Abstract
Cinnamyl alcohol dehydrogenase (CAD) catalyzes the last step of monolignol biosynthesis. The rice genome contains 12 CAD-like genes, and whereas the proteins encoded by OsCAD2 and OsCAD7 are known to function in monolignol biosynthesis, the degree to which these enzymes contribute to this process and the involvement of the enzymes encoded by the remaining ten genes is unclear. This paper investigates the role of OsCAD2 and the nine other OsCAD-like proteins in monolignol biosynthesis. Among the OsCAD genes analyzed, OsCAD2, an enzyme belonging to the bona fide CAD phylogenetic group, was the most abundantly expressed gene in the uppermost internode, and was expressed at levels that were more than seven times greater than those of the second most abundantly expressed gene, OsCAD1. Promoter-GUS analysis of OsCAD2 (pCAD::GUS) in the internode, sheath, and roots revealed that GUS expression was strong in tissues that accumulated high levels of lignin. Furthermore, expression always preceded lignin accumulation, showing the tight correlation between OsCAD2 expression and monolignol biosynthesis. Additionally, expression of pCAD::GUS was well synchronized with that of rice caffeic acid 3-O-methyltransferase (OsCOMT::GUS), suggesting that the two enzymes function cooperatively during monolignol biosynthesis. Co-expression network analysis of eight OsCAD genes further revealed that, among the OsCAD genes, expression of OsCAD2 was most tightly associated with the transcription of lignin biosynthesis-related genes. These results suggest that OsCAD2 is largely responsible for monolignol biosynthesis in rice, which is similar to that indicated for the predominant role of other plant bona fide CAD protein to monolignol biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
To ensure the availability of sustainable energy supplies, considerable effort is being made to utilize plant biomass as biofuel. Although the production of ethanol from starch and sugars is industrialized, the use of corn grain, sugarcane, and sugar beet as feedstock compete with food supply and thus non-food lignocellulosic biomass usage is favored (Yuan et al. 2008). Since lignocellulosic biomass is mostly derived from plant cell walls, efficient conversion of cell wall components to ethanol is essential; however, there are still several technical problems in this process. Currently, the main issue that needs to be addressed is the cost of breaking down cell wall components to simple sugars (Weng et al. 2008).
Lignin constitute up to 20% of grass secondary cell wall (Vogel 2008) and undergoes cross-linking with cellulose and hemicelluloses to form a rigid cell wall. When effects of lignin on bio-ethanol production are considered, lignin impedes the saccharification of cellulose probably by reducing the accessibility of the saccharification enzymes to the cellulose microfibrils (Weng et al. 2008). Lignin is also considered to disturb the fermentation process of biofuel production. Consequently, manipulating the lignin content and/or composition within the cell wall is an important strategy for improving the utility of the materials in biofuel production.
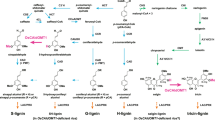
The major components of lignin are monolignols, which belong to the phenolic hydroxycinnamyl alcohol monomers, such as p-coumaryl, coniferyl, and syringyl alcohols (Boerjan et al. 2003). Monolignols are incorporated into lignin polymers in the form of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units. Attempts to change the content/quality of lignin polymers are therefore mainly focused on manipulating monolignol biosynthesis (Li et al. 2008), and one such target is the cinnamyl alcohol dehydrogenase (CAD), which catalyzes the last step of monolignol biosynthesis, in which hydroxyl-cinnamyl aldehydes are converted into their corresponding alcohols.
Arabidopsis contains nine CAD-like genes in its genome and are classified differently by three groups (Costa et al. 2003; Goujon et al. 2003a; Raes et al. 2003). Here we will refer to the classification proposed by Costa et al. (2003; AtCAD1 to 9), unless otherwise stated. Among them, AtCAD-C and -D (At3g19450 and At4g34230, respectively; nomenclature proposed by Goujon et al. 2003a) are classified as so-called bona fide CAD genes (Costa et al. 2003). Recent phylogenetic analysis suggested that the emergence of real lignin in the vascular plant lineage was associated with the origin of the bona fide CAD genes (Guo et al. 2010). The Arabidopsis cad-c/-d double mutant shows a 40% reduction in Klason lignin content in the flowering stem (Sibout et al. 2005), indicating that bona fide CAD genes are the major CAD genes involved in lignin biosynthesis. In Oryza sativa (rice), 12 CAD-like genes have been identified (Tobias and Chow 2005; Table 1), whereas only one (OsCAD2) has been identified that belongs to the bona fide CAD gene family. Curiously, however, the Klason lignin content of the OsCAD2 null mutant gh2 was reduced by only 5–6% in comparison to its original strain (Zhang et al. 2006). This may suggest that another OsCAD gene(s) besides the bona fide OsCAD2 cooperatively functions in monolignol biosynthesis in rice, or that there is another type of lignin that is independently produced by CAD.
To investigate the role of non-bona fide CADs in monolignol biosynthesis, each CAD in Arabidopsis have been studied extensively (Kim et al. 2004; Eudes et al. 2006; Kim et al. 2007). CAD enzymatic activity was detected for recombinant AtCAD2, 3, 7, and 8 (At2g21730, At2g21890, At4g37980, and At4g37990, respectively) proteins expressed in Escherichia coli, but at significantly lower levels than for bona fide AtCAD-C and -D, while AtCAD1, 6, and 9 (At1g72680, At4g37970, and At4g39330, respectively) had no detectable CAD activity (Kim et al. 2004). The expression pattern of AtCAD genes investigated by promoter-GUS analysis revealed that, like AtCAD-C and -D, AtCAD7 and 8 are expressed at sites where lignin accumulates, whereas AtCAD2 and 3 are not expressed in lignified tissues, and AtCAD1, 6, and, 9 are expressed in lignin-forming tissues despite the lack of detectable CAD activity in their encoded proteins (Kim et al. 2007). T-DNA knockout mutants of AtCAD1, 3, 6, 7, and 8 did not display apparent phenotypes, whereas the atcad9 knockout mutant had a slight reduction in Klason lignin content in the elongating stems (Eudes et al. 2006). Similarly, AtCAD-D promoter-driven AtCAD1, 2, 6, 7, and 8 were unable to complement the atcad-c/-d double mutant phenotype, while AtCAD5 promoter-driven AtCAD9 partially did (Eudes et al. 2006). Thus, although some AtCADs, such as AtCAD7, 8, and 9, may participate in lignin biosynthesis, it seems that bona fide AtCAD-C and -D act as the primary CADs in monolignol biosynthesis in Arabidopsis.
In rice, apart from the bona fide OsCAD2, the involvement of OsCAD7 in lignin biosynthesis was recently reported. OsCAD7 is the causal gene of flexible culm 1 (fc1), which shows reduced mechanical strength, and reduced cellulose and lignin content (18% reduction in Klason lignin content; Li et al. 2009). This observation supports the hypothesis that non-bona fide CADs in rice are involved in lignin biosynthesis, and suggests that the mechanism of lignin biosynthesis might differ in rice and Arabidopsis.
To investigate the possibility that non-bona fide CADs contribute to lignin biosynthesis in rice, we conducted several experiments. Although non-bona fide CADs were suggested to contribute in monolignol biosynthesis to some extent, our results suggest that the bona fide OsCAD2 plays the major role in monolignol biosynthesis, as observed for Arabidopsis AtCAD-C and -D.
Results
Phylogenetic analysis of OsCAD
According to the previous report (Tobias and Chow 2005), 12 CAD-like genes have been identified in the rice genome (Table 1). Among them, the OsCAD8 locus is assumed to have undergone recent duplication events, resulting in four tandemly localized CAD-like genes termed OsCAD8A to 8D (Tobias and Chow 2005). CAD belongs to the medium-chain dehydrogenase/reductase (MDR) superfamily, the members of which possess two Zn2+ ions per subunit and require NADPH as a cofactor (Youn et al. 2006). All of the deduced amino acid sequences of OsCAD genes contain the conserved Zn-binding signatures and the NADPH-binding domain (Supplemental Fig. 1). OsCAD5 is shorter than other OsCADs, due to a premature stop codon and is therefore probably a non-functional protein. However, none of the other OsCAD genes are apparently disrupted in their coding sequences, indicating that they may encode functional proteins. Three genes (OsCAD4, 5, and 9) were not found in the public EST database (GENBANK) or in the FULL-length database (KOME: http://cdna01.dna.affrc.go.jp/cDNA/) (Table 1), suggesting that these genes are expressed at low levels or are not expressed at all, or that their expression is restricted to certain tissues and/or stages of development.
When a phylogenetic tree was created using amino acid sequences deduced from the OsCAD genes and CAD genes from other species, OsCADs could be classified into three groups (Fig. 1). One group (bona fide CAD clade) consists of OsCAD2 (GH2; Hamberger et al. 2007). This clade contains proteins such as AtCAD-C and -D, and Zea mays (maize) CAD Bm1 (Halpin et al. 1998). Mutation of AtCAD-C/-D and Bm1 results in a 40 and 20.3% reduction in Klason lignin content, respectively (Sibout et al. 2005; Halpin et al. 1998). The second group (class II) is composed of rice CADs and does not contain Arabidopsis CADs. The causal protein of fc1, OsCAD7, exists within this clade (Li et al. 2009). The third group (Class III) includes OsCAD1, 4, and 6 and forms a clade with AtCAD1, which was previously reported to have no detectable CAD activity (Kim et al. 2004).
Phylogenetic tree of CAD proteins. A phylogenetic tree of the deduced amino acid sequences of CAD genes was created by the neighbor-joining method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. Bootstrap values were obtained by 1,000 bootstrap replicates and numbers above branches refer to bootstrap values. Values below 50 were not reported. At, Arabidopsis thaliana; Os, Oryza sativa; Eg, Eucalyptus gunni. Bm1 and Bmr6 are bona fide CADs from Zea mays (maize) and Sorghum bicolor (sorghum), respectively
Transcript abundance of the OsCAD gene in the rice internode
To investigate the expression of each OsCAD gene in rice, we examined the transcription level of OsCAD RNA in the culm. To resist the stresses imposed by water transport and to support the plant structure, the rice culm forms well-developed secondary walls in the xylem and sclerenchyma (Hirano et al. 2010), and this organ was therefore selected for analysis. Total RNA was extracted from the uppermost internode at the heading stage, a stage in which culms begin to accumulate lignin. OsCAD4, -5, and -9 were not amplified from the total RNA, and therefore expression of these genes could not be examined. Among the nine OsCADs, OsCAD2 was the most abundantly expressed (Fig. 2). OsCAD1 was also expressed in the culm; however, its copy number was more than seven times lower than that of OsCAD2. The other OsCAD genes were either not expressed or expressed at very low levels. This suggests that OsCAD2 plays a dominant role in lignin biosynthesis in rice culm.
Promoter-GUS analysis of OsCAD2 and OsCOMT
To confirm that OsCAD2 is the OsCAD gene that makes the greatest contribution to lignin biosynthesis in rice culm, we analyzed the spatial and temporal expression of this gene during lignin accumulation. For this purpose, we constructed a reporter gene (pCAD::GUS), which contained a GUS reporter fused to an OsCAD2 genome sequence spanning from 2,100 bp upstream of the start codon to 24 bp downstream of the start of the second exon, and analyzed its expression in transgenic rice plants.
We also analyzed the expression of the rice caffeic acid 3-O-methyltransferase (OsCOMT) gene to determine if the expression of OsCAD2 is synchronized with that of OsCOMT. COMT catalyzes the synthesis of sinapyl alcohol. Rice contains seven genes related to COMT (OsCOMT and OsCOMTL1 to -L6; Xu et al. 2009). Although the role of these rice genes in lignin biosynthesis has not been directly studied, phylogenetic analysis of the deduced amino acid sequence of various plant COMT-like genes has revealed that OsCOMT, but not the COMT-like genes, is categorized into the clade containing the bona fide COMT of Arabidopsis, maize (Bm3), and Sorghum bicolor (Fig. 3; Vignols et al. 1995; Goujon et al. 2003b; Bout and Vermerris 2003). Thus, we expected that the expression of OsCOMT would reflect the site of lignin biosynthesis.
Phylogenetic tree of COMT proteins. A phylogenetic tree of the deduced amino acid sequences of COMT genes was created by the neighbor-joining method. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. Bootstrap values were obtained by 1,000 bootstrap replicates and the numbers above the branches refer to bootstrap values. Values below 50 were not reported. At, Arabidopsis thaliana; Os, Oryza sativa; Lp, Lolium perenne; Sb, Sorghum bicolor; Zm, Zea mays
For the pCOMT::GUS assay, a fragment 2535 bp upstream of the OsCOMT start codon to just before the stop codon was used to create a transcriptional fusion to the GUS reporter gene. Figure 4 shows the expression of OsCAD2 and OsCOMT in the uppermost internode just before heading. Rapid elongation of the uppermost internode occurs at around 10–15 days before the heading stage. In developing rice internodes, we can divide each internode into three parts from the base to the upper portion, that is, the basal cell divisional zone, the internal cell elongation zone, and the secondary cell wall developing zone. Phloroglucinol staining, which labels lignin, reveals a gradient of development from the lower to the upper part of the internode (Fig. 4, second and fifth columns).
Histochemical staining of rice culm for OsCAD2 and OsCOMT promoter activity and lignin staining. The uppermost internode of OsCAD2 promoter::GUS or OsCOMT promoter::GUS transgenic plants was sectioned transversely to 80–100 μm thickness and stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, or with phloroglucinol. The approximate positions of the sections are shown as red lines beside the internode. The sectioned areas presented in the top, middle, and bottom panels correspond to the lines at the top, middle, and bottom of the central photograph, respectively. Note that both OsCAD2 and OsCOMT promoter-driven GUS expression is stronger in internodes that have not yet accumulated lignin, whereas GUS expression is reduced in the internodes that have accumulated lignin. Bar 100 μm
CAD and COMT expression was well synchronized in the internode regions of all transgenic lines analyzed (more than 5 lines for each constructs), suggesting that these enzymes function cooperatively in the internode (Fig. 4). At the young stage (bottom row), faint GUS expression caused by both promoters was observed in almost all tissues, including the parenchymal cells. When lignin started to accumulate (middle row), GUS expression became stronger and was restricted to tissues that accumulate lignin, such as sclerenchyma and the vascular bundle. At the developed stage (top row), GUS expression was no longer observed, even in cells stained with phloroglucinol. Thus, during these developmental processes, GUS expression always preceded lignin accumulation, suggesting that CAD and COMT may be involved in lignin synthesis.
We also observed GUS expression driven by these promoters in the sheath and roots (Figs. 5, 6, and Supplemental Fig. 2). The pattern of OsCAD2 and OsCOMT promoter-driven GUS expression was always synchronized, and was observed strongly in the vascular bundle and in the sclerenchyma tissues of both organs. Expression of these genes was again always observed before lignin accumulation, whereas no or faint GUS expression was observed in mature tissues that had already accumulated lignin. Such a tight correlation between the site of OsCAD2 expression and lignin accumulation, with some temporal differences, provides further evidence that this gene plays a dominant role in lignin biosynthesis in the culm, and might also be dominantly involved in lignin biosynthesis in the leaf and root.
Histochemical staining of rice sheath for OsCAD2 and OsCOMT promoter activity and lignin staining. The sheaths of OsCAD2 promoter::GUS or OsCOMT promoter::GUS transgenic plants was sectioned transversely to 80 μm thickness and stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid or phloroglucinol. Note that both OsCAD2 and OsCOMT promoter-driven GUS expression was strongest at the inner leaf sheath, which has not yet accumulated lignin, and reduced in the outer layer of leaf sheath, which has accumulated lignin. Bars 100 μm
Histochemical staining of rice roots for OsCAD2 and OsCOMT promoter activity and lignin staining. The roots of OsCAD2 promoter::GUS or OsCOMT promoter::GUS transgenic plants were sectioned transversely to 80 μm thickness and stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid or phloroglucinol. The approximate positions of the sections are indicated as red lines beside the roots. The sectioned areas presented in the top, middle, and bottom panels correspond to the lines at the top, middle, and bottom of the central photograph, respectively. Note that both OsCAD2 and OsCOMT promoter-driven GUS expression is stronger in the roots that have not yet accumulated lignin, whereas GUS expression is reduced in the roots that have accumulated lignin. Bars 100 μm
Co-expression network analysis of OsCAD genes
To further demonstrate that OsCAD2 is the primary OsCAD gene involved in lignin biosynthesis, co-expression network analysis of eight OsCAD genes was performed. Co-expression analysis assumes that proteins that operate in the same biochemical pathway or those that function in concert are coordinately regulated at the transcriptional level. In this study, we constructed a co-expression network of each OsCAD gene using publicly available Affymetrix microarray data obtained from various organs grown under different physiological conditions. For each analysis, mutual rank less than 30 was adopted to create the network (Obayashi and Kinoshita 2011). To evaluate whether OsCAD genes are co-expressed with other lignin biosynthesis genes, we counted the number of genes in the network that are considered to encode lignin biosynthesis genes and secondary cell wall-related cellulose synthase genes (OsCesA4, 7, and 9, Supplemental Table 1).
In the OsCAD2 network, 17 lignin biosynthesis or CesA genes appeared, which was the highest number observed among all of the OsCAD gene networks created (Table 2). The OsCAD8A and 8C network showed the second highest appearance of 2 lignin-associated genes. Unexpectedly, OsCAD7 network did not show any appearance of lignin-associated genes, despite that OsCAD7 has been reported to participate in monolignol biosynthesis. These results suggest that compared to other OsCAD genes, the transcription of OsCAD2 is more tightly associated with the transcription of lignin biosynthesis-related genes. OsCOMT also showed a high level of co-expression with lignin-associated genes (18 genes), as was expected from the promoter-GUS expression analysis. Among the lignin-related genes that are co-expressed with OsCAD2, various genes encoding lignin biosynthesis enzymes and cellulose synthase appeared, including genes related to PAL, C4H, C3H, 4CL, CCoAOMT, CCR, COMT, and CesA (Fig. 7).
Discussion
Previous phylogenetic analysis showed that OsCAD2 is the only rice gene that belongs to the bona fide CAD clade (Hamberger et al. 2007). However, other OsCADs were also suggested to contribute to rice monolignol biosynthesis, because the oscad2 null mutant showed only a slight reduction in the Klason lignin content (5.3–6.1%; Zhang et al. 2006). In contrast, atcad-c/-d, a double mutant of the Arabidopsis bona fide CADs, shows a 40% reduction in Klason lignin content in the flowering stem. This indicates that the contribution of bona fide CADs to monolignol biosynthesis might differ between rice and Arabidopsis. However, CAD/SAD (sinapyl alcohol dehydrogenase) activity of the crude extract from the gh2 mutant is significantly lower than that of the wild type (Zhang et al. 2006), which suggests that CAD/SAD activity in rice depends mostly on the activity of OsCAD2.
To clarify these seemingly contradictory results, we re-investigated the contribution of OsCAD2 to monolignol biosynthesis. Our results suggest that OsCAD2 is the predominant CAD in monolignol biosynthesis, at least in the culm. First, expression of OsCAD2 was the highest among the OsCADs in the rice internode, an organ that accumulates lignin. Although the fc1 mutant, which has a mutation of the OsCAD7 gene, shows a 13.4% reduction in Klason lignin content, expression of OsCAD7 in the culm was more than three orders of magnitude lower than that of OsCAD2. Second, promoter-GUS analysis showed that the expression pattern of OsCAD2 always synchronized with that of the bona fide COMT gene, OsCOMT. The fact that OsCAD2 expression was detected in lignin-forming tissues and always preceded lignin accumulation indicates the tight correlation between OsCAD2 and lignin biosynthesis. Third, co-expression network analysis showed that among the OsCAD genes, OsCAD2 was co-expressed with the presumed lignin biosynthesis genes with the highest frequency.
We consider two possible explanations for why the Klason lignin content of gh2 is reduced to only 5.3–6.1% of the levels found in the original cultivar. One explanation is that other OsCAD genes are mainly responsible for monolignol biosynthesis and that OsCAD2 does not play a major role in this process; however, our results and some of those presented by Zhang et al. (2006) do not support this hypothesis. The second possibility is that compounds other than hydroxycinnamyl alcohol monomers are incorporated into lignin polymers at a high degree in gh2. Indeed, although a precise composition analysis of the lignin polymer in gh2 has not been conducted, lignin in atcad-c/-d shows increased amounts of indene derivatives (which originate from β-O-4 linkage sinapaldehyde and coniferaldehyde units), dithioketal derivatives of coniferaldehyde (vanillin and syringaldehyde), and ferulic and sinapic acids (Sibout et al. 2005). An increase in indene derivatives has also been reported for the sorghum bmr6 mutant (Sattler et al. 2009). Detailed composition analysis of gh2 mutant will be important.
However, even if the incorporation of other compounds in the lignin polymer might explain the slight reduction in Klason lignin content in gh2, considerable amounts of H, G, and S monolignols are still released by thioacidolysis in the mutant (Zhang et al. 2006), implying that, other OsCADs also contribute to monolignol biosynthesis to some extent. Despite that the level of OsCAD7 transcript was low compared to that of OsCAD2 in the internode (Fig. 2), the level of monolignols released by thioacidolysis in the culm of the oscad7 null mutant was 16–36% lower than that of the wild type (Li et al. 2009), indicating that OsCAD7 does indeed affect monolignol biosynthesis. However, although expression of OsCAD7 has been reported to be highest in the uppermost internode compared to second and the third internodes, our analysis did not indicate a major role for OsCAD7 in monolignol biosynthesis at least in the uppermost internode, as stated above. It is possible that OsCAD7 possesses a higher level of enzymatic activity or a longer duration of expression than OsCAD2 in the developing internode. This possibility needs further investigation such as analysis of gh2/fc1 double mutant. Although several monocot CADs belonging to the bona fide CAD clade have been implicated in monolignol biosynthesis (Bm1 of maize, Bmr6 of Sorghum, TaCAD1 of wheat, and GH2 of rice), OsCAD7 is the only non bona fide CAD reported to have a role in monolignol biosynthesis (Halpin et al. 1998; Sattler et al. 2009; Ma 2010; Zhang et al. 2006). Thus at the moment, we do not know whether function of non bona fide CADs in monolignol biosynthesis is a general phenomenon, or OsCAD7 is unique among non bona fide CADs.
Another possibility that considerable amounts of H, G, and S monolignols are still released by thioacidolysis in the gh2 mutant is that mutation in the OsCAD2 gene leads to increased expression of other OsCAD genes, which could partially compensate monolignol biosynthesis in the gh2 mutant. Such phenomenon has been observed for Arabidopsis Cinnnamoyl-CoA reductase 1 (CCR1) mutants, in which expression of other CCR gene, CCR2, increases showing that mutation in the CCR1 induces expression of CCR2 (Mir Derikvand et al. 2008).
Although all of the bona fide CAD-deficient mutants investigated show reduced lignin content (Halpin et al. 1998; Sibout et al. 2005; Zhang et al. 2006; Sattler et al. 2009), the G:S ratio was altered in only some mutants. That is, the G:S ratio of gh2 and the maize bm1 mutant does not differ significantly from that of the wild type, suggesting that these monomers are evenly reduced in these mutants (Halpin et al. 1998; Zhang et al. 2006). In contrast, the atcad-c/-d double mutant and maize bmr6 mutant show considerable change in the G:S ratio compared to the wild type (WT: 2.6–3.2, atcad-c/-d: 11.8-13.8, and WT:1.3, bmr6 3.2) (Sibout et al. 2005; Sattler et al. 2009). There are reports that modulation of the G:S ratio could lead to advantageous alterations in cell wall properties, such as improved saccharification efficiency (Bose et al. 2009; Kishimoto et al. 2010). Therefore, further studies on enzyme activity, substrate preference, and expression pattern of each CAD isozyme, and the relationship among these enzymes, is required for an understanding of the fine-tuning of lignin content and composition.
Materials and methods
Protein sequence alignment and phylogenetic analysis
The deduced amino acid sequences of CAD and COMT were aligned with ClustalW version 1.83 with default parameters (Thompson et al. 1994; http://www.genome.jp/tools/clustalw/). Box shade (http://www.ch.embnet.org/software/BOX_form.html) was used to draw the alignments, using default parameters.
A neighbor-joining (NJ) phylogenetic tree (Saitou and Nei 1987) was obtained with Mega 4 (http://www.megasoftware.net/mega4/mega.html) using Poisson correction for amino acid distance estimates. Bootstrap analyses were performed by repeating the procedure on 1,000 datasets.
Plasmid construction
The sequences of primers used in this study are listed in Supplemental Table 2. All PCR fragments were sequenced to confirm that no mutations were introduced.
For the OsCAD2 promoter::GUS construct, the promoter region of the second exon (2100 bp upstream of the OsCAD2 start codon to 24 bp into the second exon) was amplified from cv. Kinmaze genomic DNA and cloned into the HindIII–SmaI site of the pBI-Hm2 vector (kindly provided my Kenzo Nakamura, Nagoya University) to produce pCAD2::GUS. For the OsCOMT promoter::GUS construct, the promoter region and the CDS of OsCOMT (2,535 bp upstream of the OsCAD2 start codon to just before the stop codon) was amplified from cv. Kinmaze genomic DNA and cloned into the SmaI site of the pBI-Hm2 vector in the correct orientation to produce pCOMT::GUS. For use as qRT-PCR standards, OsCAD genes were PCR amplified using full-length cDNA clones provided by the Rice Genome Resource Center (http://www.rgrc.dna.affrc.go.jp/index.html.en) or using cDNA produced from total RNA from the uppermost internode of cv. Nipponbare, and cloned into the pCR4 Blunt-TOPO vector (Invitrogen).
Plant transformation
Constructs were introduced into Agrobacterium tumefaciens strain EHA105 and used to infect rice callus of cv. Kinmaze according to Hiei et al. (1994). Transformed cells and plants were selected by hygromycin resistance, and regenerants were grown to maturity in pots in a greenhouse.
Histochemical staining
For histochemical staining, transformed plants were cut into pieces, fixed in 5% (w/v) agar dissolved in water, and sectioned with a microtome (Microslicer DTK-1000) at a thickness of 80–100 μm.
For phloroglucinol staining, sections were incubated for 5 min in phloroglucinol solution [2% in ethanol:water (95:5)], the phloroglucinol was removed, and the sections were treated with 18% HCl for 5 min before being observed with a microscope (Olympus BX51). For Mäule staining, sections were treated with 1% KMnO4 for 5 min, rinsed with water, treated with 25% HCl for 20 s, rinsed with water again, mounted in 28% NH3·H2O, and examined immediately under a microscope (Olympus BX51).
For GUS staining, sections were stained for GUS according to the method reported by Murakami and Ohashi (1992), destained with an ethanol series, and observed under a microscope (Olympus BX51).
RNA isolation and quantitative RT-PCR analysis
Total RNA was prepared from the uppermost internode of O. sativa cv. Nipponbare at the heading stage and isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The first strand of cDNA was synthesized from total RNA using an Omniscript Reverse Transcription Kit (Qiagen). Quantitative Real-time RT-PCR (qRT-PCR) was performed with the LightCycler System (Roche) and the SYBR Green PCR Kit (Qiagen). The results were confirmed using three independent biological replicates. The ubiquitin gene from rice was used as an internal standard for normalizing cDNA concentration variations. OsCAD genes cloned into pCR4 Blunt-TOPO products were used as standards for determining the transcript copy numbers of each gene.
Co-expression network analysis
Network analysis was performed using OsCAD genes as guide genes. Affymetrix Rice Genome Array datasets (GSE6893, GSE7951, GSE19024) were extracted from the GEO (Gene Expression Omnibus) database, which is maintained by NCBI (http://www.ncbi.nlm.nih.gov/geo/). For microarray data normalization, we used the RMA+ method (Harbron et al. 2007), using the Bioconductor RefPlus package (http://www.bioconductor.org/index.html). Mutual rank less than 30 was calculated based on Pearson Correlation Coefficient as co-expression measures and the networks were created. For the visualization of the OsCAD2 network, Cytoscape software (http://www.cytoscape.org/) was used. To assess whether OsCAD genes are co-expressed with lignin-related genes, P values were calculated based on hypergeometric distribution. The accession numbers of lignin-related genes are listed in Supplemental Table 1.
Accession numbers
RAP-DB accession numbers, GenBank/EMBL accession numbers and Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: OsCAD1 (AAN09864), OsCAD2 (DQ234272), OsCAD3 (AAP53892), OsCAD4 (BK003970), OsCAD5 (BK003971), OsCAD6 (CAD39907), OsCAD7 (CAE05207), OsCAD8A (NP_001063109), OsCAD8B (Q6ERW9), OsCAD8C (Q6ERW7.2), OsCAD8D (NP_001063114.1), OsCAD9 (AAN05338), OsCOMT (Os08g0157500), OsCOMTL1 (Os12g09770), OsCOMTL2 (Os04g01470), OsCOMTL3 (Os12g13800), OsCOMTL4 (Os02g57760), OsCOMTL5 (Os04g09604), OsCOMTL6 (Os04g09654), sorghum CAD (Bmr6; BAF42789), SoCAD (CAA13177), ZmCAD2 (bm1; CAA06687), AsCAD (AF217957), EgCAD2 (CAA46585), AsSAD (AF273256), LpCOMT (AAC18623.1), LpOMT1 (AAD10253), LpOMT3 (AAD10255), SbCOMT (AAO43609), ZmOMT (AAB03364.1), AtCAD1 (AT1G72680), AtCAD2 (AT2G21730), AtCAD3 (AT2G21890), AtCAD-C (AT3G19450), AtCAD-D (AT4G34230), AtCAD6 (AT4G37970), AtCAD7 (AT4G37980), AtCAD8 (AT4G37990), AtCAD9 (AT4G39330), AtCOMT (AT5G54160), AtCOMT-like1 (AT1G21100), AtCOMT-like2 (AT1G21110), AtCOMT-like3 (AT1G21120), AtCOMT-like4 (AT1G21130), AtCOMT-like5 (AT1G33030), AtCOMT-like6 (AT1G51990), AtCOMT-like7 (AT1G63140), AtCOMT-like8 (AT1G76790), AtCOMT-like9 (AT1G77520), AtCOMT-like10 (AT1G77530), AtCOMT-like11 (AT3G53140), AtCOMT-like12 (AT5G37170), AtCOMT-like13 (AT5G53810).
References
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Bose SK, Francis RC, Govender M, Bush T, Spark A (2009) Lignin content versus syringyl to guaiacyl ratio amongst poplars. Bioresour Technol 100:1628–1633
Bout S, Vermerris W (2003) A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol Genet Genomics 269:205–214
Costa MA, Collins RE, Anterola AM, Cochrane FC, Davin LB, Lewis NG (2003) An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry 64:1097–1112
Eudes A, Pollet B, Sibout R, Do CT, Séguin A, Lapierre C, Jouanin L (2006) Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225:23–39
Goujon T, Sibout R, Eudes A, MacKay J, Jouanin L (2003a) Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol Biochem 41:677–687
Goujon T, Sibout R, Pollet B, Maba B, Nussaume L, Bechtold N, Lu F, Ralph J, Mila I, Barrière Y, Lapierre C, Jouanin L (2003b) A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapyl esters. Plant Mol Biol 51:973–989
Guo DM, Ran JH, Wang XQ (2010) Evolution of the cinnamyl/sinapyl alcohol dehydrogenase (CAD/SAD) gene family: the emergence of real lignin is associated with the origin of Bona Fide CAD. J Mol Evol 71:202–218
Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA (1998) Brown-midrib maize (bm1)-a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14:545–553
Hamberger B, Ellis M, Friedmann M, de Azevedo Souza C, Barbazuk B, Douglas CJ (2007) Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can J Bot 85:1182–1201
Harbron C, Chang KM, South MC (2007) RefPlus: an R package extending the RMA Algorithm. Bioinformatics 15:2493–2494
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hirano K, Kotake T, Kamihara K, Tsuna K, Aohara T, Kaneko Y, Takatsuji H, Tsumuraya Y, Kawasaki S (2010) Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis. Planta 232:95–108
Kim SJ, Kim MR, Bedgar DL, Moinuddin SG, Cardenas CL, Davin LB, Kang C, Lewis NG (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101:1455–1460
Kim SJ, Kim KW, Cho MH, Franceschi VR, Davin LB, Lewis NG (2007) Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations? Phytochemistry 68:1957–1974
Kishimoto T, Chiba W, Saito K, Fukushima K, Uraki Y, Ubukata M (2010) Influence of syringyl to guaiacyl ratio on the structure of natural and synthetic lignins. J Agric Food Chem 58:895–901
Li X, Weng JK, Chapple C (2008) Improvement of biomass through lignin modification. Plant J 54:569–581
Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q, Wu C (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69:685–697
Ma QH (2010) Functional analysis of a cinnnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J Exp Bot 10:2735–2744
Mir Derikvand M, Sierra JB, Ruel K, Pollet B, Do CT, Thévenin J, Buffard D, Jouanin L, Lapierre C (2008) Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227:943–956
Murakami T, Ohashi Y (1992) Methods for histochemical detection of GUS reporter gene expression in transgenic plants. Shokubutu-Saibou-Kougaku 4:281–286
Obayashi T, Kinoshita K (2011) COXPRESdb: a database to compare gene coexpression in seven model animals. Nucleic Acids Res 39:D1016–D1022
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051–1071
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol Biol Evol 4:406–425
Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF (2009) A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the Sorghum brown midrib6 phenotype. Plant Physiol 150:584–595
Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17:2059–2076
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tobias CM, Chow EK (2005) Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 220:678–688
Vignols F, Rigau J, Torres MA, Capellades M, Puigdomenech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7:407–416
Vogel J (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11:301–307
Weng JK, Li X, Bonawitz ND, Chapple C (2008) Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr Opin Biotechnol 19:166–172
Xu Z, Zhang D, Hu J, Zhou X, Ye X, Reichel KL, Stewart NR, Syrenne RD, Yang X, Gao P, Shi W, Doeppke C, Sykes RW, Burris JN, Bozell JJ, Cheng MZ, Hayes DG, Labbe N, Davis M, Stewart CN Jr, Yuan JS (2009) Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinform 10(Suppl 11):S3
Youn B, Camacho R, Moinuddin SG, Lee C, Davin LB, Lewis NG, Kang C (2006) Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org Biomol Chem 4:1687–1697
Yuan JS, Tiller KH, Al-Ahmad H, Stewart NR, Stewart CN Jr (2008) Plants to power: bioenergy to fuel the future. Trends Plant Sci 13:421–429
Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140:972–983
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97–166
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Jouanin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hirano, K., Aya, K., Kondo, M. et al. OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep 31, 91–101 (2012). https://doi.org/10.1007/s00299-011-1142-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1142-7