Abstract
The scent bouquets of flowers of Nicotiana species, particularly those of section Alatae, are rich in monoterpenes, including 1,8-cineole, limonene, β-myrcene, α- and β-pinene, sabinene, and α-terpineol. New terpene synthase genes were isolated from flowers of Nicotiana bonariensis, N. forgetiana, N. longiflora, and N. mutabilis. The recombinant enzymes synthesize simultaneously the characteristic ‘cineole cassette’ monoterpenes with 1,8-cineole as the dominant volatile product. Interestingly, amino acid sequence comparison and phylogenetic tree construction clustered the newly isolated cineole synthases (CIN) of section Alatae together with the catalytically similar CIN of N. suaveolens of section Suaveolentes, thus suggesting a common ancestor. These CIN genes of N. bonariensis, N. forgetiana, N. longiflora, and N. mutabilis are distinct from the terpineol synthases (TERs) of the taxonomically related N. alata and N. langsdorfii (both Alatae), thus indicating gene diversification of monoterpene synthases in section Alatae. Furthermore, the presence of CINs in species of the American section Alatae supports the hypothesis that one parent of the Australian section Suaveolentes was a member of the present section Alatae. Amino acid sequences of the Nicotiana CINs and TERs were compared to identify relevant amino acids of the cyclization reaction from α-terpineol to 1,8-cineole.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Nicotiana belongs to the Solanaceae and comprises about 75 species classified in 3 subgenera and 13 sections (Goodspeed 1954; Knapp et al. 2004; Chase et al. 2003; Clarkson et al. 2004). About 75 % of the Nicotiana species occur naturally in America, and 25 % in Australia, while only one species has so far been found in Africa (Aoki and Ito 2000; Merxmüller and Buttler 1975). The species of section Alatae originated in the Americas, comprising to date 8 species (N. alata, N. bonariensis, N. langsdorfii, N. rastroensis, N. longiflora, N. plumbaginifolia, N. forgetiana) (Lim et al. 2006; Knapp et al. 2004) and the recently discovered Nicotiana mutabilis in southern Brazil (Stehmann et al. 2002). The flowers of the Alatae species vary in shape and color, e.g. Nicotiana longiflora and N. bonariensis bear white flowers, whereas N. forgetiana petals are red and those of N. mutabilis change from white to pink during flower aging (Goodspeed 1954; Macnish et al. 2010). The floral scent compositions are also quite variable in Nicotiana species of section Alatae (Table 1), despite the conserved and characteristic emission of a set of seven monoterpenes, the so-called ‘cineole cassette’ monoterpenes (Fig. 1; Table 1, 1,8-cineole, limonene, β-myrcene, α- and β-pinene, sabinene, α-terpineol) (Raguso et al. 2003, 2006).
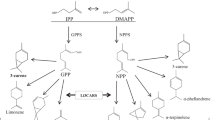
Chemical structures of monoterpenes. Schematic overview of the biosynthesis of acyclic and cyclic ‘cineole cassette’ monoterpenes and ocimene. The substrate geranyl pyrophosphate is converted into an acyclic cation and further to the cyclic precursor α-terpinyl cation. 1,8-cineole is synthesized via α-terpineol
Terpenoids are the largest group of natural products, and at least 40,000 compounds are known to date (Bohlmann and Keeling 2008). This wealth of terpenes is synthesized by terpene synthases (TPS), which catalyze the reaction of acyclic prenyl diphosphates into various acyclic and cyclic compounds. Depending on the substrate, the terpene synthases are classified as monoterpene synthases (substrate geranyl pyrophosphate), sesquiterpene synthases (substrate farnesyl pyrophosphate), diterpene synthases (substrate geranyl geranyl pyrophosphate) and are subdivided into seven subfamilies, TPS A to H (Chen et al. 2011).
Up to now, ninety monoterpene synthases have been isolated. The enzymes were found in floral and vegetative tissues of angiosperms (TPS B) and gymnosperms (TPS D) (Degenhardt et al. 2009). They produce the wide variety of terpenoids (summarized in Fähnrich et al. 2011). Some monoterpene synthases synthesize almost exclusively their name-giving compound, e.g. geraniol synthases (GES), linalool synthases (LIS), and limonene synthases (LIM), while the majority of the isolated monoterpene synthases are multiproduct enzymes. Typical multiproduct enzymes are cineole synthases (CIN), terpinene synthases (TS), terpinolene synthases (TES), bornyl diphosphate synthases (BOR), carene synthases (CAR), some myrcene synthases (MYR) and terpineol synthases (TER), which produce cyclic and acyclic compounds (Fig. 1). Several compounds, such as limonene, β-myrcene, sabinene, α/β-pinene are synthesized by several different multiproduct enzymes, while volatiles such as α-terpineol and 1,8-cineole were only synthesized by one type of monoterpene synthases (CIN or TER enzymes) (Fähnrich et al. 2011).
The ecological or biological relevance and the reason why some monoterpenes (e.g. limonene) may result from more than one enzymatic source while others terpenes are synthesized by just one enzyme are presently not known. This issue could be addressed after isolation and detailed studies of respective monoterpene synthase enzymes.
The characteristic ‘cineole cassette’ monoterpene emission in Nicotiana species of section Alatae was originally noticed by Raguso et al. (2003, 2006). Since 1,8-cineole was always the major product across the Alatae species, it was suggested that the Nicotiana CINs are similar to the CIN of Salvia officinalis (Kampranis et al. 2007). However, we observed that floral ‘cineole cassette’ emission profiles of Nicotiana species (Table 1) or the 1,8-cineole/α-terpineol product ratios of in vitro assays are not identical (Fähnrich et al. 2011), suggesting that the synthesis capacities of respective enzymes are distinct and cannot be unified. Additionally, it is presently not known which molecular features of the enzymes determine the 1,8-cineole/α-terpineol ratio, which is an indicator of the efficacy of the cyclization reaction (Fig. 1). This demands clarifying studies of respective genes and enzymes.
Another interesting observation should be added to the list of open questions. A cineole synthase with 1.8-cineole as major product was previously isolated from N. suaveolens (Roeder et al. 2007) as well as two enzymes that produce α-terpineol as major component within the ‘cineole cassette’ monoterpenes (N. alata and N. langsdorfii, Fähnrich et al. 2011). It is interesting to note that species of the Australian clade Suaveolentes typically do not emit ‘cineole cassette’ monoterpenes. Therefore, the emission of the ‘cineole cassette’ monoterpenes from N. suaveolens flowers indicates an evolutionary interesting exception and suggests the existence of a common ancestral gene prior to gene duplication and diversification.
Considering the mentioned aspects, further studies have to be undertaken to fully understand the production, catalytic reaction, and evolution of the ‘cineole cassette’ monoterpene synthases from Nicotiana flowers. As a first step in reaching these goals we present here the isolation and functional characterization of monoterpene synthases from N. bonariensis, N. forgetiana, N. longiflora, and N. mutabilis.
Materials and methods
Plant growth
Nicotiana bonariensis (TW28) (Raguso et al. 2003), Nicotiana forgetiana (TW 50, Raguso et al. 2003), Nicotiana longiflora (TW78, Raguso et al. 2003), Nicotiana mutabilis (collected by Rainee Kaczorowski in Quebra Cabo, acc. RK200813120; it is not ‘Rastroensis’) were grown on Vermiculite (Deutsche Vermiculite Dämmstoffe GmbH, Sprockhövel, Germany) in growth chambers under long day conditions (illumination: 16 h, 6 am–10 pm, 160 μEm−2 s−1, mercury vapor lamp (Osram, München, Germany) 22 °C, 8 h darkness: 8 h, 10 pm–6 am, 21 °C). Plants were watered with Hoagland’s solution (Hoagland and Arnon 1938).
Volatile collection of flowers of N. bonariensis, N. forgetiana, N. mutabilis
The collection of volatiles from whole flowers was performed using the open loop system (Heath and Manukian 1994). One flower was kept in each glass globe, and headspace volatiles of four globes were investigated individually for each species. Collection began on the day of anthesis of flowers, when they were placed into glass globes. A compressor (Schneider Werkstatt- und Maschinenfabrik, Bräunlingen, Germany) delivered a constant air flow of 5 l min−1, which was divided between the four glass globes. Headspace volatiles of flowers of N. bonariensis were collected between 6 and 8 pm, and of N. forgetiana and N. mutabilis between 10 and 12 am. The volatile-enriched air was sucked through a 100 mg SuperQ column (Alltech Associates, Deerfield, IL, USA) using a vacuum pump with 2.8 l min−1 (KNF Neuberger, Freiberg, Germany) (Effmert et al. 2008). For quantification, nonyl acetate or cis-nerolidol (5 ng μl−1) was used as internal standard and volatiles were eluted in fractions of 200 and 100 μL dichloromethane and analyzed by GC/MS (Effmert et al. 2008).
GC/MS analysis
The volatile compounds were analyzed with a Shimadzu QP5000 gas chromatograph connected to a mass spectrometer for identification (GC/MS). Separation was performed on a DB5-MS column (60 m × 0.25 mm × 0.25 mm; J+W Scientific Folsom, CA, USA) with helium as carrier gas (flow rate of 1.4 ml min−1) at a temperature gradient from 35 °C (2 min hold) to 275 °C (3.5 min hold) using a ramp of 10 °C min−1. Mass spectra were obtained by using the scan mode (total ion count, 40–280 mz−1). Compound identity was confirmed by (1) comparison of mass spectra and retention times with those of available authentic standards (terpenoid standards from Sigma-Aldrich, St. Louis, MS, USA; nonyl acetate from Roth, Karlsruhe, Germany), (2) comparison of obtained spectra with spectra in the library of the National Institute of Standards and Technology (NIST147), and (3) by comparison of the Kovats indices.
Crude protein extracts from petals
Petals were harvested every 3 h (starting at 9 am) and placed in an ice-cold mortar. Samples of 0.2 g of the petals were extracted with 1 ml buffer containing 0.1 M sodium phosphate, 0.25 mM saccharose, 5 mM MgCl2, 1 mM CaCl2, 25 mM Na2S2O5, 2 mM DTT, 5 mM ascorbate, 2 μl 2-mercaptoethanol, 0.1 g PVPP (polyvinyl polypyrrolidone) and protease inhibitor cocktail tablets (Roche, Mannheim, Germany). The crude extracts were centrifuged for 30 min at 13,000 g (4 °C). The supernatant was collected. After addition of 10 % glycerol, the crude extract was stored at −70 °C until used for Western blots. Extracts for enzyme activity determination were always freshly prepared.
RNA extraction
RNA from Nicotiana bonariensis (TW28), Nicotiana forgetiana (TW50), Nicotiana longiflora (TW78), Nicotiana mutabilis (acc. RK2008131201) plants were isolated according to Chang et al. (1993). In brief, 0.5–1 g petals were ground in liquid nitrogen and incubated for 15 min with 400 μl 2-mercaptoethanol and 15 ml CTAB buffer (2 % CTAB (hexadecyltrimethyl ammonium bromide), 2 % PVP (polyvinylpyrrolidone K40), 100 mM Tris–HCl, 25 mM EDTA, 2 M NaCl add H2O to 500 ml). Samples were extracted with 15 ml of chloroform: isoamyl alcohol (24:1) added twice and centrifuged for 20 min (10,000 g). The samples were resuspended in 0.25 M LiCl buffer to allow precipitation overnight at 8 °C. RNA was centrifuged for 30 min at 4 °C and the pellet was resuspended in 700 μl SSTE buffer (1 M NaCl, 0.5 % SDS, 10 mM Tris–HCl, 1 mM EDTA) and again precipitated with ethanol (99.8 %) for 1 h at −70 °C. The RNA pellet was gently washed twice with ethanol (70 %).
Isolation of monoterpene synthase genes
A homology-based RT-PCR strategy was used to clone genes of interest. Oligonucleotides of recently described cineole synthases were deduced (Fähnrich et al. 2011). The RT reaction was performed with the SuperScript III™ Reverse Transcriptase (Invitrogen, Karlsruhe, Germany) according to manufacturer’s recommendation. For cDNA synthesis, 2 μg RNA, 25 pmol oligo dT (GAC TGG TCA ATC AGT TAC (T)16) primer, 10 mM dNTPs, 0.1 M DTT, 5 μl 5× cDNA synthesis buffer were incubated at 58 °C for 1 h. The reaction was terminated by heating at 85 °C for 5 min. Subsequently the PCR reaction was performed by using the following components: 1 μg first strand cDNA, 5 μl 10× Pfu buffer, 1.25 units Pfu DNA polymerase (Invitrogen, Karlsruhe Germany), 25 pmol of each antisense/sense primer and sterile H20 was added to a final volume of 50 μl. To amplify the monoterpene synthase of N. bonariensis, N. forgetiana, N. longiflora and N. mutabilis, the primer combination CINS6 (5’AGA CGT TCG GGG AAT TAC CAA CCT 3′), which binds to the RR(X)8W motif, and R2 (5′GAC TGG TCA ATC AGT TAC3′), which binds to the adapter sequence of oligo dT was used. The PCR reactions were performed at standard conditions: 98 °C 2 min (1×), 98 °C 30 s, 54 °C 30 s, 68 °C 1 min kb−1 (30×) and 10 min 72 °C. Products were separated on an 1.2 % agarose gel and extracted with the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). For sequencing, the full-length monoterpene synthase cDNAs were ligated into the cloning vector pJET1.2/blunt and subsequently the plasmid was transformed into E. coli TOP 10 cells (Invitrogen, Karlsruhe, Germany). After incubation overnight on Luria–Bertani (LB) agar containing 100 μg ml−1 ampicillin, the transformants were analyzed by colony PCR using the vector primers T7 Promotor (5′TAA TAC GAC TCA CTA TAG GG3`) and pJET1.2 reverse (5′AAG AAC ATC GAT TTT CCA TGG CAG3′) by using Master mix (Qiagen, Hilden Germany) at standard conditions.
Gene expression analysis
For temporal expression analysis RT-PCR was applied using N. forgetiana and N. bonariensis. Total RNA was isolated every 3 h according to Chang et al. (1993). 2 μg of DNase I (Sigma-Aldrich, St. Louis, MS, USA) digested RNA was used for cDNA synthesis performed with the SuperScript III™ Reverse Transcriptase (Invitrogen, Karlsruhe, Germany) according to manufacturer’s recommendation. Subsequently PCR reactions were performed by using the following components: 1 μg first strand cDNA, 25 pmol of each antisense/sense primer, 14 μl Master mix (Fermentas, Thermo Fisher Scientific Inc) and sterile H20 was added to a final volume of 50 μl. To amplify a CIN gene fragment the primer combination CINS2 and R2 was used and a 1.1 kbp fragment was obtained. The PCR reactions were performed at standard conditions: 98 °C 2 min (1×), 98 °C 30 s, 50 °C 30 s, 68 °C 1 min kb−1 (20/30/40× cycles) and 10 min 72 C°. For quantification the CIN transcript was adjusted to the control (elongation factor 1α = EF − 1α, Dean et al. 2002). A 370 bp fragment of this housekeeping gene was obtained by PCR reaction using 1 μg cDNA, 14 μl Master mix (Fermentas, Thermo Fisher Scientific Inc) and 25 pmol of primer EF1 fow and EF1 rev. The PCR reactions were performed at standard conditions: 94 °C 90 s (1×), 94 °C 30 s, 50 °C 30 s, 72 °C 1 min kb−1 (20/30/40× cycles) and 10 min 72 C°. The amplifications were analyzed on an 1.2 % agarose gel. Fragments of the CIN gene of N. forgetiana and N. bonariensis were extracted with the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and sequenced to verify the correct sequence.
Heterologous protein expression
The protein was overexpressed by using the Expression Champion™ pET SUMO Protein Kit (Invitrogen, Karlsruhe, Germany). The forward primer Sumo (5′AGA CGT TCG GGG AAT TAC CAA CCT3′) and a reverse primer Sumo (5′TCA GGC TGG AGG AAT AGA TTC AAA GAC3′) without stop codon were applied to amplify a truncated monoterpene synthase. A his-tag was added to the N-terminus. To avoid formation of inclusion bodies in E. coli the truncated monoterpene synthases without target sequence were amplified (Bohlmann et al. 1998). RT-PCR reactions were performed with 1 μg first strand cDNA, 10 mM dNTPs, 25 pmol primer (each), 5 μl 10× Pfu buffer, 1.25 units Pfu DNA polymerase (Invitrogen, Karlsruhe, Germany) and sterile water added to a final volume of 50 μl. PCR products were purified according the manufacturer’s recommendation (Qiagen, Hilden, Germany), ligated into the Champion™ pET SUMO vector (Invitrogen, Karlsruhe, Germany), and sequenced to assure the correctness of the sequence. The plasmid was transformed into E. coli HMS 174 (DE3) (Novagene, Darmstadt, Germany) which were then cultivated overnight at 37 °C in 5 ml LB medium supplemented with 50 μg ml−1 kanamycin. 1 ml of an overnight preculture was transferred into 50 ml LB medium containing 50 μg ml−1 kanamycin and 1 % glucose and growth was continued at 37 °C until an OD600 of 0.6 was reached. For functional expression the cultures were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (final concentration) and growth continued for additional 48 h at 13 °C in a rotary shaker. The cells were then harvested by centifugation at 4 °C for 30 min (8,000 g) and resuspended in 2 ml lysis buffer (50 mM Tris–HCl, pH 7.5, 10 % glycerol, 10 mM β-mercaptoethanol). The cells were frozen in liquid nitrogen and immediately thawed at 42 °C. Freeze–thaw cycles were repeated three times and followed by incubation with 1 mg ml−1 lysozyme for 1 h on ice. After centrifugation at 4 °C for 30 min (8,000 g), the resulting supernatant was either transferred to a Ni–NTA column or directly used in an enzyme assay.
Overexpressed protein was purified by Ni–NTA affinity chromatography according to the manufacturer’s recommendations (Qiagen, Hilden, Germany). In brief, the supernatant was incubated for 1 h with the column matrix and then washed with 50 mM NaH2PO4, 300 mM NaCl and 40–60 mM imidazole. The protein was eluted with 500 μl elution buffer (50 mM NaH2PO4, 300 mM NaCl, 80–200 mM imidazole). The protein concentration was analyzed using Bradford reagent (Bradford et al. 1976).
SDS–polyacrylamide gel electrophoresis and Western blot analysis
Samples (15 μg) of the crude protein extract or the purified recombinant enzyme (5 μg) were loaded onto polyacrylamide gels (12.5 %), electrophoretically separated (Miniprotean, Bio-Rad Laboratories, Hercules, CA, USA), and finally transferred to PVDF (polyvinyldifluoride) membrane (Roth, Karlsruhe, Germany) by using a mini-tank blotting gel cassette (XCell II, Novex, San Diego, CA, USA). The membranes were placed in a Tris-buffered blocking solution overnight (TBS 20 mM) with 0.05 % (vv−1) Tween 20, 4 % (wv−1) skim milk, and 1 % (wv−1) BSA). After incubation with the specific antibody against the CIN from N. suaveolens (Roeder et al. 2007) (diluted 1:3,000 in blocking solution, Davids Biotechnology, Regensburg, Germany) for 1.5 h, repeated washes with TBS containing 0.05 % (vv−1) Tween 20 (TBS-Tween) were performed. The membrane was incubated with the secondary antibody (anti-rabbit alkaline phosphatase-conjugate, diluted 1:20,000 in TBS-Tween (Sigma-Aldrich, St. Louis, MS, USA) and washed again with TBS-Tween and TBS. The membrane was equilibrated in detection buffer (100 mM Tris–HCl, 150 mM NaCl, 50 mM MgCl2) and incubated with CDP-Star (Roche, Mannheim, Germany; 0.25 μM in detection buffer) in darkness and analyzed in a luminescence image analyser (LAS-1000, Fujifilm, Japan). Its luminescence after 30 min was quantified with Fujifilm Image Gauge software. After measuring the luminescence, the proteins were stained with NBT/BCIP (50 mg ml−1 in dimethylformamide, 2:1; Roche, Mannheim, Germany).
Enzyme assay
The Ni–NTA affinity chromatography purified enzyme (2 μg) or the purified supernatant was used for the enzyme assay, which was performed as described by Fähnrich et al. 2011. The assay buffer consisted of 250 mM Hepes/KOH buffer (pH 8) containing 10 % glycerol, 100 mM MgCl2 and 0.25 mM MnCl2. The putative terpene synthase was incubated with an assay buffer, 147 μM GPP (Echelon Biosciences) and 5 mM DTT (final volume of 200 μl). Crude protein extracts (100 μl) were incubated with the enzyme assay buffer, 7 μM GPP and 5 mM DTT. The assay samples were overlaid with 200 μl hexane and incubated for 3 h at 32 °C. To quantify the products of the TPS, 5 ng internal standard were added (5 ng μl−1 cis-nerolidol or nonyl acetate, Roth, Karlsruhe, Germany). The products were extracted by vortexing 2 min followed by a centrifugation for 2 min at 2,000 g. Aliquots of the hexane phase were analyzed by GC/MS.
Sequence analysis and phylogenetic tree construction
The genes were sequenced by GATC Biotech AG (Konstanz, Germany). Homologous monoterpene synthases were identified with the BLAST search tool of NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al. 1990). The complete sequences were aligned with the ClustalW program of EMBL (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Phylogenetic analysis was performed by BioEdit version 7.0.5. The phylogenetic tree was constructed by using the neighbor joining algorithm, maximum parsimony algorithm and MEGA Software v 4.0 (Tamura et al. 2007). To display the phylogenetic tree, the TreeExplorer 2.12 was used. Plant species and accession numbers were used for the tree construction: Arabidopsis thaliana CIN AY691947, Citrus unshiu CIN BAD91045, Magnolia grandiflora TER ACC66282, Nicotiana alata TER JQ346173, Nicotiana langsdorfii TER JN989317, Nicotiana suaveolens CIN EF175166, Rosmarinus officinalis CIN DQ839411, Salvia fruticosa CIN ABH07677, Salvia officinalis CIN AAC26016, Santalum album TER ACF 24767, Solanum lycopersicum MTS2 AY840092, Solanum lycopersicum MTS1 AY840091, Vitis vinifera TER AAS79351, Zea mays TER AAL59230, Abies grandis bisabolene synthase AF006194. Newly isolated gene sequences were submitted to the EMBL data databank, accession numbers are: Nicotiana forgetiana CIN JX028206, Nicotiana bonariensis CIN JX028207, Nicotiana longiflora CIN JX040448, Nicotiana mutabilis putative CIN JX040449.
3D modeling
To build the homology-based 3D structure of the N. alata TER, N. forgetiana CIN, N. longiflora CIN and N. bonariensis CIN, the limonene synthase (LIM) of Mentha spicata (Hyatt et al. 2007) was chosen because of the high sequence similarity of 96 % (recommendation ≥ 30 %). Using ClustalW or BioEdit version 7.0.5 as alignment tools, the cDNA sequence of N. alata TER was adjusted to the M. spicata LIM sequence. The program SPDBV3.7. (Swiss Institute of Bioinformatics) was used to perform and visualize the 3D structure. The pdb-file was loaded using the program Pymol (version 0.98, Delano scientific LLC) to get a better 3D view of the protein model.
Results
Floral emission of ‚cineole cassette’ monoterpenes of Nicotiana species of section Alatae
Nicotiana species of section Alatae release monoterpenes from their flowers (Raguso et al. 2003, 2006; Table 1). Here the emission of floral monoterpenes of N. bonariensis, N. forgetiana, N. mutabilis and N. longiflora was examined in detail. A complex volatile emission pattern comprising 12 identified compounds (sabinene, β-myrcene, limonene, 1,8-cineole, phenylacetaldehyde, phenylethanol, α-terpineol, 2-phenylethylacetate, cinnamylalcohol, eugenol, phenylacetate, isoeugenol) and an unidentified compound was obtained for N. bonariensis (Fig. 2a). 1,8-cineole and α-terpineol were the major compounds of the ‘cineole cassette’ with a 1,8-cineole/α-terpineol ratio of 2:1 (Fig. 2b). The emission profiles of N. forgetiana were composed of seven compounds (α-pinene, sabinene, β-myrcene, limonene, 1,8-cineole, (E)-β-ocimene and α-terpineol) with 1,8-cineole dominating the scent bouquet (Fig. 2c). A high 1,8-cineole/α-terpineol ratio of 12:1 was determined (Fig. 2d). The scent of N. mutabilis was composed of β-myrcene, limonene, 1,8-cineole, (E)-β-ocimene and α-terpineol, with a 1,8-cineole/α-terpineol ratio of 4:1 (Fig. 2e, f). Since the scent emissions of the ‘cineole cassette’ monoterpenes of N. longiflora flowers were very low (Raguso et al. 2003), we were unable to measure volatiles in the headspace. According to Raguso’s measurements, limonene is the major compound followed by 1,8-cineole and α-pinene, whereas α-terpineol was not detected (Table 1).
Floral volatiles emitted from different Nicotiana species. a Headspace volatiles of flowers of N. bonariensis were collected between 6 and 8 pm. c Headspace volatiles of flowers of N. forgetiana were collected between 10 and 12 am. e Headspace volatiles of flowers of N. mutabilis were collected between 10 and 12 am. Each glass globe contained one flower, headspace volatiles of 4 flowers were investigated for each species. Compounds were analyzed by GC/MS, identified by their retention index, by comparison of mass spectra of the library of the National Institute of Standards and Technology (NIST147), and by comparison with the authentic standards. (1) α-pinene, (2)-sabinene, (3) β-pinene, (4) myrcene, (5) limonene, (6) cineole, (7) terpineol, (8) E-β ocimene, (9) phenylacetaldehyde, (10) phenylethanol, (11) phenylethylacetate, (12) cinnamylalcohol, (13) eugenol, (14) phenylacetate, (15) isoeugenol. (IS) internal standard (5 ng cis-nerolidol or nonyl acetate). b, d, f Quantification of emitted monoterpenes from flowers of N. bonariensis, N. forgetiana, N. mutabilis, respectively. The volatiles were collected for 2 h, amounts were calculated in ng per flower and per h, n = 3. Statistical analyses were performed using ANOVA and t test calculated with Sigma Plot (*significance p < 0.05)
Synthesis of monoterpenes in Nicotiana flowers
As a next step we determined terpene synthase activities in petal raw extracts. After addition of the substrate GPP to petal protein extracts, five and eight monoterpenes were synthesized by N. bonariensis and N. forgetiana, respectively (Fig. 3). Almost identical product spectra were obtained when neryl pyrophosphate (NPP) was applied as substrate (Fig. S3). In the white flowers of N. bonariensis (Fig. 3a), the enzyme activities increased continuously from day 1 to day 5, reaching a specific activity of 0.42 nkat 1,8-cineole/mg protein at day 5 (Fig. 3b). The same patterns were observed for the other monoterpenes of the ‚cineole cassette’ (maximum activities at day 5 were: 0.2 nkat α-terpineol/mg protein, 0.09 nkat β-myrcene/mg protein, 0.05 nkat sabinene/mg protein, 0.06 nkat limonene/mg protein; total activity 0.8 nkat/mg protein) (Fig. 3b). The cineole and terpineol synthase activities were significantly higher at day 5 compared to day 1–4. Enzyme activity levels peaked with 0.33 nkat 1,8-cineole/mg protein at day 3 in the red flowers of N. forgetiana (Fig. 3d, e). The enzyme activities regarding other monoterpene products were lower (0.12 nkat α-terpineol/mg protein, 0.14 nkat β-myrcene/mg protein, 0.04 nkat sabinene/mg protein, 0.04 nkat limonene/mg protein, total activity 0.6 nkat/mg protein at day 3).
Monoterpene synthase activities during flower development and during the day. The precursor geranyl pyrophosphate (GPP) was provided as substrate to the newly isolated monoterpene synthases. a, d The development of the flowers of Nicotiana bonariensis and Nicotiana forgetiana respectively is depicted. Day of anthesis (day 0), senescence (day 6). b Monoterpene synthase activities were determined in petal extracts of N. bonariensis flowers throughout flower development (day 1–day 5). The specific enzyme activities (pkat/mg total protein) were calculated for sabinene (gray), β-myrcene (green), limonene (yellow) 1,8-cineole (red) and α-terpineol (black), n = 3. c Determination of the specific enzyme activities for sabinene (gray), β-myrcene (green), limonene (yellow) 1,8-cineole (red) and α-terpineol (black) in N. bonariensis flower petals at different time points during the day (6 am, 9 am, noon, 3 pm, 6 pm, 9 pm and midnight), n = 3. e Monoterpene synthase activities were determined in petal extracts of N. forgetiana flowers throughout flower development (day 1–day 5). The specific enzyme activities (pkat/mg total protein) were calculated for α-pinene (orange), sabinene (gray), β-pinene (violet), β-myrcene (green), limonene (yellow) 1,8-cineole (red), (E)-β-ocimene (cyan), and α-terpineol (black), n = 3. f Determination of the specific enzyme activities for α-pinene, (orange), sabinene (gray), β-pinene (violet), β-myrcene (green), limonene (yellow) 1,8-cineole (red), (E)-β-ocimene (cyan), and α-terpineol (black) in N. forgetiana flower petals at different time points during the day (6 am, 9 am, noon, 3 pm, 6 pm, 9 pm and midnight), n = 3. Statistical analyses were performed using ANOVA and t test calculated with Sigma Plot (*significance: p < 0.05)
The activities of the terpene synthases in petal raw extracts not only vary during flower development but also throughout the day. Exemplified analysis of the pattern of the major product 1,8-cineole revealed higher monoterpene synthase activities at the beginning of the night phase (6 and 9 pm) compared to lower levels during the day in N. bonariensis flowers (Fig. 3c), while N. forgetiana did not exhibit enzyme activity alterations throughout the day (Fig. 3f). Maximum enzyme activities of 0.12 nkat 1,8-cineole/mg protein were reached in petal extracts of N. bonariensis at 6 pm, while the enzyme activities in flowers of N. forgetiana leveled around 0.23 nkat 1,8-cineole/mg protein. The specific activity of N. mutabilis petals was investigated only once at day 3 post-anthesis at 9 am, and the enzyme activity was highest for 1,8-cineole (0.167 nkat/mg protein) while activities with other monoterpenes were very low (sabinene 0.01 nkat/mg protein, β-myrcene 0.005 pkat/mg protein, β-pinene 0.02 pkat/mg protein, limonene 0.01 pkat/mg protein, α-terpineol 0.057 pkat/mg protein).
1,8-cineole is synthesized from α-terpineol by one reaction step (Fig. 1). The 1,8-cineole/α-terpineol ratio therefore expresses the performance capability of the cyclization reaction. In N. bonariensis and N. forgetiana, the 1,8-cineole/α-terpineol ratios varied (only) between 2:1 and 3:1 during flower development and at different time points of the day (Table S1). These constant compound ratios, also supported by correlation coefficient of R = 0.9 (Table S2), were evaluated as an indication of precisely regulated catalysis rather than randomized product formation.
Expression of TPS in N. forgetiana
The presence of monoterpene synthases in flower extracts of the investigated Nicotiana species was further underpinned by determination of the cineole synthase specific mRNAs. The mRNAs were isolated from N. forgetiana flowers at different time points during the day and RT-PCR reactions were performed (Fig. S1). The PCR products of the expected size of circa 1200 nucleotides were obtained with specific cineole synthase primers. The mRNA levels were high between 2, 6 and 8 pm in N. forgetiana prior to the elevated enzyme activities.
Taken together, these examinations support the presence of cineole monoterpene synthases in petals of these Nicotiana species of section Alatae.
Gene isolation and sequence analysis of ‚cineole cassette’ monoterpene synthases from Nicotiana species of section Alatae
Via RT-PCR we successfully isolated putative terpene synthases from petals of N. bonariensis, N. forgetiana, N. longiflora and N. mutabilis. The amino acid sequences were aligned with the previously isolated terpineol synthases of N. alata and N. langsdorfii and cineole synthase of N. suaveolens (Fig. 4) with which they share high sequence identity (89–99 %, Table S3). The newly cloned coding sequences comprised 1,572 bp and encode mature proteins of 524 amino acids. The N-terminus of all four enzymes contained the typical RR(X)8W-motif (1), and other motifs (2–7) characteristic of monoterpene synthases were also present (Fig. 4). The DDXXD-motif is known to be involved in divalent metal ion binding (Starks et al. 1997; Bohlmann et al. 1998), while the NALV sequence directed the formation of different product amounts and probably influences the enzyme specificity of Salvia monoterpene synthases (Kampranis et al. 2007). Since monoterpene synthases are functionally active in plastids, they should harbor a transit peptide upstream of the RR(X)8W-motif (RR-motif). The transit sequences were not cloned for the four new genes. The RR-motif is involved in binding the substrate GPP and isomerization of the initial carbocation to the linalyl diphosphate intermediate by recapture of the diphosphate (Williams et al. 1998). This motif was conserved in the N-terminal region of all Nicotiana monoterpene synthases. The RWW- and CYMNE-motifs were also found in these terpene synthase sequences, but their functions remain unknown (Roeder et al. 2007). Another highly conserved RXR-motif was localized circa 35–40 amino acids upstream of the DDXXD-motif. The function of this motif is related to the complexation of the diphosphate after ionization of the substrate preventing nucleophilic attack on any of the carbocationic intermediates (Starks et al. 1997).
Sequence of Nicotiana monoterpene synthases. Alignment of the newly isolated monoterpene synthase genes from N. bonariensis, N. forgetiana, N. longiflora, N. mutabilis with the terpineol synthase (TER) sequences of N. alata, N. langsdorfii and the cineole synthase (CIN) sequence of N. suaveolens (BLAST Search, Altschul et al. 1990, alignment with Clustal W). Conserved sequence motifs are indicated: (1) RR(X)8W-motif, (2) RWW-motif, (3) RXR-motif, (4) NALV-motif, (5) DDXXD-motif, (6) CYMNE-motif. Indicated in gray: Sequence differences between TER/CIN sequences. Amino acids involved in formation of the active pocket are marked with filled dots: a = arg263, b = trp272, c = asn293, d = asp300-asp304, e = ile395, f = leu436-glu446. Lines above the sequence compilation (α1–α22) indicate amino acids involved in the formation of helices
Amino acid alterations within the new and known Nicotiana monoterpene synthases are highlighted in gray (Fig. 4). Not a single amino acid alteration was observed in the sequence motifs or within the active pocket (amino acids indicated with black dots in Fig. 4). In total, eleven amino acid positions were altered in one of the four newly isolated genes compared to the TER of N. alata. To get a better idea of the location of these altered amino acids, we have shown them in a homology-based protein model constructed on the 3-D crystal structure of the limonene synthase from Mentha spicata (Hyatt et al. 2007) (Fig. 5). Four of the eleven altered amino acids of N. bonariensis (Fig. 5a; Table S4A) were located close to conserved motifs: gly259 and ile 376 were found near the DDXXD motif, thr368 was near RWW and NALV motifs, met393 was close to the DTE motif. One of the eight altered amino acids of N. forgetiana (Fig. 5b; Table S4B) was closely located to conserved motifs DDXXD and RWR. One (ile 376) of the seven altered amino acids of N. longiflora (Fig. 5c; Table S4C) was located close to conserved motifs DDXXD and RWW. One (ile 376) of the ten altered amino acids of N. mutabilis (Fig. 5d; Table S4D) was located close to conserved motifs DDXXD and RWR. This analysis highlights the altered amino acids at position 259 and 376, because they are in close vicinity to the catalytically important motif DDXXD and may influence the chemical reaction of the enzyme. It is interesting to note that at position 376 the TER genes of N. alata and N. langsdorfii display the non-polar amino acid isoleucine, while all isolated CIN genes carry the basic amino acid arginine (Fig. 4; Table S4).
Localization of altered amino acids of newly isolated cineole synthases compared to Nicotiana alata. The 3D structure of N. alata was modelled using Swiss Pdb Viewer (Fähnrich et al. 2011). The crystal structure of the limonene synthase of Mentha spicata was a master sequence (gaps in the alignment were hidden). All amino acids were eliminated except those of the motifs (blue: RR(X)8W, beige: RWR, green: DTE, violet: RWW, pink: NALV, yellow: DDXXD, orange: CYMNE) and the amino acids that are different to the N. alata sequence. Presented is the perspective of the 3D structure where the distances of the altered amino acids are short to the motifs. a Altered amino acids of CIN of N. bonariensis compared to TER N. alata: ile99, pro112, tyr 219, gly259, thr368, ile376, met393, val496, ser508. b Altered amino acids of CIN of N. forgetiana compared to TER N. alata: val191, gln246, ile376, val496, arg536, leu541. c Altered amino acids of CIN of N. longiflora compared to TER N. alata: val191, gln246, ile376, val496, arg536. d Altered amino acids of CIN of N. mutabilis compared to TER N. alata: his129, val191, ile226, ile245, gln246, ile376, asn495, val496
The newly isolated terpene synthase genes together with the known CIN and TER genes of other plant species were used to construct a phylogenetic tree (Fig. 6, neighbor joining method). This analysis clustered the genes of N. bonariensis, N. forgetiana, N. longiflora and N. mutabilis to the closely related CINs and TERs of other Nicotiana species. Interestingly, the newly isolated sequences of the species of section Alatae are more closely related to the CIN of N. suaveolens, which belongs to the Australian section Suaveolentes, than to the TERs of N. alata and N. langsdorfii of the American section Alatae. The same result was obtained when the parsimony algorithm was applied, which supports the conclusion that apparently the catalytical capabilities rather than the taxonomical relationships were decisive for the clustering.
Plant cineole and terpineol terpene synthases. Phylogenetic relationship of newly isolated cineole synthase genes (CIN) from N. bonariensis, N. forgetiana, N. longiflora, N. mutabilis with related monoterpene synthases of other plant species. Rooted neighbor joining phylogenetic (100 % bootstrap) tree construction based on amino acid sequence similarities. The bisabolene synthase (a sesquiterpene synthase) was used as outgroup. The tree was created with MEGA 4.0 and displayed using TreeView. Gaps (Clustal W) and the target sequence upstream of the RR(X)8W motif of the alignment was removed. Plant species used for the tree construction (accession numbers see “Materials and methods”)
Functional characterization of monoterpene synthases from Nicotiana species of section Alatae
To ultimately determine their biological functions, the newly isolated enzymes were overexpressed in E. coli and purified via his-tag affinity chromatography (Fig. S2, e.g. N. forgetiana). The enzymes were cloned using PCR-techniques specifically designed to only introduce the RR(X)8W truncated version of the enzyme into E. coli in order to get the highest expression and to prevent inclusion body formation (Williams et al. 1998; Peters et al. 2000). The purified enzymes were tested with the substrates GPP and NPP, and the volatiles were analyzed and identified by GC/MS (Figs. 7a, c, e, S4). The recombinant enzymes of N. bonariensis, N. forgetiana and N. longiflora reached enzyme activities in the same range (0.12–0.27 nkat 1,8-cineole/mg protein) and produced the seven compounds of the ‚cineole cassette’ (Figs. 7b, d, f, S4). While α- and β-pinene, sabinene, β-myrcene and limonene were minor products, contributing between 1 and 10 % to the synthesized blend, 1,8-cineole and α-terpineol were the major compounds of the volatile profiles. All three enzymes synthesized Two- to threefold more 1,8-cineole than α-terpineol and were therefore named cineole synthases (CIN). These similar ratios were supported by high correlation coefficients (Table S2). The quantitative product profiles were almost identical regarding the products 1,8-cineole, α-terpineol, sabinene, limonene, and α- and β-pinene. The contribution of β-myrcene was nearly identical for both enzymes of N. forgetiana and N. longiflora, whereas β-myrcene synthesis by the N. bonariensis enzyme was lower. Furthermore, this high similarity of the product profiles (of N. forgetiana and N. longiflora) correlates with the close correspondence of these enzymes at the level of amino acid identities (99 %). However, both enzymes have a distinct protein sequence compared to the CIN of N. bonariensis and a different product pattern.
GC chromatograms of overexpressed cineole synthases of Nicotiana species. The recombinantly overexpressed his-tagged enzymes were purified via Ni–NTA affinity chromatography (Fig. S2). Volatiles synthesized in enzyme assays were analyzed by GC–MS. Product identification was based on comparison with authentic standard compounds and with mass spectra of the NIST147 library. a GC chromatogram of the cineole synthase of N. bonariensis. c GC chromatogram of the cineole synthase of N. forgetiana. e GC chromatogram of the cineole synthase of N. longiflora. (1) α-pinene, (2) sabinene, (3) β-pinene, (4) β-myrcene, (5) limonene, (6) 1,8-cineole, (7) α-terpineol. (IS) internal standard (5 ng cis-nerolidol), n.i.: not identified. b, d, f Specific activities of recombinant cineole synthases from N. bonariensis, N. forgetiana, N. longiflora were calculated for each of the seven ‚cineole cassette’ monoterpenes. GPP was provided as substrate. n = 3 (*significance: p < 0.05)
Discussion
Many species of the genus Nicotiana developed scented flowers, of which several emit the characteristic monoterpenes of the ‘cineole cassette’. This set of monoterpenes has primarily been found in species of section Alatae (Raguso et al. 2003, 2006; 1,8-cineole, limonene, β-myrcene, α-pinene, β-pinene, sabinene, and α-terpineol). However, the contributions of the individual monoterpenes to the emission profiles vary in the different species (Table 1). These variations might result from intrinsic features of the enzymes catalyzing the synthesis of these compounds or might be due to different physiological and cellular conditions in the petals. To investigate the first issue in more detail we isolated and characterized ‘cineole cassette’ monoterpene synthases from N. bonariensis, N. forgetiana, N. longiflora, and N. mutabilis. It turned out that the recombinant enzymes exhibit very similar properties with respect to product profiles, efficiencies of the cyclization reaction, maximum enzyme activities, and amino acid sequences.
Emission profiles versus synthesis profiles
Floral scents are usually traits to communicate with the environment, for example, by attracting pollinators, or protecting against herbivores and pathogens via direct or indirect defense mechanisms (Pichersky and Gershenzon 2002; Raguso et al. 2003). Different Nicotiana species exhibit various flower colors and flower morphologies to interact with nocturnal hawkmoths as well as diurnal hummingbirds and bees (Kaczorowski et al. 2005; Knudsen et al. 2004; Raguso et al. 2003; Stehmann et al. 2002). The flowers of N. alata and N. bonariensis open around dusk and N. alata is pollinated by the hawkmoths Agrius cingulatus and Eumorpha labruscae (Sphingidae) and N. bonarienis is pollinated by small perching moths (Kaczorowski et al. 2005). Therefore it comes as no surprise that species-specific daily emission patterns as well as enzyme activities were observed for N. bonariensis and N. alata (Figs. 2, 3; Fähnrich et al. 2011), which correlated with the pollination of night-active organisms (Kaczorowski et al. 2005). For N. forgetiana, hummingbirds and small hawkmoths have been documented as pollinators before dusk (Ippolito et al. 2004), which is in accordance with the continuous emission profile (Fig. 3).
The scent profiles of Nicotiana flowers described by Raguso et al. (2003, 2006) were verified here, with particular focus on the presence of the ‘cineole cassette’ monoterpenes for N. bonariensis, N. forgetiana, and N. mutabilis. While these three species emitted relatively high levels of volatiles, in the headspace of N. longiflora only small amounts or none (α-terpineol) were detected (Raguso et al. 2003). Despite these very low emission levels of N. longiflora flowers, we were able to isolate a terpene synthase from this and the other three Nicotiana species. The synthesis profiles of these four overexpressed enzymes turned out to be very similar, they produced predominantly 1,8-cineole followed by α-terpineol, and the side products occurred in significantly smaller amounts. These recombinant enzymes always synthesized in vitro twice as much 1,8-cineole as α-terpineol (Tables S1, S2). An exception was N. forgetiana, which emitted 12-fold 1,8-cineole compared to α-terpineol. The latter is in discrepancy to the in vitro capacity of the enzyme, but presently we have no sound explanation for this.
In the GC/MS profiles of the cineole synthases of N. bonariensis, N. forgetiana and N. longiflora neither no (E)-β-ocimene nor linalool was detected (Fig. 7). Since (E)-β-ocimene was present in the floral scents of N. forgetiana, N. longiflora and N. mutabilis and linalool in N. longiflora and N. mutabilis, it is very likely that beside the CIN additional genes were expressed in petals. (E)-β-ocimene does not belong to the monoterpenes of the ‘cineole cassette’ as described by Raguso et al. (2006). Our observation in this investigation as well as the profiles of the previously isolated terpineol synthases of N. alata and N. langsdorfii support the notion that (E)-β-ocimene is not a member of the ‘cineole cassette’ monoterpenes (Fähnrich et al. 2011). (E)-β-Ocimene has also not been demonstrated as a product of other CIN or TER multiproduct enzymes of various other plant species (summarized in Fähnrich et al. 2011). The only known exception is the CIN of N. suaveolens (Roeder et al. 2007). In conclusion, for the synthesis of (E)-β-ocimene additional enzymes with the capability to synthesize (E)-β-ocimene must exist in respective Nicotiana species (Table 1; Fähnrich et al. 2011).
Molecular analysis of the cineole synthases of Nicotiana species
In each case investigated so far, the emission of ‘cineole cassette’ monoterpenes was due to the presence of multiproduct enzymes. 1,8-cineole was the major compound of the enzymes isolated from N. bonariensis, N. forgetiana and N. longiflora, while α-terpineol was the major compound of the enzymes isolated from the related species N. alata and N. langsdorfii (Fähnrich et al. 2011). The bicyclic 1,8-cineole is synthesized by a reaction of the hydroxyl group of α-terpineol with the double bond to form a second cycle (Fig. 1). When both compounds appear simultaneously in the product profile, it is reasoned that either the addition reaction is not efficient or a premature termination reaction occurs (Peters and Croteau 2003; Iijima et al. 2004). The conversion of the intermediate to the final product was apparently particularly limited in the TER genes of N. alata and N. langsdorfii, while the cyclization reaction was more pronounced by enzymes of N. bonariensis, N. forgetiana, N. longiflora, and N. suaveolens (Roeder et al. 2007). The ratio 1,8-cineole/α-terpineol (expressing the efficacy of the cyclization reaction) of the newly isolated genes was 2:1 and ranged between those of the TERs of N. alata and N. langsdorfii (0.5:1 and 0.9:1, respectively) and the CIN of N. suaveolens (7.8:1) (Fähnrich et al. 2011).
It remains to be explained why the CIN enzymes more efficiently perform the cyclization reaction than the TER enzymes. During the search for key amino acids important for the catalytic reaction mechanisms, amino acids of the active pocket have often been considered, but recently it was shown that also amino acids distant to the active pocket influence the product spectrum (Hyatt and Croteau 2005; Greenhagen et al. 2006; Yoshikuni et al. 2006). Both alternative possibilities might also explain the different efficacies in the Nicotiana species. Therefore, the amino acid variations of TERs/CINs of Nicotiana were localized in the amino acid sequences (Fig. 4) and it turned out that neither amino acids in conserved motifs nor of the active pocket were altered in respective Nicotiana species (Fig. 4). Kampranis et al. (2007) indicated amino acids at positions 338 and 327 in region 1 of monoterpene synthases of Salvia fruticosa and Salvia pomifera, respectively, and at 436 (S. fruticosa) in region 2 to be relevant for the 1,8-cineole and α-terpineol production. It was also concluded that serine 436 prevents 1,8-cineole production and therefore limits the cyclization reaction. However, the amino acids shown to be important in Salvia species were conserved in the enzymes of all isolated CINs and TERs of Nicotiana species (N. alata: asn292 and ala293 in the NALV motif, ser396) and therefore could not be decisive for the efficiency of the cyclization reaction in the Nicotiana species. We therefore searched for amino acids that were closely positioned to those conserved motifs or the active pocket that may influence catalysis (Fig. 5). The amino acid isoleucine at position 376 in the two TER enzymes was altered to arginine in all CIN enzymes. It is possible that this change from a non-polar to a basic amino acid had an impact on the catalytically active motif DDXXD of Nicotiana enzymes. At the equivalent position (#412) S. fruticosa’s isoleucine resulted in a 1,8-cineole/α-terpineol ratio of 10:1, while S. pomifera harbors a positively charged lysine, which favors α-terpineol emission (ratio 0.3:1). These apparent controversial results indicate once more that amino acids relevant in one species may not play the same role in another species, even if the enzymes are related. Future research in the Nicotiana enzymes will focus on the amino acids (ile and arg at position 376) to evaluate their impact on the α-terpineol to 1,8-cineole cyclization reaction.
Phylogenetic and evolutionary analysis of ‘cineole cassette’ monoterpene synthases from related Nicotiana species
The genus Nicotiana is the fifth largest group within the Solanaceae and comprises 75 species. Goodspeed (1954) provided detailed information on the taxonomy, cytology, biogeography, and the floral and vegetative morphologies. The data indicated that N. sec. Suaveolentes is monophyletic due to the tree generated from internal transcribed spacers (Chase et al. 2003) and RFLP analysis of the plastid matK gene (Aoki and Ito 2000; Clarkson et al. 2004). Based on ancient chromosome hybridizations and the present chromosome number (amphidiploidy) Goodspeed (1954) hypothesized that one parent of the Australian section Suaveolentes is a member of the present section Alatae and the other parent either a member of the section Acuminatae (= Petunioides) or N. sect. Noctiflorae. We isolated and functionally analyzed related CINs from Nicotiana species of section Alatae and compared them with TER enzymes of N. alata and N. langsdorfii of the same section as well as CIN of N. suaveolens of section Suaveolentes. The three isolated enzymes of N. bonariensis, N. forgetiana and N. longiflora (section Alatae) were clearly cineole synthases (Fig. 7) and clustered together with the CIN of N. suaveolens (Fig. 6), supporting the hypothesis that one parent of the Australian Suaveolentes descended from the American Alatae (Goodspeed 1954). Furthermore, our results are in agreement with the phylogenetic trees that position Alatae next to Suaveolentes (Chase et al. 2003) or where Noctiflorae, Repandae, and Sylvestres are located between the sections Alatae and Suaveolentes (Clarkson et al. 2004). To understand the evolution of the TER/CIN genes of Nicotiana species in more detail, a future goal is to find and characterize a progenitor TER/CIN enzyme.
A novel finding of this research is that the newly isolated CINs of Alatae do not cluster with the taxonomically more closely related TERs of Alatae but with the catalytically similar CIN of N. suaveolens, which apparently indicates an amino acid sequence-based phylogeny rather than the taxonomy-based phylogeny. This is unusual for enzymes of the specialized metabolism (e.g. terpene synthases) but is common for enzymes of plant primary metabolism (Martin et al. 1993). A plausible explanation may apply: since N. suaveolens is an amphidiploid species with one assumed parent from Alatae tribe it can be speculated that the CIN is inherited from this parent. In this scenario, the isolated CINs of N. bonariensis, N. forgetiana, N. longiflora kept the original cyclization capability, while the TERs of N. alatae and N. langsdorfii diverged during evolution to establish a less efficient cyclization reaction.
Abbreviations
- TPS:
-
Terpene synthase
- CIN:
-
Cineole synthase
- TER:
-
Terpineol synthase
References
Altschul SF, Gish W, Miller W, Byers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:402–410
Aoki S, Ito M (2000) Molecular phylogeny of Nicotiana (Solanaceae) based on the nucleotide sequence of the matK gene. Plant Biol 2:316–324
Bohlmann J, Keeling CI (2008) Terpenoid biomaterials. Plant J 54:656–669
Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95:4126–4133
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Biochem 72:248–254
Chang S, Puryear J, Caiurney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11:113–116
Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko Y, Joseph J, Savolainen V, Parokonny AS (2003) Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann Bot 92:107–127
Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66:212–229
Clarkson JJ, Knapp S, Garcia VF, Olmstead RG, Leitch AR, Chase MW (2004) Phylogenetic relationships in Nicotiana (Solanaceae) inferred multiple plastid DNA regions. Mol Phylogenet Evol 33:75–90
Dean JD, Goodwin PH, Hsiang T (2002) Comparison of relative RT-PCR and northern blot analyses to measure expression of β-1,3-glucanase in Nicotiana benthamiana infected with Colltotrichum destructivum. Plant Mol Biol Rep 20:347–356
Degenhardt J, Köllner TG, Gershenzon J (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70:1621–1637
Effmert U, Dinse C, Piechulla B (2008) Influence of green leaf herbivory by Manduca sexta on floral volatile emission by Nicotiana suaveolens. Plant Physiol 146:1996–2007
Fähnrich A, Krause K, Piechulla B (2011) Product variability of the, cineole cassette’ monoterpene synthases of related Nicotiana species. Mol Plant 4:965–984.
Goodspeed TH (1954) The genus Nicotiana. Chronica Bot Co 16(1/6):1–536
Greenhagen BT, O’Maille PE, Noel JP, Chappell J (2006) Identifying and manipulating structural determinates linking catalytic specificities in terpene synthase. Proc Natl Acad Sci USA 103:9826–9831
Heath RR, Manukian A (1994) An automated system for use in collecting volatile chemicals released from plants. J Chem Ecol 20:593–608
Hoagland DR, Arnon DL (1938) The water-culture method of growing plants without soil. Calif Agric Exp Stn Circ 374:1–39
Hyatt DC, Croteau RB (2005) Mutational analysis of a monoterpene synthase reaction: altered catalysis through directed mutagenesis of (-)-pinene synthase from Abies grandis. Arch Biochem Biophys 439:222–233
Hyatt DC, Younn B, Zhao Y, Santhamma B, Coates RM, Croteau RB, Kang CH (2007) Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc Nat Acad Sci USA 104:5360–5365
Iijima Y, Gang DR, Fridman E, Lewinsohn E, Pichersky E (2004) Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol 134:370–379
Ippolito A, Fernandes GW, Holtsford TP (2004) Pollinator preferences for Nicotiana alata, N. forgetiana, and their F1 hybrids. Evolution 58:2634–2644
Kaczorowski RL, Gardener MC, Holtsford TP (2005) Nectar traits in Nicotiana section Alatae (Solanaceae) in relation to floral traits, pollinators and mating system. Am J Bot 92:1270–1283
Kampranis SC, Ioannidis D, Purvis A, Mahrez W, Ninga E, Katerelos NA, Annsour S, Dunwell JM, Degenhardt J, Majris AM, Goodenough PW, Johnson CB (2007) Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: structural insights into the evolution of terpene synthase function. Plant Cell 19:1994–2005
Knapp S, Chase MW, Clarkson JJ (2004) Nomenclatural changes and a new sectional classification in Nicotiana (Solanaceae). Taxon 52:73–82
Knudsen JT, Tollsten L, Groth I, Bergstrom G, Raguso RA (2004) Trends in floral scent chemistry in pollination syndromes: floral scent composition in hummingbird-pollinated taxa. Bot J Linn Soc 146:191–199
Lim KJ, Kovarik A, Matyasek R, Chase MW, Knapp S, McCarthy E, Clarkson JJ, Leitch AR (2006) Comparative genomics and repetitive sequence divergence in the species of diploid Nicotiana section Alatae. Plant J 48:907–919
Macnish AJ, Jiang CZ, Negre-Zakharov F, Reid MS (2010) Physiological and molecular changes during opening and senescence of Nicotiana mutabilis flowers. Plant Sci 179:267–272
Martin W, Brinkmann H, Savonna C, Cerff R (1993) Evidence for a chimeric nature of nuclear genomes: eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. PNAS 90:8692–8696
Merxmüller H, Buttler KP (1975) Nicotiana in der afrikanische Namib: ein pflanzengeographisches und phylogenetisches Rätsel. Mitteilungen der Botanischen Staatssammlung München 12:91–104
Peters RJ, Croteau RB (2003) Alternative termination chemistries utilized by monoterpene cyclases: chimeric analysis of bornyl diphosphate, 1,8-cineole, and sabinene synthase. Arch Biochem Biophys 417:203–211
Peters RJ, Flory JE, Jetter R, Ravn MM, Lee HJ, Coates RM, Croteau RB (2000) Abietadiene synthase from grand fir (Abies grandis): characterization and mechanism of action of the “pseudomature” recombinant enzyme. Biochemistry 39:15592–15602
Pichersky E, Gershenzon J (2002) The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5:237–243
Raguso RA, Levin RA, Foose SE, Holmberg MW, Mc Dade LA (2003) Fragrance chemistry, nocturnal rhythms and pollination “syndromes” in Nicotiana. Phytochemistry 63:265–284
Raguso RA, Schlumpberger BO, Kaczorowski RL, Holtsford TP (2006) Phylogenetic fragrance patterns in Nicotiana sections Alatae and Suaveolentes. Phytochemistry 67:1931–1942
Roeder S, Hartmann AM, Effmert U, Piechulla B (2007) Regulation of simultaneous synthesis of floral scent terpenoids by the 1,8-cineole synthase of Nicotiana suaveolens. Plant Mol Biol 65:107–124
Sallaud C, Rontein D, Onillon S, Jabe’s F, Duffe’ P, Giacalone C, Thorava S, Escoffier C, Herbette G, Leonhardt N, Causse M, Tissier A (2009) A Novel Pathway for sesquiterpene biosynthesis from Z, Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21:301–317
Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E (2009) Monoterpenes in glandular trichomes of tomato are synthesized from neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106:10865–10870
Starks CM, Back K, Chappell J, Noel JP (1997) Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolchene synthase. Science 277:1815–1820
Stehmann JR, Semir J, Ippolito A (2002) Nicotiana mutabilis (Solanaceae), a new species from southern Brazil. Kew Bull 57:639–646
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Williams DC, McGarvey DJ, Katahira EJ, Croteau RB (1998) Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37:12213–12220
Yoshikuni Y, Ferrin TE, Keasling JD (2006) Designed divergent evolution of enzyme function. Nature 440:1078–1082
Acknowledgments
The authors thank Robert Raguso (Cornell University, Ithaca, NY, USA) for providing the seeds of Nicotiana species, Dr. Uta Effmert (University of Rostock) for many helpful suggestions, and the DFG for financial funding (Pi153/22) as well as the University of Rostock.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Expression of cineole synthase genes of Nicotiana forgetiana. RNA was isolated from flower petals at different time points of the day (6 am, 8 am, noon, 2 pm, 6 pm and midnight). RT-PCR reactions with 20, 30 and 40 cycles were performed using gene specific primers (S2/R2). cDNA fragments were analyzed on 1.2 % agarose gels (TIFF 5011 kb)
Figure S2
Isolation of overexpressed recombinant cineole synthase of Nicotiana forgetiana. The recombinant monoterpene synthase of N. forgetiana was overexpressed in E. coli and the his-tagged enzyme purified via Ni-NTA affinity chromatography. Proteins of the chromatographic fractions were separated on a 12.5 % SDS polyacrylamide gel and the overexpressed enzyme was detected with anti his antibodies (Western blot). M: marker proteins, S: supernatant, WF: wash fraction, E: eluted proteins with 200 mM imidazole. (JPEG 24 kb)
Figure S3
Monoterpene synthase activities during flower development and during the day with the substrate NPP. The stereoisomer of GPP, neryl pyrophosphate (NPP), was provided as substrate to the newly isolated monoterpene synthases (Sallaud et al. 2009, Schilmiller et al. 2009). (A) Monoterpene synthase activities were determined in petal extracts of N. bonariensis flowers throughout flower development (day 1 – day 5). The specific enzyme activities (pkat/mg total protein) were calculated, (n =3). (B) Determination of the specific enzyme activities for N. bonariensis flower petals at different time points during the day (6 am, 9 am, noon, 3 pm, 6 pm, 9 pm and midnight),.( n=3). (C) Monoterpene synthase activities were determined in petal extracts of N. forgetiana flowers throughout flower development (day 1 – day 5). The specific enzyme activities (pkat/mg total protein) were calculated, (n =3). (D) Determination of the specific enzyme activities in N. forgetiana flower petals at different time points during the day (6 am, 9 am, noon, 3 pm, 6 pm, 9 pm and midnight), (n=3). α-pinene (orange), sabinene (gray), β-pinene (violet), β-myrcene (green), limonene (yellow) 1,8-cineole (red), (E)-β-ocimene (cyan), and α-terpineol (black). Statistical analyses were performed using ANOVA and t-test calculated with Sigma Plot (*significance: p < 0.05).(JPEG 4343 kb)
Figure S4
Specific activities of recombinant cineole synthases from N. forgetiana and N. longiflora with NPP as substrate.For each of the seven ‘cineole cassette’ monoterpenes enzyme activities were calculated, n=1.(JPEG 2046 kb)
Rights and permissions
About this article
Cite this article
Fähnrich, A., Brosemann, A., Teske, L. et al. Synthesis of ‘cineole cassette’ monoterpenes in Nicotiana section Alatae: gene isolation, expression, functional characterization and phylogenetic analysis. Plant Mol Biol 79, 537–553 (2012). https://doi.org/10.1007/s11103-012-9933-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-012-9933-y