenotypic characterization between the NIL-CEF1 and NIL-cef1. a Three-month-old plant of NIL-CEF1 and NIL-cef1. Bar = 8 cm. b the cef1 culms are easily broken. c Triple measurements of the break force of culms. d–g Scanning electron micrographs of the sclerenchyma cell walls of the NIL-CEF1 (d) and NIL-cef1 (e). f, g are enlargements of the red boxed areas in d and e, respectively. Bars = 25 μm (d, e) and 5 μm in (f, g). H-M, Contrasting grain yield of NIL-CEF1 and NIL-cef1. h Plant height. i Tiller number. j 1000-grain weight. k Panicle length. l Number of grains of panicle. m Seed setting rate. All phenotypic data measured from plants grown with 20 × 20 cm spacing in paddies under normal cultivation conditions. Error bars represent SE (n = 50)
To understand the underlying cause of the observed brittleness, we used scanning electron microscope (SEM) to observe the second internodes cross-sections of NIL-CEF1 and NIL-cef1 plants. The observations revealed that the sclerenchyma cell wall of NIL-cef1 was extremely thinned (Fig. 3e, g), compare with the NIL-CEF1 (Fig. 3d, f). On the other hand, no significant difference in parenchyma cell wall was observed (Fig. 3d, e) between the pair of NIL. These results demonstrated the culm easily fragile phenotype in NIL-cef1 was due to the defect in thickening in sclerenchyma cell wall.
To figure out whether the thinner parenchyma cell walls and the reduced mechanical strength in cef1 plants was combined with altering cell wall composition, we next examined the cell wall composition in the second internodes of NIL-CEF1, NIL-cef1 and NIL-cef1 pCEF1::CEF1 plants at mature stage. As shown in Table 1, the cellulose content was dramatically decreased by approximately 28 % in NIL-cef1 relative to NIL-CEF1 plants, whereas the contents of Ara and Xyl, the two major sugars of arabinoxylan, were substantially increased by approximately 37 and 41 %, respectively (Table 1). The level of glucose was increased by approximately 18 % (Table 1). The contents of other neutral sugars altered only slightly between NIL-cef1 and NIL-CEF1 samples.
Taken together, the cef1 defects in mechanical strength was correlated with the thinned parenchyma cell walls and lower cellulose content in the mutant.
CEF1 is mainly expressed in internodes and panicles
To examine expression pattern of CEF1, we used quantitative real-time (qRT)-PCR to analyze the expression level of CEF1 in various organs from NIL-CEF1 plants. The results suggested that CEF1 is ubiquitously expressed in all organs while it was predominantly expressed in internodes and panicles. However, the expression level of CEF1 is relatively low in leaves, sheaths and roots, during heading stage and seedling stage (Fig. 4). The expression pattern was consistent with the culm easily fragile phenotype of the Wandao60cef1.
CEF1 encodes a R2R3-MYB family transcription factor
The coding sequence of the CEF1 is 1080 bp in length and encodes a protein of 359 amino acid residues with a predicted molecular mass of approximately 40 kDa. A search in the National Center for Biotechnology Information (NCBI) Conserved Domains database and the Simple Modular Architecture Research Tool (SMART) to analysis CEF1 showed that it belongs to R2R3-MYB family member and has two MYB DNA binding domains at N-terminus, and four low complexity domains at C- terminus (Fig. S1A).
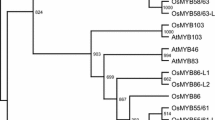
A BLASTP search for CEF1 homologs in Oryza sativa (japonica) and Arabidopsis thaliana genomes identified 17 and 25 closer homologs, respectively. Phylogenetic analysis revealed that the closest homolog of CEF1 is an Arabidopsis protein, AtMYB103 (At1g63910) (Fig. S1B). They are highly conserved in the R2- and R3-MYB DNA-binding domains, shown by protein sequence alignment.
One of the significant features of MYB family members is nuclear localization. To determine the subcellular localization of CEF1/MYB103L, a green fluorescent protein (GFP) tagged CEF1/MYB103L protein was generated by fusing GFP to the C-terminus of CEF1/MYB103L in a construct pActin::CEF1-GFP which was driven by Rice Actin promoter (Fig. 2f). The localization of GFP positive CEF1/MYB103L was observed using confocal laser scanning microscope. Our results indicated that the MYB103L-GFP fusion protein was located predominantly in nucleus (Fig. S1C).
C-terminal domains have strong transcriptional activation activity
To investigate the potential transcriptional activity of MYB103L, we used a yeast assay system to investigate full length MYB103L, N-terminal and C-terminal truncations. The growth of transformants containing pBD-MYB103L on selective medium (SD/-Trp) and (SD/-Trp, His, Ade) indicated that the MYB103L protein has transcriptional activity (Fig. S2A). To elucidate which domain of MYB103L has transcriptional activity, a series of N-terminal and C-terminal truncations (Fig. 5a, left panel) were made and cloned into the pGBKT7. The results indicated that C-terminal domains have the transcriptional activity (Fig. 5a, right panel).
Transcriptional activation of CEF1/MYB103L. a Transactivation activity of MYB103L in a yeast assay. Schematic representation (left panel) and transactivation activity in a yeast assay (right panel) of truncated OsMYB103L, OsMYB103L-M1 to OsMYB103L-M12 represent constructs with different truncated versions of OsMYB103L. Vertical broken lines from the left-hand side to the right-hand side indicate positions of DNA-binding domain and activation domain. The middle panel and the right panel indicate yeast growth on SD-Trp and SD-Trp, His, Ade medium, respectively. b Transcriptional activation ability of OsMYB103L and OsMYB103L-M1 to OsMYB103L-M12 as revealed by relative LUC activity
We also used the dual-luciferase reporter (DLR) assay system in the rice protoplast to test the transcriptional activation of full length MYB103L and different truncations (Fig. 5a, left panel). The constructs were illustrated in Fig. S2B. Compared with the GAL4-BD negative control, the OsMYB103L-M4 (1-280Aa), OsMYB103L-M5 (281-359Aa), OsMYB103L-M6 (251-359Aa), OsMYB103L-M7 (201-359Aa), OsMYB103-M8L (119-359Aa), OsMYB103L-M9 (1-323Aa), OsMYB103L-M10 (deletion at 251-280Aa), OsMYB103L-M11 (deletion at 281-323Aa) strongly activated the reporter gene, while the OsMYB103L-M1 (1-118Aa), OsMYB103L-M2 (1-200Aa), OsMYB103L-M3 (1-250Aa), OsMYB103L-M12 (deletion at 251-323Aa) did not activate the LUC gene (Fig. 5b). These results suggested that the two C-terminal domains (251-280Aa and 281-323Aa) of OsMYB103L have strong activation ability. The proteins containing 281-323Aa (OsMYB103L-M5~M10) have higher activation ability than those only containing 251-280Aa (OsMYB103L-M4, M11).
Expression of BC1, OsCESA4, OsCESA7 and OsCESA9 are down-regulated in NIL-cef1
The lower cellulose content in NIL-cef1 was the main cause of the culm easily fragile phenotype, therefore we measured cellulose synthesis relative genes expression level in NIL-CEF1, NIL-cef1 and NIL-cef1 p35S::myc-MYB103L transgenic plants. qRT-PCR analysis showed that the BC1, OsCESA4, OsCESA7, OsCESA9 genes were down-regulated more than twofold in NIL-cef1, comparing to that of NIL-CEF1 (Figs. 6, S3A). Interestingly, these genes were up-regulated by several fold in overexpression (OX) transgenic plants (NIL-cef1 p35S::myc-MYB103L) and the increased expression levels were coordinate with that of MYB103L (Figs. 6, S3B, S4). The results suggested that these genes were downstream of MYB103L.
Transcription of BC1, CESA4, CESA7 and CESA9 is directly activated by MYB103L
To address whether MYB103L can directly activate transcriptions of BC1 and three CESA 4, 7, 9 genes, we used yeast one-hybrid system to verify. The yeast one-hybrid assay showed that the pB42AD-MYB103L strongly activated the LacZ, which were driven by BC1 or three CESA genes promoters while the pB42AD without MYB103L did not activate the LacZ expression or to a lesser extent (Fig. 7b).
CEF1/MYB103L directly regulate BC1, CESA4, CESA7, CESA9 expression. a Diagram of the effector construct and the reporter constructs used in b. b Yeast one-hybrid assay showing the activity of LacZ reporters driven by BC1, CESA4, CESA7, CESA9 promoters and activated by activation domain (AD) fusion effectors. The empty pB42AD and pLacZi were used as negative control. c Diagram of the effector construct and the reporter constructs used in d. d Luciferase activities in protoplasts contransfected with the constructs shown in effectors and reporters. The transactivation activity was monitored by assaying the luciferase activities, with the activity in protoplasts transfected with an empty effector construct defined as 1. Error bars, SE of three biological replicates
We also used the dual-luciferase reporter (DLR) assay system in rice protoplasts. The CaMV 35S promoter drive MYB103L as an effector plasmid (Fig. 7c) and the firefly luciferase reporter gene was driven by BC1 and three CESA genes promoter (2 kb) as a reporter plasmid (Fig. 7c). The effector plasmid without MYB103L was used as negative control. The Renilla LUC driven by the 35S promoter was used as an internal control. These plasmids were coexpressed in rice protoplasts. Transactivation activity analysis showed that the luciferase activity in the protoplasts coexpressing an effector containing MYB103L and a reporter containing the BC1 and three CESA genes promoter driving luciferase was significant higher than the negative control (Fig. 7d). The results indicated MYB103L can directly activate transcription of BC1, CESA4, 7, 9.
MYB103L can regulate expression of other transcription factors
Hierarchical TFs involved in secondary wall biosynthesis exist in Arabidopsis thaliana. Therefore we tested if Hierarchical TFs regulating secondary wall biosynthesis exist in rice and if MYB103L can regulate expression of other TFs. We selected a number of TFs which are coexpressed with MYB103L and tested expression level of these TFs in NIL-CEF1, NIL-cef1 and NIL-cef1 p35S::myc-MYB103L overexpression transgenic plants. The qRT-PCR assay showed that the expression of OsSND2 (LOC_Os05g48850), OsMYB42/85 (LOC_Os09g36250) and OsMYB52/54 (LOC_Os03g51110) were down-regulated in NIL-cef1 plants and up-regulated in the transgenic overexpression lines (Fig. S4) with increased expression level of MYB103L (Fig. 8). The results suggested these TFs may be downstream of MYB103L. To address whether MYB103L can directly regulate expression of SND2, MYB42/85 and MYB52/54, we also used yeast one-hybrid system to verify. The yeast one-hybrid assay showed that the pB42AD-MYB103L cannot activate the LacZ expression (Fig. S5), so these TFs are regulated indirectly by MYB103L.
SLR1 interacts with MYB103L for secondary wall formation
As previously reported, gibberellin (GA) promotes cellulose biosynthesis. It functions through relieving the interaction between SLENDER RICE1 (SLR1), a DELLA repressor of GA signaling, and NACs, the crucial transcription factors for secondary wall formation. The SLR1-NACs interaction can effectively block the NACs-MYB-CESAs hierarchical regulation pathway (Huang et al. 2015). Therefore we were interested to test if the exogenous GA treatment can rescue the cef1 plants brittle culm phenotype. We treated the NIL-CEF1 and NIL-cef1 plants with exogenous GA and examined CESAs and BC1 expression in the shoots. The CESAs and BC1 expression of NIL-CEF1 was dramatically upregulated by GA treatment, comparing to the NIL-CEF1 without GA treated. More interestingly, the CESAs and BC1 expression of NIL-cef1 was only slightly upregulated (Fig. 9). Furthermore, the exogenous GA treatment cannot rescue the brittle culm phenotype (not shown). Hence the expression of MYB103L was required for the GA regulated cellulose biosynthesis pathway.
SLR1 is a key repressor to mediate the downstream responses through protein interactions. To address whether MYB103L can interact with SLR1, we used yeast two-hybrid (Y2H) system to verify the physical interaction of MYB103L and SLR1 in vitro (Fig. 10a). Bimolecular fluorescence complementation (BiFC) analysis in Arabidopsis revealed a direct interaction between MYB103L and SLR1 in the nuclei of living plant cells (Fig. 10c). Using MYB103L-HA or BC1-HA and SLR1-Flag transient coexpression in Arabidopsis protoplasts, we also confirmed the physical interaction between MYB103L and SLR1 by coimmunoprecipitation in vivo (Fig. 10b).
MYB103L interacts with the SLR1. a MYB103L interacts with SLR1 in yeast. MYB103L, a prey plasmid containing full length cDNA of OsMYB103L; SLR1, a bait plasmid containing full length cDNA of SLR1; pGADT7, empty prey plasmid; pGBKT7, empty bait plasmid; SD/-T, L, SD medium lacking tryptophan and leucine; SD/-T, L,H, A, SD medium lacking tryptophan, leucine, histidine, and adenine; b MYB103L interacts with SLR1 in Arabidopsis protoplasts. MYB103L-HA and BC1-HA were expressed in WT Arabidopsis protoplasts, along with SLR1-Flag, immunoprecipitated with anti-Flag antibody, and the bound protein was detected by immunoblot with the indicated antibodies. c BiFC analysis of the interaction between MYB103L and SLR1 in Arabidopsis protoplasts
To further understand the mechanism how the MYB103L-SLR1 complex regulate CESAs and BC1 gene expression. We used the dual-luciferase reporter (DLR) assay system in Arabidopsis protoplasts. These effectors and reporters plasmids were coexpressed in Arabidopsis protoplasts (Fig. 10a). Transactivation analysis showed that luciferase activity in the cells coexpressing reporters containing the CESAs or BC1 promoter driven luciferase and an effector containing MYB103L was significantly repressed by the additional coexpression of SLR1. This repression was relieved by the exogenous GA treatment (Fig. 11b), suggesting that SLR1 inhibits the MYB103L regulating CESAs and BC1 expression. Thus GA promoting cellulose biosynthesis not only through disrupting the interaction between SLR1 and NACs, but also through the relieving of the SLR1/MYB103L complex.
SLR1-MYB103L Interaction regulates CESAs and BC1 expression. a Diagrams of the effector and reporter constructs used in b. b Luciferase activities in protoplasts cotransfected with the reporter and different combinations of effectors. The transactivation activity was monitored by assaying the luciferase activities, with the activity in protoplasts transfected with an empty effector construct defined as 1. Error bars indicate the SE of three biological repeats
Discussion
In this study, we found a new rice variety (Wandao60) that showed its culm easily fragile in breeding, its straws were smashed by rice combine and evenly distribute into the field and not affect its lodging, although its break force reduced to approximately 35 % compared with that of the NIL-CEF1. We studied the phenotype of the cef1 plants, CEF1 gene mapping and its expression pattern. We confirmed CEF1 gene lead to the phenotype of cef1 through functional complementation assay. We also further investigated CEF1 protein function and downstream genes of CEF1.
CEF1 is a member of the MYB family
In plants, the MYB proteins are the one of the largest of transcription factor families (Riechmann 2000; Romero et al. 1998). They can be classified into four subfamilies based on the sequence and number of adjacent repeats in the MYB domain (R1, R2R3, 3R, 4R) (Dubos et al. 2010; Jin and Martin 1999). Among them, R2R3-MYB subfamily is the largest and plays important roles in processes including plant development, secondary metabolism, and stress responses (Stracke et al. 2001). In rice, at least 155 MYB genes distribute into different chromosomes and R2R3-MYB accounts for approximately 56.77 % of all MYB genes (Katiyar et al. 2012). CEF1 encodes a putative MYB-like DNA-binding domain containing protein. Phylogenetic analysis indicates that AtMYB103 is the most closely homolog of CEF1 (Fig. S1B). AtMYB103 is a typical R2R3-MYB protein (Ohman et al. 2013; Zhong et al. 2008). It is similar to AtMYB103, CEF1 has two DNA-binding domain at N-terminus (Fig. S1A), belongs to the R2R3-MYB subfamilies, and located in the nucleus (Fig. S1C). In R2R3-MYB subfamilies, highly conserved at MYB domains, while the C-terminal regions are high variable (Stracke et al. 2001). Go analysis of MYB proteins suggested that 98.7 % OsMYB involved in transcription activation (Katiyar et al. 2012). Transactivation analysis indicated that CEF1/OsMYB103L has strong transcriptional activation activity at C-terminal domains (Fig. 5).
CEF1/OsMYB103L plays an important role in secondary cell wall synthesis
Secondary cell walls providing the mechanical support and protection to the plant body, are mainly composed of cellulose, xylan and lignin. The complicated assembly causes problems for the straw disposal. Therefore, understanding the formation of secondary cell walls is critical for the straw disposal. In the past decade, many studies were focus on the biosynthesis of cell walls, however up to date only three rice TFs have been reported to be associated with secondary cell walls formation (Hirano et al. 2013; Yang et al. 2014; Zhong et al. 2011). We reported in this study that CEF1 is a TF regulating secondary cell walls formation. Loss-of-function mutation in CEF1 can reduce cellulose content (Table 1) and decrease the thickness of sclerenchyma cell walls (Fig. 3d, g). Even the cef1 plant exhibits easily broken in clum, it is different from other brittle mutants reported previously. The cef1 has no morphological abnormalities such as dwarfism, low fertility and withering of leaf apex. Hence the discovery of CEF1 gene may have great contribution to genetic modification of rice.
In rice, three synthase (CESA4, CESA7, CESA9) are necessary for cellulose production in the secondary cell walls (Tanaka 2003). Mutation of these CESA genes can induce brittle culm phenotype and decrease the cellulose content (Kotake et al. 2011; Song et al. 2013; Tanaka 2003; Zhang et al. 2009). CEF1/OsMYB103L as a transcription factor can regulate expression of these genes (Fig. 7b, d). The BC1 is also involved in cellulose synthesis, and plays an important role in the biosynthesis of the secondary cell walls of mechanical tissues (Li 2003; Liu et al. 2013a). We also demonstrated that CEF1/OsMYB103L regulates BC1 expression (Fig. 7b, d). AtMYB46, a master switch for secondary cell wall formation in Arabidopsis, can function as a direct regulator of all three secondary wall-associated cellulose synthase genes: AtCESA4, AtCESA7 and AtCESA8 (Kim et al. 2013). Dominant repression or RNA interference inhibition of AtMYB46 resulted in thinner secondary cell walls and a concomitant collapsed vessel phenotype (Zhong et al. 2007a). Mutation of the any of the AtCESA4/IRX5, AtCESA7/IRX3 and AtCESA8/IRX1 also caused severe reduction in cellulose content, secondary cell walls thickness and a collapsed vessel phenotype (Taylor et al. 1999, 2000, 2003). Consistent with these findings in Arabidopsis, CEF1/OsMYB103L regulated the cellulose content through mediating the expression of cellulose synthases. Therefore mutation of CEF1/OsMYB103L resulted in the brittle clum phenotype.
In the cef1 plants, in addition to decreased cellulose content, other cell wall components were altered. The content of arabinose and xylose, the two major sugars of arabinoxylan, were substantially increased by approximately 37 and 41 %, respectively (Table 1). Many brittle culm mutants, such as bc1 (Li 2003), bc11 (Zhang et al. 2009), bc13 (Song et al. 2013) and bc15 (Wu et al. 2012), also showed the decreased cellulose content and increased content of arabinose and xylose, corresponding with their wild type controls. It may have a feedback mechanism to compensate the decreased cellulose to maintain whole plant carbon balance in these brittle culm mutants.
The cef1 is a tissue specific brittle culm plant
The cef1 internodes are easily broken, but compare with other rice brittle culm mutants, the leaves are not fragile. The specific expression pattern of CEF1 in internodes and panicles (Fig. 4) was important to this phenotype in culm and panicle. This unique expression pattern have also been reported in Arabidopsis. In Arabidopsis the NAC and MYB family transcription factors regulate secondary cell wall biosynthesis in various secondary wall-forming cell types. The SND1, NST1 and NST2 are expressed specifically in fibers (Zhong et al. 2006, 2008). VND6 and VND7 are specially expressed in metaxylem and protoxylam, respectively. The MYB46 and MYB83 are expressed in both vessels and fibers (McCarthy et al. 2009; Zhong et al. 2007a). Dominant repression of these genes result in severe decreased in secondary cell wall thickening in corresponding cells and tissues.
Overexpression of OsMYB103L resulted in leaf rolling phenotype in rice (Yang et al. 2014), it may be caused by ectopic deposition of secondary cell walls in leaves. In Arabidopsis, overexpression of MYB46 also caused the leaf rolling phenotype (Kim et al. 2013). Overexpression of SND1 also exhibited a prominent visual phenotype that shows small rosette size and stunted growth of leaves with severely upward-curling blades, it mainly caused by massive deposition of secondary walls in cells that are normally nonsclerenchymatous (Zhong et al. 2006). Due to the expression of CEF1 in specific tissue, the cef1 plants result in brittle phenotype in internodes, but not in leaves. Similar to these reports, the overexpression lead to CEF1/OsMYB103L high expressed in all organs and caused the leaf rolling phenotype (Yang et al. 2014).
Hierarchical TFs involved in secondary wall biosynthesis may exist in rice
In Arabidopsis, the coordinate activation of secondary cell wall biosynthesis genes is controlled by a transcriptional network. In this transcriptional network, the secondary wall NAC families (SWNs) are the top-level master switches that can activate a battery of downstream transcription factors, including SND2, SND3, MYB20, MYB42, MYB46, MYB83, MYB103 and KNAT7 (Zhong et al. 2008). The MYB46 acts as the secondary-level master switch to regulate downstream transcription factors and the expression of the biosynthesis genes for cellulose, xylan and lignin (Zhong et al. 2007a). In rice, hierarchical TFs involved in secondary wall biosynthesis have been reported is rarely (Huang et al. 2015). In this study, we find some secondary wall-associated TFs, such as OsSND2, OsMYB42/85 and OsMYB52/54 are downstream of CEF1/OsMYB103L. We propose that CEF1/OsMYB103L may act as a master switch to regulate these TFs. Further illustration of the transcriptional network for secondary wall biosynthesis can help to depict the hierarchy of TFs in rice.
GA regulates cellulose biosynthesis also via SLR1-MYB103L-CESAs pathway
GA is an important hormone for plant growth and development, it mainly promotes the degradation of DELLAs and consequently activate the downstream responsive processes (Fu 2002). Previous study shows that GA regulates cellulose biosynthesis via SLR1-NAC Signaling Cascade (Huang et al. 2015). In the presence of GA, the GA-induced degradation of SLR1 released the suppression of MYB103L, which further upregulated the CESAs and BC1 expression (Fig. 11b). Comparing to the significant increase of CESAs and BC1 genes in NIL-CEF1, only slight increase of these genes were found in the NIL-cef1 plants (Fig. 9). Although the SLR1-NACs-MYB-CESAs signaling cascade has been reported in rice (Huang et al. 2015), the SLR1-NACs-MYB and SLR1-MYB103L may be together involved in GA regulates secondary cell wall synthesis. The OsMYB103L also is downstream of NAC family TFs (Huang et al. 2015). Hence, GA may regulates MYB103L expression via SLR1-NACs signaling cascade. Interestingly, the loss-of-function of MYB103L impaired regulation of CESAs and BC1 genes by GA (Fig. 9), GA regulates cellulose biosynthesis via SLR1-NACs signaling cascade may dependent on MYB103L. Therefore the MYB103L is a central factor in GA regulates cellulose biosynthesis pathway.
The cef1 has a potential prospect for residue management
The processing of rice straw has been a challenge for breeders because the rice straws cannot decompose in short-term. Straw burning is preferred by farmers, as it is economic and convenient, but it causes environmental problems. The high cellulose content of cell wall of rice straw was the major cause of slow decomposition (Tian et al. 1992). The brittle stem rice mutants have faster residue decomposition due to the lower cellulose content and finer breakage during threshing (Cabiles et al. 2008; Johnson et al. 2006). However, these mutants have concomitant phenotypes such as dwarfism, low fertility and withering of leaf apex, made them inadequate for breeding. The cef1 plants were different from other brittle culm mutants, since it has no morphological abnormalities. Under field conditions, the grain yield of NIL-cef1 and NIL-CEF1 plants was indistinguishable (Fig. 3h–m). Hence Wandao60cef1 is an ideal material to resolve the straw processing problem, and the mutation site of CEF1/OsMYB103L can also be introduced to other high-yielding varieties.
References
Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40:419–427
Cabiles DMS, Angeles OR, Johnson-Beebout SE, Sanchez PB, Buresh RJ (2008) Faster residue decomposition of brittle stem rice mutant due to finer breakage during threshing. Soil Tillage Res 98:211–216
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Fu X (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell Online 14:3191–3200
Gunnarsson S, Marstorp H (2002) Carbohydrate composition of plant materials determines N mineralisation. Nutr Cycl Agroecosyst 62:175–183
Hirano K, Kotake T, Kamihara K, Tsuna K, Aohara T, Kaneko Y, Takatsuji H, Tsumuraya Y, Kawasaki S (2010) Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis. Planta 232:95–108
Hirano K, Aya K, Kondo M, Okuno A, Morinaka Y, Matsuoka M (2012) OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep 31:91–101
Hirano K, Kondo M, Aya K, Miyao A, Sato Y, Antonio BA, Namiki N, Nagamura Y, Matsuoka M (2013) Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol 54:1791–1802
Huang D, Wang S, Zhang B, Shang-Guan K, Shi Y, Zhang D, Liu X, Wu K, Xu Z, Fu X, Zhou Y (2015) A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27:1681–1696
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Johnson SE, Angeles OR, Brar DS, Buresh RJ (2006) Faster anaerobic decomposition of a brittle straw rice mutant: implications for residue management. Soil Biol Biochem 38:1880–1892
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom 13:544
Keegstra K (2010) Plant cell walls. Plant Physiol 154:483–486
Kim WC, Ko JH, Kim JY, Kim JM, Bae HJ, Han KH (2013) MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J 73:26–36
Ko JH, Jeon HW, Kim WC, Kim JY, Han KH (2014) The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann Bot 114:1099–1107
Kotake T, Aohara T, Hirano K, Sato A, Kaneko Y, Tsumuraya Y, Takatsuji H, Kawasaki S (2011) Rice Brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J Exp Bot 62:2053–2062
Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19:1855–1860
Li Y (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell Online 15:2020–2031
Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, Pauly M, Cheng Z, Zhou Y (2009a) Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J 60:1055–1069
Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q, Wu C (2009b) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69:685–697
Li J, Jiang J, Qian Q, Xu Y, Zhang C, Xiao J, Du C, Luo W, Zou G, Chen M, Huang Y, Feng Y, Cheng Z, Yuan M, Chong K (2011) Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23:628–640
Liu L, Shang-Guan K, Zhang B, Liu X, Yan M, Zhang L, Shi Y, Zhang M, Qian Q, Li J, Zhou Y (2013a) Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet 9:e1003704
Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM (2013b) BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA 110:6205–6210
McCarthy RL, Zhong R, Ye ZH (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50:1950–1964
McCarthy RL, Zhong R, Fowler S, Lyskowski D, Piyasena H, Carleton K, Spicer C, Ye ZH (2010) The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol 51:1084–1090
Ohman D, Demedts B, Kumar M, Gerber L, Gorzsas A, Goeminne G, Hedenstrom M, Ellis B, Boerjan W, Sundberg B (2013) MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J 73:63–76
Ohta M, Ohme-Takagi M, Shinshi H (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22:29–38
Rahn C, Bending G, Turner M, Lillywhite R (2003) Management of N mineralization from crop residues of high N content using amendment materials of varying quality. Soil Use Manag 19:193–200
Raineri D, Bottino P, Gordon M, Nester E (1990) Agrobacterium-mediated transformation of rice (Oryza sativa L.). Nat Biotechnol 8:33–38
Riechmann JL (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J (1998) More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 14:273–284
Song XQ, Liu LF, Jiang YJ, Zhang BC, Gao YP, Liu XL, Lin QS, Ling HQ, Zhou YH (2013) Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants. Mol Plant 6:768–780
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Sun H, Qian Q, Wu K, Luo J, Wang S, Zhang C, Ma Y, Liu Q, Huang X, Yuan Q, Han R, Zhao M, Dong G, Guo L, Zhu X, Gou Z, Wang W, Wu Y, Lin H, Fu X (2014) Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet 46:652–656
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tanaka K (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133:73–83
Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11:769–780
Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12:2529–2540
Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100:1450–1455
Tian G, Kang B, Brussaard L (1992) Biological effects of plant residues with contrasting chemical compositions under humid tropical conditions—decomposition and nutrient release. Soil Biol Biochem 24:1051–1060
Updegraff DM (1969) Semimicro determination of cellulose inbiological materials. Anal Biochem 32:420–424
Vega-Sanchez ME, Verhertbruggen Y, Christensen U, Chen X, Sharma V, Varanasi P, Jobling SA, Talbot M, White RG, Joo M, Singh S, Auer M, Scheller HV, Ronald PC (2012) Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol 159:56–69
Wadsworth GJ, Redinbaugh MG, Scandalios JG (1988) A procedure for the small-scale isolation of plant RNA suitable for RNA blot analysis. Anal Biochem 172:279–283
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954
Wu B, Zhang B, Dai Y, Zhang L, Shang-Guan K, Peng Y, Zhou Y, Zhu Z (2012) Brittle culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol 159:1440–1452
Xiong G, Li R, Qian Q, Song X, Liu X, Yu Y, Zeng D, Wan J, Li J, Zhou Y (2010) The rice dynamin-related protein DRP2B mediates membrane trafficking, and thereby plays a critical role in secondary cell wall cellulose biosynthesis. Plant J 64:56–70
Yan C, Yan S, Zeng X, Zhang Z, Gu M (2007) Fine mapping and isolation of Bc7(t), allelic to OsCesA4. J Genet Genomics 34:1019–1027
Yang C, Li D, Liu X, Ji C, Hao L, Zhao X, Li X, Chen C, Cheng Z, Zhu L (2014) OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol 14:158
Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, Huang H, Xia HJ, Yang ZN (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J 52:528–538
Zhang B, Deng L, Qian Q, Xiong G, Zeng D, Li R, Guo L, Li J, Zhou Y (2009) A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol Biol 71:509–524
Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, Mengiste T, Zhang Y, Zhou JM (2010a) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7:290–301
Zhang M, Zhang B, Qian Q, Yu Y, Li R, Zhang J, Liu X, Zeng D, Li J, Zhou Y (2010b) Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J 63:312–328
Zhang S, Fang Z, Zhu J, Gao J, Yang Z (2010c) OsMYB103 is required for rice anther development by regulating tapetum development and exine formation. Chin Sci Bull 55:3288–3297
Zhang B, Liu X, Qian Q, Liu L, Dong G, Xiong G, Zeng D, Zhou Y (2011) Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc Natl Acad Sci USA 108:5110–5115
Zhao Q, Dixon RA (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci 16:227–233
Zhong R, Ye ZH (2012) MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53:368–380
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170
Zhong R, Richardson EA, Ye ZH (2007a) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–2792
Zhong R, Richardson EA, Ye ZH (2007b) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225:1603–1611
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782
Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH (2011) Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol 52:1856–1871
Zhou Y, Li S, Qian Q, Zeng D, Zhang M, Guo L, Liu X, Zhang B, Deng L, Liu X, Luo G, Wang X, Li J (2009) BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J 57:446–462
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (Grant 31301297), The Science and Technology Service program of Chinese Academy of Sciences (Grant KFJ-EW-STS-083), the State Key Laboratory of Plant Cell and Chromosome Engineering(Grant PCCE-KF-2014-03) and the Ministry of Agriculture of China for Transgenic Research (Grant 2014ZX08009-35B).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yafeng Ye and Binmei Liu have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1
CEF1 is a R2R3-MYB family transcription factor. A, Prediction of the domain structure of CEF1. B, Phylogenetic tree of the CEF1 homologs in Arabidopsis and rice. CEF1 and AtMYB103 have a very high homology. C, CEF1/MYB103L is a Nuclear-localized protein. A rice protoplast cell expressing MYB103L-GFP, indicating that CEF1/MYB103L is a Nuclear-localized protein. DAPI is a Nuclear dye (TIFF 542 kb)
Supplemental Figure 2
Transcriptional activation of CEF1/MYB103L. A, Transactivation activity of MYB103L in a yeast assay. Transformants harbouring pBD-MYB103L, the positive control pGAL4 and the negative control pBD were streaked onto SD-Trp or SD-Trp, His, Ade medium to determine growth. B, Effector and reporter constructs used in the Arabidopsis protoplast transient assay (TIFF 645 kb)
Supplemental Figure 3
Expression of cellulose synthesis related genes. A, the expression of OsCESA genes in NIL-CEF1 and NIL-cef1. B, the expression of CESA4, 7, 9 in NIL-CEF1 and NIL-cef1 p35S::myc-MYB103L transgenic plants. The Actin1 was used as internal control. All data given as mean ± SE (n = 3) (TIFF 436 kb)
Supplemental Figure 4
CEF1/OsMYB103L expression in overexpression (OX) transgenic plants (NIL-cef1 p35S::myc-MYB103L) as determined by qRT-PCR. The Actin was used as internal control. Error bars, SE of three biological replicates (TIFF 157 kb)
Supplemental Figure 5
MYB103L can’t binding SND2, MYB42/85, MYB52/54 promoters. Yeast one-hybrid assay showing the activity of LacZ reporters driven by BC1, SND2, MYB42/85, MYB52/54 promoters and activated by activation domain (AD) fusion effectors. The empty pB42AD and pLacZi were used as negative control (TIFF 469 kb)
Rights and permissions
About this article
Cite this article
Ye, Y., Liu, B., Zhao, M. et al. CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice. Plant Mol Biol 89, 385–401 (2015). https://doi.org/10.1007/s11103-015-0376-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0376-0