Abstract
Lectin receptor-like kinases (LecRLKs) are members of RLK family composed of lectin-like extracellular recognition domain, transmembrane domain and cytoplasmic kinase domain. LecRLKs are plasma membrane proteins believed to be involved in signal transduction. However, most of the members of the protein family even in plants have not been functionally well characterized. Herein, we show that Pisum sativum LecRLK (PsLecRLK) localized in plasma membrane systems and/or other regions of the cell and its transcript upregulated under salinity stress. Overexpression of PsLecRLK in transgenic tobacco plants confers salinity stress tolerance by alleviating both the ionic as well the osmotic component of salinity stress. The transgenic plants show better tissue compartmentalization of Na+ and higher ROS scavenging activity which probably results in lower membrane damage, improved growth and yield maintenance even under salinity stress. Also, expression of several genes involved in cellular homeostasis is perturbed by PsLecRLK overexpression. Alleviation of osmotic and ionic components of salinity stress along with reduced oxidative damage and upregulation of stress-responsive genes in transgenic plants under salinity stress conditions could be possible mechanism facilitating enhanced stress tolerance. This study presents PsLecRLK as a promising candidate for crop improvement and also opens up new avenue to investigate its signalling pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stresses arise when plants are subjected to sub-ideal conditions of growth, either due to excess or deficit in physical or chemical environment. The most widespread abiotic stresses which decrease the yield of major crop plants by more than 50 % are extreme temperature, salinity and drought (Mahajan and Tuteja 2005). Conservation of agricultural land affected by environmental stresses causes an enormous financial burden on governments worldwide. Among abiotic stresses, salinity stress affects 20 % of the irrigated land encompassing 45 Mha area and the affected land area continues to grow at a rate of 20,000 km2/year (Munns and Tester 2008). Salinity stress has an osmotic component and an ionic component. The osmotic or the physical component of salinity decreases the soil porosity, leading to decreased oxygen and water movement to the root (Munns and Tester 2008). The ionic effect comes to picture when the concentration of salt inside the plant/cell starts approaching the toxic levels. High Na+ content in plant also hampers the formation of new leaves, while in older leaves the process of senescence is initiated, thus decreasing the rate of photosynthesis as well as nutrient availability (Munns and Tester 2008), which in turn affects the biomass and the yield.
Plants lack somatic gene flexibility due to absence of combinatorial diversification; therefore they have evolved mechanisms to respond to general as well as evolving stresses like biotic and abiotic stresses (Afzal et al. 2008). In wake of wide range of environmental insults, plants have developed adaptations at the physical, cellular and molecular level (Zhu 2001). Induction of stress is first perceived by sensors [such as, G-protein couple receptors-like protein(s), receptor-like kinases (RLKs) or histidine kinases] which then activate secondary signaling molecules. Followed by this, changes in the concentration of cytosolic [Ca2+] and reactive oxygen species (ROS) lead to phosphoprotein cascade and the activation of stress responsive genes (such as the genes involved in biosynthesis of osmolytes, antioxidant enzymes and transporters, etc.) via activation of transcription factors (Xiong et al. 2002). Membranes act as the first site of signal/stress perception via receptor proteins embedded in it (Tuteja and Sopory 2008). Plant RLKs are a typical example of membrane embedded, stress perceiving proteins in plants, where the extracellular domain or the recognition domains have diversified to recognize the ever increasing environmental challenges. These proteins have been implicated in a wide variety of roles ranging from development (Becraft 1998), signaling (Morris and Walker 2003; Tichtinsky et al. 2003), biotic and abiotic stress tolerance (Chinnusamy et al. 2004; Lehti-Shiu et al. 2009).
One of the RLK families, whose role in environmental stress conditions has been a topic of research in recent years, is lectin receptor-like kinase (LecRLK) family. LecRLK is an extensively duplicated family with 173 and 75 members in rice and Arabidopsis, respectively (Vaid et al. 2012). LecRLKs are plasma membrane proteins with an extracellular region resembling lectin proteins, followed a single-pass transmembrane region and a kinase domain (Vaid et al. 2013). The kinase domain, mediating the downstream signaling, is highly conserved among the whole RLK family while the extracellular lectin domain which is the signal perception/recognition domain, is variable even within the LecRLK family (Vaid et al. 2012). The lectin region also differs from the lectin proteins in the fact that the lectin domain of LecRLKs has lost its mono/oligo-saccharide binding capacity, but probably retains its capacity to bind to hydrophobic compounds (such as hormones) and complex saccharides (Barre et al. 2002; Hervé et al. 1999). On the basis of domain organization and identity, the LecRLK family has been subdivided in L-type, G-type and C-type LecRLKs (Bouwmeester and Govers 2009). Though, many studies have been carried out on evolution and characterization of this gene family (André et al. 2005; Barre et al. 2002; Bouwmeester and Govers 2009; Gouget et al. 2006; Hervé et al. 1999) and recently on biotic stress tolerance (Bonaventure 2011; Chen et al. 2006; Desclos-Theveniau et al. 2012; Gilardoni et al. 2011; Kanzaki et al. 2008; Singh et al. 2012), very few reports are available on the putative role of LecRLKs in abiotic stress tolerance, and specifically on salinity stress tolerance (Deng et al. 2009; He et al. 2004; Huang et al. 2013; Joshi et al. 2010; Sun et al. 2013). Also, most of these reports lack mechanistic insight to explain the salinity tolerance phenotype.
In the present study, we have studied an L-type LecRLK from Pisum sativum (garden pea) (PsLecRLK), which, in an earlier study, was identified during a functional screen for salinity stress tolerance in E. coli (Joshi et al. 2010). This study demonstrated that PsLecRLK is a catalytically active kinase which could phosphorylate general substrate proteins, such as myelin basic protein and casein, and upon expression in E. coli, could provide it salinity stress tolerance by avoiding intracellular accumulation of Na+. As a promising candidate for crop improvement, we have studied PsLecRLK mediated salinity stress tolerance by overexpression study in model plant tobacco, and analyzed the mechanism at gross molecular level. Our study indicates that PsLecRLK overexpression in tobacco renders it salinity stress tolerant. Also, PsLecRLK overexpression mitigates salinity stress induced osmotic stress as well as ionic toxicity which translates downstream as lowered ROS content and reduced membrane damage leading to tolerance phenotype. Also, PsLecRLK overexpression modulates the expression of various transporters and several other genes that are essential for maintaining cellular homeostasis under abiotic stress conditions. Most importantly, this tolerant phenotype is exhibited without significant yield penalty, thus warranting further investigation of PsLecRLK mediated salinity stress tolerance and its employment for crop improvement.
Materials and methods
Plant materials, growth conditions and stress treatments
Pea (Pisum sativum) seeds were grown as described earlier (Misra et al. 2007). Two week old plants were subjected to various stresses/treatments (as listed in Supplementary Table 3) and harvested at different time points for RNA isolation. Tobacco seeds (wild type (WT) (Nicotiana tabacum USA) or transgenic) were surface sterilized by washing with 4 % sodium hypochlorite solution for 30 s followed by a 30 s wash with 70 % ethanol. The seeds were then rinsed with sterile water 5–6 times to remove excess bleach. The surface sterilized seeds cultured on MS medium (Duchefa) without phytohormones in Petri dishes were incubated at 26 °C under 16/8 h photoperiod.
RNA isolation and real-time PCR
Total RNA was isolated from 0.1 g plant tissue. cDNA was prepared with oligo dT primers using AccuScript High Fidelity 1st Strand cDNA Synthesis Kit (Agilent technologies, USA) as per manufacturer’s instructions.
Real-time PCR was carried out in Applied Biosystems 7500 Real-time PCR System using Agilent Brilliant II SYBR Green qRT-PCR master mix kit as per the manufacturer’s instructions. Pea tubulin gene was taken as endogenous control. The sequences of primers used throughout in this study have been listed in Supplementary Table 4.
Subcellular localization of PsLecRLK
The coding sequence of PsLecRLK gene was fused with GFP and cloned into the BamH1/XbaI restriction sites of the pMBPII-GFP expression vector under the control of the CaMV 35S promoter. Localization of PsLecRLK was performed on the onion epidermal cells as described earlier (Tuteja et al. 2014).
Generating tobacco transgenics for PsLecRLK
PsLecRLK gene (Accession number: EU041719) was cloned in binary vector pCAMBIA1301. For functional validation of PsLecRLK gene under salinity stress conditions, pCAMBIA1301 PsLecRLK construct and empty vector pCAMBIA1301 (as an ideal control) was transformed in Nicotiana tabacum (cv. USA Petite Havana) using the agrobacterium co-cultivation method. Regenerating calli were subjected to two rounds of antibiotic selection, after which the regenerated plantlets were transferred to vermiculite pots in the green house and maintained under high humidity conditions for 4–5 days and then transferred to soil pots, maintained under green-house condition till maturity. As a control, wild type tobacco plants were also generated using the same procedure. The transgenic lines were selected by growth on antibiotic selection, reporter gene (GUS) analysis, transcript analysis as well as western blot analysis. T1 and T2 generation seeds from single copy transgenic lines, as tested by southern blot analysis, were harvested, dried and used for further analysis.
Salinity stress tolerance analysis of transgenic tobacco plants
Leaf disk senescence assay was performed as described earlier (Misra et al. 2007). Chlorophyll, measured (from disks derived from same leaves) prior to stress treatments (200 mM NaCl or 200 mM Mannitol), was taken as the initial chlorophyll content. Initial and final readings of extract were taken at 470, 645 and 662 nm and chlorophyll content quantified as described earlier (Dere et al. 1998). The result has been depicted as percentage reduction in the pigment contents.
Plate assay: To assess the effect of salinity stress on germination potential, surface sterilized T2 transgenic and control seeds were placed on MS plate supplemented with 30 g/L sucrose and 200 mM NaCl or 200 mM Mannitol (for osmotic stress tolerance analysis). The plates were then incubated at 26 °C under 16/8 h photoperiod. The seeds were analyzed for a period of 21 days for germination and greening of cotyledons. At the end of this period root length, shoot length, bio-mass and chlorophyll content of transgenic and control seedlings (wild type and empty vector seedlings) was analyzed.
Salinity stress in vermiculite/soil: 4 week old plantlets grown under normal condition were transferred to vermiculite trays or potted soil. Plants were subsequently irrigated with 300 mM (high stress) or 100 mM NaCl (low stress) solution at a regular interval of 2 days for a total of 14 days or end of life cycle (for short term high stress or long term low stress conditions), respectively. The experiment was conducted in triplicates (with 10 plants in each set for high salinity stress in vermiculite tray). The plants were analyzed for their growth pattern and yield. The yield was quantified as total weight of seeds from the transgenic or the control plants under non-stressed or stressed conditions. The young leaves, mature leaves, shoot and roots (from plants subjected to long term low stress) were harvested for Na+ and K+ measurements.
Estimation of water uptake: Water uptake was measured in 4 week old hydroponically grown tobacco plants that were supplied with either water (for unstressed conditions) or 50 mM NaCl (for salinity stress conditions). The experimental set-up was modified to minimize surface evaporation of the solutions. The reduction in solution level was noted and the solutions were replenished every day. The experiment was carried out on three sets of five plants each.
Qualitative and quantitative Na+ content
Qualitative estimation of Na+ content in tobacco seedlings subjected to salinity stress was measured using CoroNa Green fluorescent dye (Invitrogen) following the protocol described earlier (Park et al. 2009). Briefly, 7 days old tobacco seedlings were transferred to hydroponic medium containing Hoagland solution. After 2 days of acclimatization, Hoagland solution was replaced with 100 mM NaCl solution. Following salinity stress for 48 h, the seedlings were rinsed in milli-Q grade water three times and transferred to 50 μM CoroNa green dye solution for 3 h or in 1 μg/ml propidium iodide (PI) solution (in PBS buffer) for 5 min at room temperature under dark conditions. Immediately after incubation, seedlings were again rinsed in milli-Q grade water three times. Root tip sections were observed under confocal microscope (Olympus FluoView). The excitation and emission wavelengths for CoroNa green stained seedlings and propidium iodide stained seedlings were 488, 520 and 545, 645 nm, respectively. During the image capture the settings of photomultiplier tube (PMT) were not changed.
For quantitative assessment of Na+ and K+ content, different tissues were processed as described earlier (Sanan-Mishra et al. 2005). The results were calculated as Na+/K+ ratios. To verify if PsLecRLK could modulate the expression of different transporters, transcript analysis of various transporters that are potentially involved in alleviating salinity stress, was carried out.
ROS content and membrane damage in salinity stressed transgenic plants
H2O2 measurement and staining of superoxide radicals: The level of H2O2 was assessed as described earlier (Velikova et al. 2000) and expressed as μM of H2O2 g−1 fresh weight of plant tissue. Quantitative analysis of catalase and ascorbate peroxidase (APX) activity was carried out as described previously by Tuteja et al. 2013. Qualitative measurement of H2O2 and superoxide accumulation was carried out in salinity stressed plants using 3,3′-diaminobenzidine (DAB) staining and nitrobluetetrazolium (NBT) staining as described earlier (Dong et al. 2009). To analyze if the superior ROS status in PsLecRLK overexpression plants was achieved due to overexpression of the ROS scavenging enzymes, real-time PCR analysis of the same was carried out.
To measure electrolyte leakage and lipid peroxidation (LP), 300 mg young and visibly healthy leaf tissue of same age was taken from different transgenic and control plants and immersed in 200 mM NaCl solution for 24 h. Electrolyte leakage and LP was assessed as per previously described protocol (Tuteja et al. 2013).
Microarray analysis and data analysis
For microarray analysis, RNA from 4 week old plants (PsLecRLK overexpression plants and empty vector transgenics as control plants, subjected to 200 mM salt stress for 48 h) were sent to Genotypic technology Pvt. Ltd (Banglore, India), where prior to cDNA synthesis and microarray analysis, quality analysis of RNA samples was carried out. The microarray analysis was carried out on Agilent microarray chip for Nicotiana tabaccum (4x44K format; Product number: G2514F). The data generated was normalized with GeneSpring GX11.5 software using percentile shift normalization. The fold change in the expression was calculated in terms of logbase2. Additionally, Chi squared test was performed to increase the statistical accuracy of the results. The genes with p < 0.05 were included for further analysis. The results of the microarray analysis were verified by performing Real-time PCR analysis on chosen candidate genes.
Results
In a previous study, PsLecRLK was identified as one of the genes whose overexpression could provide salinity stress tolerance to bacteria (Joshi et al. 2010). Aiming at crop improvement, the current study explores if PsLecRLK overexpression could render model plant tobacco, salinity stress tolerance.
PsLecRLK exhibits strongest response to NaCl stress
In order to evade stress conditions, plants manipulate expression of several stress responsive genes which aid in maintenance of structural and functional integrity of the cellular components. Using real-time PCR, the response of PsLecRLK under different stress conditions such as salt stress (NaCl, LiCl and KCl), temperature stress (heat and cold), wounding, ROS induction and treatment to hormones was examined at different time points (Fig. 1a–c, Supplementary Fig. S1). Although the expression analysis shows multi-stress response of PsLecRLK, strongest expression was exhibited in case of NaCl stress wherein 80 fold up-regulation was observed as compared to the control plants (Fig. 1a). Induction of PsLecRLK expression by osmotic stress has been reported previously (Joshi et al. 2010). Also, a strong but late response (12–24 h post treatment) was observed under temperature stresses, wounding and ABA treatment (Supplementary Fig. S1a–c, e). In synchrony with the transcript analysis under salinity stress, the promoter of PsLecRLK also exhibited strong induction when Arabidopsis plants harbouring the PsLecRLK::GUS fusion were subjected to salinity stress (Fig. 1d).
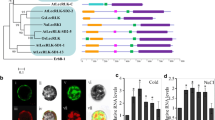
Response of PsLecRLK to different stresses. a–c PsLecRLK transcript analysis of pea plants subjected to NaCl (a), KCl (b) and LiCl (c) stresses. The fold change was normalized to the transcript level of the unstressed pea plants. Blue and red bars indicate relative transcript accumulation in shoots and roots, respectively. d GUS staining of Arabidopsis plant transformed with PsLecRLK::GUS fusion under salt stress. WT wild type Arabidopsis Col-0, EV Arabidopsis plant transformed with promoter-less binary vector pCAMBIA1391Z, Mock PsLecRLK::GUS fusion line treated with water, L1, L2 two independent transgenic lines of PsLecRLK::GUS fusion. All the plants except mock were treated with 100 mM NaCl for 6 h. e Subcellular localization of GFP-fused PsLecRLK protein in onion epidermal cells. The upper panel shows 35S:GFP signal spread throughout the cell. Lower panel shows the GFP signals from cell expressing PsLecRLK which is localized in the plasma membrane systems and/or other regions of the cell. Around 10 cells were examined for localization and a representative image is shown. Left panel shows the merged image; middle panel shows only GFP signal and the right panel is showing corresponding bright field image
Subcellular localization of PsLecRLK protein
Previous in silico studies have shown that PsLecRLK is membrane localized (Joshi et al. 2010). In the onion epidermal cells, the GFP fused constructs [CaMV35S-GFP (control) and PsLecRLK-GFP] were transiently expressed and examined after 48 h of transformation by confocal microscopy. In control the GFP expression was detected in the nucleus and cytosol while expression of the PsLecRLK-GFP fusion protein was exclusively observed in the plasma membrane systems and/or other regions of the cell (Fig. 1e).
PsLecRLK overexpressing plants are salinity stress tolerant
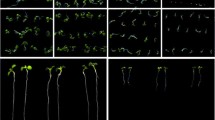
To examine the role of PsLecRLK under salinity stress conditions, overexpression lines of constitutive promoter (CaMV35S) driven PsLecRLK [vector construct (Supplementary Fig. S2a)] were generated in model plant tobacco. The T2 generation was tested for gene copy number [by southern blot analysis (Supplementary Fig. S2b)] and transgenic protein production [by western blot analysis (Supplementary Fig. S2c)]. T2 seeds of single copy PsLecRLK expressing lines (S6 and S8) were compared with the control plants [tobacco wild-type (WT) and the empty binary vector control (EV) harbouring tobacco transgenics] for their salinity stress tolerance. While under normal conditions PsLecRLK transgenics showed similar growth as the control plants, under salinity stress conditions, PsLecRLK transgenics germinated (marked by radical emergence) 3 days earlier than the control plants and exhibited 90 % germination as compared to 40–60 % germination exhibited by the control lines (Fig. 2a). Similar to the germination assay, the PsLecRLK transgenics exhibited greening of cotyledons 3 days prior to the control plants under salinity stress conditions (Fig. 2b). PsLecRLK overexpression plants also showed higher rate of germination and greening of cotyledon under osmotic stress (Supplementary Fig. S3a–b). PsLecRLK overexpressing plants exhibited higher biomass, growth and pigment content as compared to the control plants when grown under salinity stress conditions (Fig. 2c–f). The salinity stress tolerant phenotype was also clearly exhibited in the leaf-disk senescence assay, where leaf disks derived from PsLecRLK transgenics retained much higher pigment content than the leaf disks from the control plants (Fig. 2g). Similarly, PsLecRLK transgenic leaf discs retained more pigment as compared to control, after 48 h of osmotic, heat and oxidative stresses (Supplementary Fig. 3c–e, g). PsLecRLK overexpressing plants, however, exhibited similar cold stress tolerance as control plants, as assessed by leaf disc assay (Supplementary Fig. 3f). PsLecRLK overexpressing plants also exhibited higher tolerance than the control plants when subjected to extremely high salinity stress conditions (300 mM). Although when subjected to 300 mM salinity stress, growth in PsLecRLK transgenic lines was completely stalled, the control plants rapidly showed necrosis and death (Fig. 2h). In addition, lower dosage of salinity stress (100 mM) over prolonged period of time led to a relatively stunted growth, delayed flowering and reduced yield in control plants as compared to the PsLecRLK transgenic plants (Fig. 2i, j).
Various parameters to assess salt stress tolerance of PsLecRLK overexpressing tobacco plants. a–f Growth analysis of seeds and plantlets grown on MS media supplemented with 200 mM NaCl. a Germination rate analysis, b greening of cotyledons, c–f analysis of 14-day old seedlings grown on salt supplemented media, c phenotypic analysis, d pigment content, e average biomass, f shoot and root length. g Percentage retention of the photosynthetic pigment after 48 h of salt stress to leaf discs of different plants, h 300 mM NaCl treatment to 21 day old tobacco plantlets for 1 week followed by recovery period of 1 week, i, j 100 mM NaCl treatment to tobacco plants (from 3 weeks to senescence stage); i growth phenotype, j yield analysis. The yield was quantified as total weight of seeds from the transgenic or the control plants under non-stressed (U) or stressed conditions (S). Significant differences are marked with asterisks: *p < 0.05, **p < 0.01, ***p < 0.005. WT wild type tobacco plants, EV tobacco transgenics transformed with pCAMBIA1301 empty vector, S6, S8 two independent transgenic lines of tobacco plants overexpressing PsLecRLK
PsLecRLK overexpression prevents ROS accumulation and membrane damage
Stress, both biotic as well as abiotic, is accompanied by disturbances in delicate redox state leading to accumulation of ROS. On one hand this acts as activator of stress signaling pathways but over-accumulation of ROS, on the other hand, could cause damage to almost all cellular macromolecules. To draw an estimate of comparative ROS content in PsLecRLK overexpressing plants and control lines under salinity stress, peroxide and superoxide radicals were qualitative assayed by DAB and NBT staining, respectively. As compared to the PsLecRLK overexpressing lines, the control plants accumulated higher hydrogen peroxide and exhibited lower superoxide dismutase activity leading to higher superoxide content (Fig. 3a, b). Relatively lower H2O2 level is maintained in PsLecRLK overexpressing plants from 1 h post stress (hps) to 24 hps (Fig. 3c), probably owing to higher catalase and APX activity in PsLecRLK overexpressing plants (Fig. 3d, e). Relative transcript analysis (by real-time PCR), on the other hand, did not reveal much perturbation in the expression of ROS scavengers in the PsLecRLK overexpressing lines as compared to the control plants (Supplementary Fig. S4a).
Assessment of reactive oxygen species (ROS) and their effect on PsLecRLK overexpressing tobacco plants under salt stress. a–e ROS content in PsLecRLK overexpressing plants under salt stress. a DAB staining assay to assess hydrogen peroxide content, b NBT staining to assess superoxide dismutase activity, c Hydrogen peroxide content after 1 and 24 h of stress treatment, d catalase activity after 1 and 24 h of stress treatment, e ascorbate peroxidase (APX) activity after 1 and 24 h of stress treatment. One unit of enzyme activity is defined as 1 µmol of ascorbate oxidized per min. f, g effect of ROS on PsLecRLK overexpressing plants after 1 and 24 h of salt stress treatment. f Assessment of membrane damage by MDA assay, g electrolyte leakage measurement to assess membrane damage. Significant differences are marked with asterisks: *p < 0.05, **p < 0.01, ***p < 0.005. WT wild type tobacco plants, EV tobacco transgenics transformed with pCAMBIA1301 empty vector, S6, S8 two independent transgenic lines of tobacco plants overexpressing PsLecRLK
One of the direct consequence of excess ROS accumulation during several abiotic stresses, such as salinity stress, cold stress etc., is membrane damage. Estimation of malondialdehyde (MDA), the end product of lipid peroxidation, revealed its higher accumulation (2.5-fold higher at 24 hps) in control lines as compared to the PsLecRLK overexpression lines (Fig. 3f). Consequently, leakage of electrolytes from cells due to membrane damage was approximately twofold higher in control plants as compared to the PsLecRLK overexpression lines when subjected to salinity stress (Fig. 3g), thus establishing that under salinity stress, PsLecRLK overexpression could maintain lower ROS content thus preventing membrane damage in plants.
In an attempt to identify the hormonal pathway modulated by the PsLecRLK overexpression, transcript analysis of different hormone responsive genes was carried out. The analysis revealed up-regulation of ethylene (PR4, prb1b), salicylic acid (ICS1, PR2) and ABA (such as, PP2C, phi2, P5CS etc.) responsive genes (in descending order). The marker genes for jasmonic acid signaling pathway, which are otherwise known to be majorly involved in biotic stress tolerance, were relatively unperturbed (Supplementary Fig. S4b). This probably points towards cross-talk between different hormonal pathways in response to PsLecRLK overexpression. The details of the marker genes have been enlisted in supplementary Table 4.
PsLecRLK overexpression prevents accumulation of Na+ in plants
Salinity stress can manifest itself both as ionic as well as osmotic stress (Munns and Tester 2008). In response to osmotic stress, which is the first phase of salinity stress, plants limit water uptake from soil (Munns and Tester 2008). Indeed, growth retardation in salinity stressed plants is often attributed to limited potential of the roots to uptake water from the soil and transport it to the aerial parts of the plant (Navarro et al. 2008). To identify the component(s) (ionic or osmotic) towards which PsLecRLK extends its protection, we estimated water uptake by PsLecRLK overexpression and the control plants. The results indicated significant differences in water uptake under unstressed conditions as well under salinity stress conditions among the control and PsLecRLK overexpressing plants (Fig. 4a). This probably indicates that PsLecRLK overexpression somehow contributes towards maintenance of osmotic water permeability in roots. On the other hand, ionic stress, which is the late phase of salinity stress, leads to accumulation of toxic ions, mainly Na+ and Cl− and depletion of K+ from cells (Munns and Tester 2008). To assess if PsLecRLK contributes towards ionic stress tolerance, intracellular Na+ accumulation was visualized by CoroNa green dye (Invitrogen) in a qualitative test. The PsLecRLK overexpressing lines showed lower CoroNa dye fluorescence as compared to the control lines which corresponds to lower Na+ accumulation in PsLecRLK overexpression lines (Fig. 4b). Also, same seedlings when stained with propidium iodide (which permeates through the membrane of non-viable cells and fluoresces on intercalating in nucleic acids) exhibited higher fluorescence in the control lines as compared to the PsLecRLK overexpression lines. This indicates that PsLecRLK overexpression protects the plants from ionic component of the salinity stress and that lower cell death is observed in the plants overexpressing PsLecRLK, when they are subjected to salinity stress.
Deducing the mechanism of salt stress tolerance in PsLecRLK overexpressing tobacco plants. a Water uptake by plants under unstressed (U) and stressed (S) condition for four days. Significant difference (p < 0.05) are marked with asterisks. b Na+ content and cell death assessment in plants after 48 h of salt stress using CoroNa green and propidium iodide (PI), respectively, c assessment of Na+/K+ ratio in different tissues (OL old leaves, NL new leaves), d Real-time PCR analysis of different transporters implicated in salt stress tolerance in various reports. The fold change of transcript from PsLecRLK overexpressing line (S6) was normalized to the transcript level of the salinity stressed empty vector control plants. Significant differences are marked with asterisks: *p < 0.05, **p < 0.01
Maintenance of low Na+/K+ ratio is considered as an important characteristic of the salinity stress tolerant plant. While lower Na+/K+ ratio was observed in new leaves, roots and shoots of PsLecRLK overexpression lines, the older leaves have almost similar Na+/K+ ratio as the control plants (Fig. 4c). To check the possibility if the accumulation of Na+ ions was controlled by the altered Na+ transporter activity, real-time PCR analysis was carried out to assess transcript accumulation of various transporters (Details of the transporters whose transcript was analyzed have been enlisted in Supplementary Table 1). The results revealed that PsLecRLK overexpression induces the expression of SOS1, NHX1, NHX2, AVP1, PIP2 and HKT1 while SOS2, AQP1 and PIP1 showed only marginal change (Fig. 4d).
PsLecRLK overexpression upregulates ER-stress response pathway and several components of G-protein signalling pathway
In order to further gain insight on the effect of PsLecRLK overexpression on tobacco plant, microarray analysis was carried out. Of a total of 43,803 gene probes available on the microarray chip, 1567 genes exhibited differential up-regulation, which were further downsized to 300 genes using strict statistical tools. Similarly for the downregulated genes, 208 genes were shortlisted. Pie-chart of the upregulated and downregulated genes categorized according to the GO terms is given in Fig. 5a. A striking feature observed was the relative overexpression of the ER stress response pathway genes namely protein disulphide isomerase 2 (PDI2), Bax-1 inhibitor protein, several chaperons such as glycine rich protein, a homologue of RNA chaperon, cyclophilins, luminal binding protein (BiP) and heat shock protein (HSP20). Also, several genes of the G-protein signalling pathway namely, G protein-β subunit (Gβ), Rab proteins, PLA2 (a downstream target of Gβ) were also found to be differentially upregulated. Several of these genes have been documented to be players of stress response pathways. Details of top 50 genes (excluding the unknown genes) showing maximum up-regulation and down-regulation have been given Supplementary Table 2. Real-Time PCR of the selected genes verified the up-regulation of ER-stress response genes and G-protein signalling pathway genes (Fig. 5b) thus validating the microarray analysis.
Microarray analysis of the PsLecRLK overexpressing tobacco plants and its validation. a Pie chart depicting classification of upregulated and downregulated genes, based on biological processes. The differential regulation of the genes was observed by microarray analysis and represent the differential fold change in expression of genes in LecRLK overexpression lines (in tobacco) over the empty vector lines in response to 200 mM NaCl stress. b Real-time PCR validation of the microarray results. The fold change of transcript from PsLecRLK overexpressing line (S6) was normalized to the transcript level of the salinity stressed empty vector control plants. Purple bars represent various genes from the ER stress response pathway while the green bars represent genes from G-protein signaling pathway
Discussion
The conservation of basic cellular stress response among different kingdoms of life (Kültz 2003) prompted the identification of genes that could be involved in salinity stress tolerance pathways (Joshi et al. 2010). In the present study, we have undertaken molecular analysis of one of the candidate genes identified in the aforementioned study, PsLecRLK from point-of-view of developing salinity stress tolerant crop varieties.
In pea plant, PsLecRLK showed up-regulation when subjected to salinity stress, specifically NaCl treatment. The PsLecRLK protein was found to be localized in plasma membrane systems and/or other regions of the cell. Overexpression lines of PsLecRLK in tobacco, when subjected to various documented salinity stress tolerance assays (such as pigment content, biomass, growth, yield etc.), exhibited comparatively higher tolerance to salinity stress than the empty vector control and the tobacco wild type plants used as controls. Similar salinity stress tolerant phenotype has also been reported for Arabidopsis and Glycine soja LecRLKs (Deng et al. 2009; Sun et al. 2013). Under normal conditions, the constitutive overexpression of PsLecRLK led to a slight yield penalty (statistically, non-significant), as also observed in case of drought stress responsive OsNAC6 overexpression in rice plants (Nakashima et al. 2007). However, under salinity stress conditions, a significant maintenance of yield was observed in PsLecRLK overexpressing tobacco plants. Therefore, as in case of OsNAC6 (Nakashima et al. 2007), the use of native PsLecRLK promoter or other salinity stress responsive promoters (such as Arabidopsis RD29B promoter) (Msanne et al. 2011) could circumvent the problem of yield penalty under non-stress conditions, thus making PsLecRLK a promising candidate for crop improvement in future.
The effector molecules of stress adaptation involve proteins that; (1) mediate ion homeostasis, (2) enhance water transport, (3) are involved in osmolyte biosynthesis, (4) mediate ROS scavenging, (5) are involved in maintenance of general homeostasis (Hasegawa et al. 2000). The first two categories of the effector proteins involved in ion homeostasis and water channels are interrelated. In halophytes (plants that can thrive in saline environment) there is neither a unique special feature nor is there a special metabolic machinery to endure extreme salinity (Flowers et al. 1977; Glenn et al. 1999). Halophytes maintain root Na+ level to minimum (Adams et al. 1992) and mobilize the excess Na+ to leaves so as to decrease the osmotic potential in the upper parts of the plant thus facilitating water uptake from roots. Similarly, PsLecRLK overexpressing plants showed lowest Na+/K+ in roots. Also water channels, such as aquaporins (AQP) and plasma membrane intrinsic proteins (PIPs), which are believed to have immense importance for plant water relation, especially under stressed conditions (Hasegawa et al. 2000) were found to be overexpressed in PsLecRLK overexpression lines. PIP1 and PIP2 overexpression has been previously reported to enhance water uptake (Mahdieh et al. 2008). AQP, PIP1 and PIP2 overexpression probably leads to enhanced water uptake in PsLecRLK overexpression plants as well (supported by water uptake analysis). It is noteworthy that even under normal conditions, PsLecRLK overexpression lines displayed higher water uptake. The reason behind this could be constitutive overexpression of water channels or activation of other genes which, under stress, act synergistically with PsLecRLK in combating osmotic component of salt stress. The enhanced water uptake, although accompanied with Na+ (Apse and Blumwald 2007), is beneficial as the excess Na+, when compartmentalized in vacuoles, serves as an effective osmolyte (Hasegawa et al. 2000). We show that in comparison to the control plants PsLecRLK overexpressing plants have much higher expression of transporters such as NHXs, HKT1, AVP1, SOS1 etc. As a management step to counter excess Na+, upregulated SOS1 in PsLecRLK overexpressing plants probably leads to increased Na+ extrusion from roots and leaves and its unloading to xylem vessels (Shi et al. 2000). At this point, increased activity of HKT1 could help to remove excess Na+ from xylem stream by diverting it to the leaves(Apse and Blumwald 2007; Davenport et al. 2007) where NHX1 and NHX2 could compartmentalize the Na+ from cytoplasm to vacuole (Apse et al. 1999; Venema et al. 2002) thus preventing ion toxicity in cytosol of photosynthetic tissues(Munns and Tester 2008; Zhu 2001). Recent publications also indicate that K+, and not Na+, is the main substrate for NHXs (Bassil et al. 2011; Barragán et al. 2012; Andrés et al. 2014). This proves added advantage due to higher accumulation of K+ which would further lower Na+/K+ ratio in plant tissues. Also, ATP binding cassette (ABC) transporters, whose essential role in cation transport has been elucidated in yeast (Miyahara et al. 1996), was found to be overexpressed in the microarray analysis of PsLecRLK overexpressing tobacco plants.
Fourth type of effector proteins are the ones involved in ROS maintenance. As elucidated in previous studies, overexpression of FeSOD, Cu/ZnSOD, catalase etc. enhances stress tolerance in plants (Gupta et al. 1993; Shikanai et al. 1998). On the other hand, silencing of catalase gene in tobacco made the plant more susceptible to oxidative stress (Willekens et al. 1997). In PsLecRLK overexpressing plants, the transcript analysis of genes involved in ROS scavenging pathway did not show much perturbations. However, the plants had much lower peroxide as well as superoxide accumulation and relatively higher activity of ROS scavenging enzymes which leads to lower oxidative damage to plants during salinity stress. Higher activity of ROS scavenging enzymes albeit lower transcript abundance probably indicate post-transcriptional control of the antioxidant enzyme activity (Zhang et al. 2009).
Lastly, several of proteins involved in general homeostasis, such as heat shock proteins and molecular chaperons, were found to be differentially expressed in PsLecRLK overexpressing plants. Chaperons are amongst the key proteins contributing to maintenance of homeostasis both under optimal and stressed conditions (Wang et al. 2004). In PsLecRLK overexpressing lines, a glycine-rich protein homologue of an RNA chaperon exhibited up-regulation. Similarly, cyclophilins are ubiquitous proteins which serve the function of molecular chaperon. Their role in transcription regulation, protein trafficking and cellular signal transduction under stress conditions including salinity stress has been reported (Price et al. 1994; Trivedi et al. 2013). Heat shock proteins (HSP) are classically known for their role in stress tolerance. HSP20 was up-regulated in response to PsLecRLK overexpression. It belongs to ATP-independent chaperons that have been reported to help in maintenance of cellular protein structures under salinity stress (Lee et al. 1997).
Apart from the effector proteins, role of several regulatory proteins could not be underestimated(Hasegawa et al. 2000). Stress signaling pathways are often designed to be modulated by several signaling molecules, many of which are plant hormones such as jasmonic acid, salicylic acid, ethylene etc. (Lam et al. 1999). To ascertain the signaling pathway involved in activation of PsLecRLK mediated response, transcript analysis of several hormone regulated genes could not answer our question, most probably due to a complex cross-talk between various hormonal pathways (Kunkel and Brooks 2002). Nevertheless, microarray analysis of PsLecRLK overexpressing tobacco plants pointed towards a possible modulation of at least two pathways that are essential in maintaining cellular homeostasis. First is the G protein-signaling pathway whose several components, including the ones that were differentially upregulated, have been documented to be important in salinity (or general) stress tolerance. G-protein β subunit (Gβ), whose role in heat and oxidative stress has been studied previously (Misra et al. 2007), was differentially expressed in PsLecRLK overexpressing lines under salinity stress. In another study, Gβ was reported to interact with PR proteins and a MAP kinase (Bhardwaj et al. 2011). Further, PsMAPK itself can functionally complement yeast MAPK, Hog1p, helping the mutant in surviving under salinity stress conditions (Pöpping et al. 1996). This putative signaling cascade would be examined in future studies.
Rab proteins are members of Ras superfamily of small G-proteins which function in regulation of membrane traffic and the exocytosis pathway (Cheng et al. 2002) were also amongst the up-regulated genes in the present study. It was also found to be differentially up-regulated in the Thellungiella halophila halophyte over its glycophyte relative Arabidopsis, when both plants were subjected to salinity stress (Kosová et al. 2011) indicating its potential importance in salinity stress tolerance.
Secondly, PsLecRLK overexpression also seems to activate endoplasmic reticulum (ER) stress machinery [or the Unfolded Protein Response (UPR)]. Under UPR response, luminal binding protein (BiP) is highly up-regulated and in concert with other ER resident proteins and nuclear transcription factors, it leads to activation of several stress responsive genes to protect plant from further damage (Tajima et al. 2008). BiP is an ER localized chaperon protein and is a member of Hsp70 family (Maruyama et al. 2010). It interacts with protein disulphide isomerase 2 (PDI2) (Cho et al. 2011) and takes part in ER mediated protein quality control wherein the misfolded/unassembled proteins are degraded by ER associated degradation machinery. Both BiP and PDI2 were overexpressed upon PsLecRLK overexpression. Another protein that showed up-regulation in PsLecRLK overexpressing lines was Bax-1 inhibitor protein, which is highly induced in ER stress response (Watanabe and Lam 2008). Its expression is unrelated to other ER stress responsive genes and it plays role in inhibiting H2O2 and SA induced cell death (Kawai-Yamada et al. 2004).
Salinity stress tolerance could be provided by alleviating ionic toxicity (as in case of SOS pathway (Shi et al. 2000; Zhu 2001), osmotic stress [example, MAPK pathway members (Pöpping et al. 1996; Zhu 2001)] or both [example, AtRab7 (Mazel et al. 2004)]. PsLecRLK seems to fall in the last category where tolerance towards both ionic toxicity and osmotic stress is imparted. Lower Na+ content, high K+ (due to NHXs overexpression) and thus lower Na+/K+ ratio indicates towards ionic stress tolerance while better compartmentation of Na+ to vacuoles (as indicated by transporter’s transcript analysis) thus leading to continued water uptake indicates towards positive osmotic adjustment.
The role of lectin domain which is believed to be the sensory domain of the protein is worth mentioning here. Salinity stress tolerance phenotype from LecRLKs from plant species such as, Glycine soja (Sun et al. 2013), Arabidopsis thaliana (Huang et al. 2013; Deng et al. 2009; He et al. 2004) has been demonstrated. Previous study on PsLecRLK showed that lectin-domain was essential to provide salinity stress tolerance to bacteria as kinase-domain alone could not ensure their survival under salinity stress (Joshi et al. 2010). However, the exact function of this domain in perception of salt stress is unknown. Recent publication has elucidated an Arabidopsis LecRLK (DORN1, At5g60300) as a sensor for extracellular ATP (eATP) (Choi et al. 2014), while another publication has reported eATP release upon application of salinity stress (Sun et al. 2012). The authors also linked eATP release with Na+ compartmentation, regulation of ROS signaling and expression of several salinity stress responsive genes (Sun et al. 2012). As our study also demonstrates that PsLecRLK provides salinity stress tolerance by all the above mentioned features, it is highly plausible that the lectin domain of PsLecRLK is involved in perception of eATP (or similar stress signals) while the kinase domain of the gene translates the perceived signal to maintain cellular homeostasis. However, further analysis on the hypothesis would be required in the future.
To summarize, PsLecRLK overexpression in model plant tobacco conferred it salinity stress tolerance. This tolerance, marked by growth and yield maintenance, was mainly due to lower Na+ and its better compartmentalization which resulted in reduced ROS accumulation and consequently to lower membrane damage and cell death in these plants. Also, microarray analysis indicates overexpression of several G-protein signalling pathway and ER stress response pathway genes that might assist PsLecRLK in achieving homeostasis.
Although the present study identifies key features of PsLecRLK mediated salinity stress tolerance, much research is required to identify the exact molecular function and the underlying signalling pathway of PsLecRLK. Till date, neither information regarding ligands or downstream targets of LecRLKs is available nor are the governing factors for their activation/inactivation known. Therefore, much research needs to be carried out to understand the role of this vast gene family.
References
Adams P, Thomas JC, Vernon DM, Bohnert HJ, Jensen RG (1992) Distinct cellular and organismic responses to salt stress. Plant Cell Physiol 33:1215–1223
Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol Plant Microbe Interact 21:507–517. doi:10.1094/mpmi-21-5-0507
André S, Siebert H-C, Nishiguchi M, Tazaki K, Gabius H-J (2005) Evidence for lectin activity of a plant receptor-like protein kinase by application of neoglycoproteins and bioinformatic algorithms. Biochim Biophys Acta (BBA) (General Subjects) 1725:222–232. doi:10.1016/j.bbagen.2005.04.004
Andrés Z, Pérez-Hormaeche J, Leidi EO, Schlücking K, Steinhorst L, McLachlan DH, Schumacher K, Hetherington AM, Kudla J, Cubero B, Pardo JM (2014) Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc Natl Acad Sci 111(17):E1806–E1814
Apse MP, Blumwald E (2007) Na+ transport in plants. FEBS Lett 581:2247–2254. doi:10.1016/j.febslet.2007.04.014
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24(3):1127–1142
Barre A, Hervé C, Lescure B, Rougé P (2002) Lectin receptor kinases in plants. Crit Rev Plant Sci 21:379–399. doi:10.1080/0735-260291044287
Bassil E, Tajima H, Liang Y-C, M-a Ohto, Ushijima K, Nakano R, Esumi T, Coku A, Belmonte M, Blumwald E (2011) The Arabidopsis Na +/H + antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell Online 23(9):3482–3497
Becraft PW (1998) Receptor kinases in plant development. Trends Plant Sci 3:384–388
Bhardwaj D, Sheikh AH, Sinha AK, Tuteja N (2011) Stress induced beta subunit of heterotrimeric G-proteins from Pisum sativum interacts with mitogen activated protein kinase. Plant Signal Behav 6:287–292
Bonaventure G (2011) The Nicotiana attenuata LECTIN RECEPTOR KINASE 1 is involved in the perception of insect feeding. Plant Signal Behav 6:2060–2063
Bouwmeester K, Govers F (2009) Arabidopsis L-type lectin receptor kinases: phylogeny, classification, and expression profiles. J Exp Bot 60:4383–4396
Chen X et al (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46:794–804. doi:10.1111/j.1365-313X.2006.02739.x
Cheng H et al (2002) Role of the Rab GTP-binding protein Ypt3 in the fission yeast exocytic pathway and its connection to calcineurin function. Mol Biol Cell 13:2963–2976
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55:225–236
Cho E, Yuen CL, Kang B-H, Ondzighi C, Staehelin LA, Christopher D (2011) Protein disulfide isomerase-2 of Arabidopsis mediates protein folding and localizes to both the secretory pathway and nucleus, where it interacts with maternal effect embryo arrest factor. Mol Cells 32:459–475. doi:10.1007/s10059-011-0150-3
Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science 343:290–294
Davenport RJ, MuÑOz-Mayor A, Jha D, Essah PA, Rus ANA, Tester M (2007) The Na+ transporter AtHKT1; 1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant, Cell Environ 30:497–507. doi:10.1111/j.1365-3040.2007.01637.x
Deng K, Wang Q, Zeng J, Guo X, Zhao X, Tang D, Liu X (2009) A lectin receptor kinase positively regulates ABA response during seed germination and is involved in salt and osmotic stress response. J Plant Biol 52:493–500. doi:10.1007/s12374-009-9063-5
Dere S, Güneş T, Sivaci R (1998) Spectrophotometric determination of chlorophyll-A, B and total carotenoid contents of some algae species using different solvents. Turk J Bot 22:13–17
Desclos-Theveniau M, Arnaud D, Huang T-Y, Lin GJ-C, Chen W-Y, Lin Y-C, Zimmerli L (2012) The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog 8:e1002513 doi:10.1371/journal.ppat.1002513
Dong C-H, Zolman BK, Bartel B, B-h Lee, Stevenson B, Agarwal M, Zhu J-K (2009) Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Mol Plant 2:59–72
Flowers TJ, Troke PF, Yeo AR (1977) The mechanism of salt tolerance in halophytes. Annu Rev Plant Physiol 28:89–121. doi:10.1146/annurev.pp.28.060177.000513
Gilardoni PA, Hettenhausen C, Baldwin IT, Bonaventure G (2011) Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell Online 23:3512–3532
Glenn EP, Brown JJ, Blumwald E (1999) Salt tolerance and crop potential of halophytes. Crit Rev Plant Sci 18:227–255. doi:10.1080/07352689991309207
Gouget A et al (2006) Lectin receptor kinases participate in protein–protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol 140:81–90
Gupta AS, Heinen JL, Holaday AS, Burke JJ, Allen RD (1993) Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc Natl Acad Sci 90:1629–1633
Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499. doi:10.1146/annurev.arplant.51.1.463
He XJ, Zhang ZG, Yan DQ, Zhang JS, Chen SY (2004) A salt-responsive receptor-like kinase gene regulated by the ethylene signaling pathway encodes a plasma membrane serine/threonine kinase. Theor Appl Genet 109:377–383. doi:10.1007/s00122-004-1641-9
Hervé C, Serres J, Dabos P, Canut H, Barre A, Rougé P, Lescure B (1999) Characterization of the Arabidopsis lecRK-a genes: members of a superfamily encoding putative receptors with an extracellular domain homologous to legume lectins. Plant Mol Biol 39:671–682. doi:10.1023/a:1006136701595
Huang P, Ju H-W, Min J-H, Zhang X, Kim S-H, Yang K-Y, Kim CS (2013) Overexpression of L-type lectin-like protein kinase 1 confers pathogen resistance and regulates salinity response in Arabidopsis thaliana. Plant Sci 203–204:98–106. doi:10.1016/j.plantsci.2012.12.019
Joshi A, Dang HQ, Vaid N, Tuteja N (2010) Pea lectin receptor-like kinase promotes high salinity stress tolerance in bacteria and expresses in response to stress in planta. Glycoconj J 27:133–150. doi:10.1007/s10719-009-9265-6
Kanzaki H, Saitoh H, Takahashi Y, Berberich T, Ito A, Kamoun S, Terauchi R (2008) NbLRK1, a lectin-like receptor kinase protein of Nicotiana benthamiana, interacts with Phytophthora infestans INF1 elicitin and mediates INF1-induced cell death. Planta 228:977–987. doi:10.1007/s00425-008-0797-y
Kawai-Yamada M, Ohori Y, Uchimiya H (2004) Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell Online 16:21–32
Kosová K, Vítámvás P, Prášil IT, Renaut J (2011) Plant proteome changes under abiotic stress—contribution of proteomics studies to understanding plant stress response. J Proteomics 74:1301–1322. doi:10.1016/j.jprot.2011.02.006
Kültz D (2003) Evolution of the cellular stress proteome: from monophyletic origin to ubiquitous function. J Exp Biol 206:3119–3124
Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5:325–331. doi:10.1016/S1369-5266(02)00275-3
Lam E, Pontier D, del Pozo O (1999) Die and let live—programmed cell death in plants. Curr Opin Plant Biol 2:502–507. doi:10.1016/S1369-5266(99)00026-6
Lee S, Suh S, Kim S, Crain RC, Kwak JM, Nam H-G, Lee Y (1997) Systemic elevation of phosphatidic acid and lysophospholipid levels in wounded plants. Plant J 12:547–556. doi:10.1046/j.1365-313X.1997.00547.x
Lehti-Shiu MD, Zou C, Hanada K, Shiu S-H (2009) Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol 150:12–26
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158. doi:10.1016/j.abb.2005.10.018
Mahdieh M, Mostajeran A, Horie T, Katsuhara M (2008) Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum plants. Plant Cell Physiol 49:801–813
Maruyama D, Endo T, S-i Nishikawa (2010) BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci 107:1684–1689
Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134:118–128
Misra S, Wu Y, Venkataraman G, Sopory SK, Tuteja N (2007) Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): role in salinity and heat stress and cross-talk with phospholipase C. Plant J 51:656–669. doi:10.1111/j.1365-313X.2007.03169.x
Miyahara K, Mizunuma M, Hirata D, Tsuchiya E, Miyakawa T (1996) The involvement of the Saccharomyces cerevisiae multidrug resistance transporters Pdr5p and Snq2p in cation resistance. FEBS Lett 399:317–320. doi:10.1016/S0014-5793(96)01353-1
Morris ER, Walker JC (2003) Receptor-like protein kinases: the keys to response. Curr Opin Plant Biol 6:339–342. doi:10.1016/S1369-5266(03)00055-4
Msanne J, Lin J, Stone J, Awada T (2011) Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234:97–107. doi:10.1007/s00425-011-1387-y
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. doi:10.1146/annurev.arplant.59.032607.092911
Nakashima K et al (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630. doi:10.1111/j.1365-313X.2007.03168.x
Navarro A, Vicente MJ, Martínez-Sánchez JJ, Franco JA, Fernández JA, Bañón S (2008) Influence of deficit irrigation and paclobutrazol on plant growth and water status in Lonicera implexa seedlings. Acta Hort (ISHS) 782:299–304
Park M, Lee H, Lee J-S, Byun M-O, Kim B-G (2009) In planta measurements of Na+ using fluorescent dye CoroNa Green. J Plant Biol 52:298–302. doi:10.1007/s12374-009-9036-8
Pöpping B, Gibbons T, Watson M (1996) The Pisum sativum MAP kinase homologue (PsMAPK) rescues the Saccharomyces cerevisiae hog1 deletion mutant under conditions of high osmotic stress. Plant Mol Biol 31:355–363. doi:10.1007/bf00021795
Price ER, Jin M, Lim D, Pati S, Walsh CT, McKeon FD (1994) Cyclophilin B trafficking through the secretory pathway is altered by binding of cyclosporin A. Proc Natl Acad Sci 91:3931–3935
Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA 102:509–514
Shi H, Ishitani M, Kim C, Zhu J-K (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci 97:6896–6901
Shikanai T, Takeda T, Yamauchi H, Sano S, Tomizawa K-I, Yokota A, Shigeoka S (1998) Inhibition of ascorbate peroxidase under oxidative stress in tobacco having bacterial catalase in chloroplasts. FEBS Lett 428:47–51. doi:10.1016/S0014-5793(98)00483-9
Singh P et al (2012) The lectin receptor kinase-VI.2 is required for priming and positively regulates arabidopsis pattern-triggered immunity. Plant Cell Online 24:1256–1270
Sun J, Zhang X, Deng S, Zhang C, Wang M, Ding M, Zhao R, Shen X, Zhou X, Lu C, Chen S (2012) Extracellular ATP signaling is mediated by H2O2 and cytosolic Ca2+ in the salt response of Populus euphratica cells. PLoS ONE 7(12):e53136. doi:10.1371/journal.pone.0053136
Sun X-L et al (2013) GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J Plant Physiol 170:505–515. doi:10.1016/j.jplph.2012.11.017
Tajima H, Iwata Y, Iwano M, Takayama S, Koizumi N (2008) Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem Biophys Res Commun 374:242–247. doi:10.1016/j.bbrc.2008.07.021
Tichtinsky G, Vanoosthuyse V, Cock JM, Gaude T (2003) Making inroads into plant receptor kinase signalling pathways. Trends Plant Sci 8:231–237
Trivedi D, Ansari M, Dutta T, Singh P, Tuteja N (2013) Molecular characterization of cyclophilin A-like protein of Piriformospora indica for its potential role to abiotic stress tolerance in E. coli. BMC Res Notes. doi:10.1186/1756-0500-6-5552014
Tuteja N, Sopory SK (2008) Plant signaling in stress: G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal Behav 3:79–86
Tuteja N, Sahoo RK, Garg B, Tuteja R (2013) OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64). Plant J 76:115–127. doi:10.1111/tpj.12277
Tuteja N, Sahoo RK, Huda KMK, Tula S, Tuteja R (2014) OsBAT1 augments salinity stress tolerance by enhancing detoxification of ROS and expression of stress-responsive genes in transgenic rice. Plant Mol Biol Report. doi:10.1007/s11105-014-0827-9
Vaid N, Pandey PK, Tuteja N (2012) Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol Biol 80:365–388. doi:10.1007/s11103-012-9952-8
Vaid N, Macovei A, Tuteja N (2013) Knights in action: lectin receptor-like kinases in plant development and stress responses. Mol Plant 6:1405–1418
Velikova V, Yordanov I, Edreva A (2000) Oxidative stress and some antioxidant systems in acid rain-treated bean plants: protective role of exogenous polyamines. Plant Sci 151:59–66. doi:10.1016/S0168-9452(99)00197-1
Venema K, Quintero FJ, Pardo JM, Donaire JP (2002) The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J Biol Chem 277:2413–2418
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252
Watanabe N, Lam E (2008) BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J Biol Chem 283:3200–3210
Willekens H et al (1997) Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J 16:4806–4816. doi:10.1093/emboj/16.16.4806
Xiong L, Schumaker KS, Zhu J-K (2002) Cell signaling during cold, drought, and salt stress. Plant Cell Online 14:S165–S183
Zhang Z, Zhang D, Zheng Y (2009) Transcriptional and post-transcriptional regulation of gene expression in submerged root cells of maize. Plant Signal Behav 4:132–135
Zhu J-K (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Acknowledgments
Work on signal transduction and plant stress signaling in NT’s laboratory is partially supported by Department of Science and Technology (DST) and Department of Biotechnology (DBT), Government of India. We thank Miss Irum Rizvi for her help in localization experiment. Neha Vaid duly acknowledges Council of Scientific and Industrial Research (CSIR) for financial assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vaid, N., Pandey, P., Srivastava, V.K. et al. Pea lectin receptor-like kinase functions in salinity adaptation without yield penalty, by alleviating osmotic and ionic stresses and upregulating stress-responsive genes. Plant Mol Biol 88, 193–206 (2015). https://doi.org/10.1007/s11103-015-0319-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0319-9