Abstract
The white flowers of N. suaveolens emit a complex bouquet of fragrance volatiles. The dominant compounds are benzenoids (e.g. methyl benzoate, methyl salicylate, benzyl benzoate and benzyl salicylate), monoterpenes (1,8-cineole, limonene, sabinene, E-β-ocimene, β-β-myrcene, α- and β-pinene and α-terpineole) and sesquiterpenes (e.g. caryophyllene), which are all emitted at higher levels during the night. Here, we show that the simultaneous nocturnal emission of most monoterpenes is realized by a single floral-specific multi-product enzyme (1,8-cineole synthase, CIN), which synthesizes the monoterpenes of the “cineole cassette”. Interestingly, N. suaveolens is the only known taxon of the Suaveolentes section to have a flower emitting “cineole cassette of monoterpenes” which is otherwise typical for the Alatae section. Gene sequence analysis of CIN has revealed the highest similarities to other angiosperm monoterpene synthases from Vitis vinifera, Quercus ilex, Citrus unshiu and C. limon, which cluster in the same branch of the terpene synthase B subfamily. However, based on its synthesized products, N. suaveolens CIN shares similarity with enzymes of the Arabidopsis thaliana root and Salvia officinalis leaf. The N. suaveolens CIN gene is only expressed in the stigma/style tissue and petals. Thin sections of petals present the enzyme primarily in the adaxial and abaxial epidermis; this facilitates the comprehensive emission of volatiles in all spacial directions. The oscillation of monoterpene emission is a consequence of the regulation of the CIN gene by the circadian clock, with oscillations occurring at the level of transcript and protein accumulations and of enzyme activity. Light/dark or dark/light transition signals synchronize the slow-running endogenous clock. Two strategies for synchronized scent emission have been established in N. suaveolens flowers: (i) the synthesis of volatile organic compounds by a multi-product enzyme and (ii) the coordination of biosynthetic pathways by a circadian clock.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In addition to colour and shape, scent is an important directory for pollinators in the process of flower localization (Dobson 2006, Andersson 2006, Ayasse 2006). Many plants emit variable floral scents with respect to quality (composition) and quantity (Knudsen and Gershenzon 2006, Knudsen et al. 2006). Parameters that determine this variability are specific metabolic features or physiological conditions of the floral organ, the developmental stage of the flower, the time of day and environmental cues (temperature, light intensity, etc.). Benzenoids, fatty acid derivatives and terpenoids are the major components of floral scents (Knudsen et al. 2006). In particular, monoterpenes, the C10 members of the terpenoid family, are common floral scent compounds. Terpenoids are generally known to fulfil protective functions during plant development in vegetative and reproductive organs of angiosperms and gymnosperms (Bohlmann et al. 2000), but such terpenoids probably adopted another function during evolution and became essential components for pollinator attraction (Chen et al. 2003).
Terpenoids are synthesized by a special class of enzymes, the terpene synthases (Trapp and Croteau 2001). A large group of genes that encode such enzymes have been characterized from vegetative tissues of several angiosperms and gymnosperms (Bohlmann et al. 1998, Davis and Croteau 2000, Aubourg et al. 2002). All terpene synthases exhibit similar physical and chemical properties, for example, requiring divalent metal ions as cofactor for catalysis, and all operate by an unusual electrophilic reaction mechanism. However, the tremendous range of possible variations in the carbocationic reactions (cyclization, hydride shifts, rearrangements, termination steps) permits the production of essentially all feasible skeletal types, isomers and derivatives (Trapp and Croteau 2001). A characteristic of many terpene synthases is the simultaneous formation of many products (multi-product enzymes).
Although terpenoids comprise a compound class that is emitted by many flowers, the number of genes/enzymes known to synthesize floral terpenoids is presently limited (Knudsen et al. 2006, Bohlmann et al. 1998, Dudareva et al. 2003). The first isolated gene of this group was that for linalool synthase (LIS) from Clarkia breweri, which uses geranyl pyrophosphate (GPP) to synthesize a single acyclic monoterpene alcohol, linalool (Pichersky et al. 1995). The reaction catalyzed by LIS is considerably simpler than the coupled isomerization-cyclization reaction catalyzed by many monoterpene cyclases. The germacrene D synthase from Rosa hybrida is a sesquiterpene synthase, which uses farnesyl pyrophosphate as substrate to synthesize germacrene D (Guterman et al. 2002). One (E)-β-ocimene synthase and two β-myrcene synthases have been isolated from floral tissue of snapdragon (Antirrhinum majus), each synthase being responsible for the production of the single olefin monoterpenes (E)-β-ocimene and β-myrcene (Dudareva et al. 2003). Two flower-specific terpene synthases (CIN, OCI) have recently been isolated from satsuma mandarins (Citrus unshiu Marc), one synthase producing 1,8-cineole and the other (E)-β-ocimene (Shimada et al. 2005). Together, these presently known floral monoterpene synthases (LIS, CIN, OCI and MYR) are single-product enzymes, and only one floral multi-product enzyme/gene from A. thaliana (At3g 25810) has been mentioned (Chen et al. 2004).
The genus of Nicotiana comprises more than 70 species, several of them being night-blooming and hawkmoth-pollinated, such as N. suaveolens, N. sylvestris, N. benthamiana and N. alata (Goodspeed 1954, Raguso et al. 2003 and 2006). Detailed lists of the scent composition of N. suaveolens have been previously established (Loughrin et al. 1990, 1991, 1993, Raguso et al. 2003). The whole plant emits 25 compounds during the day and, during the night, the compound number increases to 41. The most complex mixture of 31 constituents are emitted from N. suaveolens flowers during the night, many of them being methylated compounds such as methyl benzoate and methyl salicylate, but respectable amounts of mono- and sesquiterpenes are also emitted, e.g. the monoterpenes 1,8-cineole, limonene, sabinene, β-β-myrcene, (E)-β-ocimene, α-pinene, β-pinene and α-terpineole and the sesquiterpenes β-caryophyllene, α-humulene, (E)-β-farnesene and (E,E)-β-farnesene. Although the quantitative and qualitative results determined by the Loughrin and Raguso research groups are different, similarities can be found regarding N. suaveolens scent composition: (i) the major monoterpene in the floral scent of N. suaveolens is 1,8-cineole and (ii) a set of at least five monoterpenes accumulate simultaneously to much higher levels at night (Loughrin et al. 1993, Raguso et al. 2003). Different scenarios may explain such synchronized terpenoid emission patterns: either synchronized transcriptional and/or post-transcriptional regulation of the expression of several individual single-product monoterpene synthases manifest coordinated emission or one multi-product enzyme exists that synthesizes a set of monoterpenes simultaneously. To investigate which possibility occurs in the flowers of N. suaveolens, we have isolated monoterpene synthase(s) from a N. suaveolens petal cDNA library and have undertaken a biochemical characterization of the enzyme overexpressed in Escherichia coli. Additional studies concerned the tissue and cellular localization of the enzyme in the flowers and the elucidation of regulation steps involved in the precise timing of emission.
Materials and methods
Plant material and plant growth
Nicotiana suaveolens plants were grown on Vermiculite (Deutsche Vermiculite Dämmstoffe GmbH, Sprockhövel, Germany) in growth chambers under long-day conditions (16 h illumination at 160 μEm−2 s−1 and 22°C, 8 h darkness at 18°C). Plants were watered with Hoaglands solution. To determine the flower age at the time of harvest, flowers were marked when flower buds opened at 6 pm.
For the cDNA library construction, flowers were harvested from approximately 3-month-old plants at intervals of 3 h over a period of 2 days (1st and 2nd day after flower opening) and then pooled.
To determine the transcript levels in different organs and tissues, plant material (flowers, flower parts, stems, leaves, roots) were harvested at 6 pm from 3-month-old plants, frozen in liquid nitrogen and stored at −70°C.
In order to examine the expression profiles of plants grown under long-day conditions (16 h L/8 h D; 10 pm to 6 am darkness), four flowers (approximately 460 mg fresh weight) were harvested at indicated time points at days 1–6 after flower opening. At days 5 and 6, the flowers showed obvious signs of senescence. Plants were also grown under varied light regimes (constant illumination LL, constant darkness DD, and shifted LD cycles). After growth under long-day conditions, the plants were transferred into LL conditions for four days, after which they were returned to long-day conditions for three days. The light was then switched off and the plants were kept under DD for three days. Thereafter, light conditions were again shifted (16 L/8 h D; 4 pm till midnight).
RNA isolation and Northern blot analysis
RNA was isolated according to Cheng and Seemann (1998) and analysed by Northern blot. Samples of 400 mg of frozen plant material was ground with liquid nitrogen and 3.5 ml extraction buffer and rinsed with 1 ml extraction buffer (250 g guanidinium isothiocyanate, 293 ml double-distilled H2O, 17.6 ml 0.75 M sodium citrate, 26.4 ml 10% (w/v) lauryl sarcosinate solubilized at 65°C; pH 7.0; shortly before use, 36 μl β-mercaptoethanol was added to 5 ml extraction buffer). Each sample was centrifuged (20 min, 4°C, 13,000 rpm) and the supernatant was mixed with 4 ml acidic phenol, 0.5 ml chloroform/isoamyl alcohol and 0.4 ml 2 M sodium acetate and incubated for 15 min at 4°C. Centrifugation (20 min at 4°C, 10,000 rpm) yielded a protein-free supernatant. One volume of 2 M potassium acetate was added, incubated for 30 min at 4°C and centrifuged (25 min, 4°C, 4,400g). RNA was precipitated by the addition of 0.6 volumes of cold isopropanol (45 min at −20°C) and centrifugation (20 min, 4°C, 5,000 rpm), resuspended in 400 μl RNase-free double-distilled H2O and 100 μl 10 M LiCl and incubated at 4°C overnight. RNA was precipitated at 20 min, 4°C, and 12,000 g, resuspended in double-distilled H2O and stored at −70°C.
Samples of 10 μg total RNA were separated by formaldehyde denaturing gel electrophoresis and transferred to nylon membranes (Roche Diagnostics GmbH, Mannheim, Germany). The RNA was fixed to the membrane by UV-crosslinking. The 379-bp DNA probe (forward primer for terpene synthase from Nicotiana suaveolens: NsTps1forw, and reverse primer for terpene synthase from Nicotiana suaveolens: NsTps1rev; see below) was labelled with digoxygenin-11-dUTP according to the protocol of the manufacturer (Roche Molecular Biochemicals, Mannheim, Germany). Hybridization and detection with CDP-Star were also performed as described in the manufacturer’s manual (Roche Molecular Biochemicals, Mannheim, Germany). Chemiluminescence signals were detected and quantified with an luminescent image analyser (LAS-1000) and a software image gauge (Fujifilm, Tokyo, Japan). CIN mRNAs were normalized to rRNA levels and the CIN mRNA/rRNA ratios were calculated. The sample with the highest expression level within a given experiment was set to 100%. The results were based on one hybridization of two independent RNA preparations (experiments regarding plant organs, flower development, and long-day conditions) or depended on two hybridizations of one RNA preparation (experiments with varied light regimes). Standard protocolls were used for reverse transcription/polymerase chain reactions (RT-PCRs) to determine CIN mRNA accumulation (NsTps1forw: 5′TAGTGAAACTCATACTGAGGA3′, NsTps1rev: 5′ATGGCAGATTCTCTGCTAAG3′).
RNA was extracted according to Chang et al. (1993) when used for the construction of the cDNA library because this method did not rely on a phenol extraction. Samples of 0.5 to 1 g frozen plant material were ground with liquid nitrogen and 15 ml preheated (65°C) extraction buffer (2% cetyltrimethylammonium bromide (CTAB), 2% polyvinylpyrolidone (PVP) 40, 100 mM Tris–HCl pH 8.0, 25 mM EDTA, 2 M sodium chloride), after which 0.4% β-mercaptoethanol was added. The extract was then incubated for 10 min at 65°C, twice extracted with 15 ml chloroform/isoamyl alcohol (24:1, v/v) and centrifuged (20 min, room temperature, 10,000 rpm). A 1/4 volume of 10 M LiCl was added to the supernatant, which was subsequently incubated overnight on ice and centrifuged (30 min, 4°C, 10,000 rpm). The pellet was resuspended in 0.7 ml pre-warmed (60°C) SSTE buffer (1 M sodium chloride, 0.5% SDS, 10 mM Tris–HCl pH 8.0, 1 mM EDTA) for 5 min at 60°C. Two extractions with 0.7 ml chloroform/isoamyl alcohol were performed and the products were centrifuged (10 min, room temperature, 10,000 rpm); 2 volumes ethanol were added to the supernatant to precipitate the RNA (60 min at −70°C, centrifugation at 30 min, 4°C, 13,000 rpm), which was resuspended in RNase-free double-distilled H2O and stored at −70°C (yield: 1.15 mg RNA/3.6 g petal fresh weight).
Poly A+RNA was isolated from flower total RNA by using the oligotex-dT mRNA kit (Qiagen, Hilden, Germany). Purification was performed according to the manufacturer’s protocol, except that column purification was repeated twice (500 μg total RNA yielded ca. 10 μg poly A+ RNA).
cDNA libray construction and gene isolation
To synthesize cDNA, a ZAP Express® cDNA Synthesis Kit (Stratagene, Amsterdam, The Netherlands) was used. A total of 5 μg polyA+RNA was used for first-strand synthesis. The cDNA preparation was separated on a 1 ml Sepharose CL®-2B column (STE buffer, 0.1 M sodium chloride, 20 mM Tris-HCl pH 7.5, 10 mM EDTA) and fractions of three drops were taken. Fractions were extracted with phenol/chloroform (1:1; v:v) and precipitated with ethanol overnight (centrifugation, 60 min, 4°C, 13,000 rpm). Samples of 200 ng DNA of 1.5–3.5 kbp was ligated into pBK-CMV and transformed into E. coli XL-Blue MRF’ according to the manufacturer’s protocol. For packaging, the ZAP Express® Gigapack® III Gold Cloning Kit was used and the experiment was performed strictly as described in the instruction manual. The titer of the primary library was 5 × 105 pfu/ml. This library was amplified (1 × 108 pfu/ml) in phages to produce a stable cDNA library, which was stored in 1 ml aliquots at −70 °C, after addition of dimethylsulphoxide to a final concentration of 7%.
cDNA library aliquots of approximately 20,000 pfu per plate were incubated with E. coli host cells in top Agar for 8 h. Nylon membranes (Roche Diagnostics, Mannheim, Germany) were placed onto the agar for 2 min or 4 min and filter lifts and hybridisation was performed according to the DIG System User’s Guide for Filter Hybridisation (Roche Diagnostics, Mannheim, Germany). A 483-bp fragment of a potential monoterpene synthase of N. suaveolens was used as a probe (primer N.alaRT1 and N.alaRT2), and the obtained clones were further investigated.
The excision of the pBK CMV phagemid was performed with the E.coli strain XLOR and the helper phage R 408 (Stratagene, Amsterdam, The Netherlands) and the resulting bacterial colonies were stored at −70°C.
Gene sequencing and tree construction
Genes were sequenced by using the LI-COR DNA-Sequencer (MWG-Biotech, Ebersberg, Germany) and a professional sequencing service (AGOWA, Berlin, Germany). Complete sequences were compared with the sequences present in the NCBI data bank by using Clustal W/Align X. Phylogenetic trees were generated by using PAUP 4.0 b10 and visualized with TreeView. Data base searches were performed in March 2007. Only functionally verified and assigned monoterpene synthases are included in the tree construction.
Heterologous expression and preparation of antibodies
The CIN gene of N. suaveolens was amplified by RT-PCR and cloned into the pENTR Directional TOPO vector (Invitrogen, Karlsruhe, Germany). By using the Gateway LR-Clonase Enzyme Mix (Invitrogen), the gene was recloned into the pHIS9 vector, which carries a N-terminal polyhistidine (6xHis) tag. The E. coli BL21[DE3]Codon Plus strain was used for the heterologous expression of the protein and the induction was performed at 16°C overnight with 0.5 mM isopropyl thiogalactoside. Cells from a 50-ml induced culture were harvested at 4°C and 6,000 g, resuspended in 5 ml extraction buffer (50 mM Tris–HCl, pH 7.5, 10% glycerol, 10 mM β-mercaptoethanol), disrupted by sonication (3 × 1 min, 50% power level) and centrifuged at 6,000 g at 4°C for 20 min. Plasmid was transformed into the E. coli overexpression strain BL21[DE3]. The overexpressed protein was purified by Ni-NTA affinity chromatography according to the manufacturer’s recommendations (Qiagen, Hilden, Germany) and eluted with 500 μl extraction buffer containing 250 mM imidazol. Polyclonal antibodies against the purified protein were produced by Biotrend (Cologne, Germany) and Davids Biotechnology (Regensburg, Germany) and were highly specific to the CIN of N. suaveolens.
GC-MS analysis and SPME
The purfied protein was used for enzyme assay with GPP (Echelon Research Laboratories, Salt Lake City). Each assay was carried out in a volume of 1 ml, containing 100 μl purified protein, 60 μM GPP, 20 mM MgCl2, 0.5 mM MnCl2, 2.5 mM dithiotreitol (DTT), 10% glycerol, 50 mM HEPES-KOH, pH 8.0. After incubation for 3 h at 30°C, a solid phase micro extraction PDMS-100 fibre (Supelco, Bellefonte, PA) was inserted into the tube and incubated for 15 min at 42°C. The solid phase micro extraction fibre was injected into a Shimadzu QP5000 gas chromatograph coupled to a mass spectrometer for analysis (GC-MS). Separation was performed on a DB5-MS column (60 m × 0.25 mm × 0.25 mm; J + W Scientific Folsom, CA, USA) with helium as the carrier gas (flow rate of 1.4 ml/min) on a temperature gradient from 50°C (2 min hold) and a ramp of 10°C/min to 275°C (3.5 min hold). Mass spectra were obtained by using the scan modus (total ion count, 40–280 m/z). Compound identity was confirmed by comparison of mass spectra and retention times with those of available standards, by comparison of the obtained spectra with spectra in the library of the National Institute of Standards and Technology (NIST 147) and by comparison of Kovats indices.
Enzyme assay
His-tagged purified enzyme (8 μg) was used for the enzyme assay. The enzyme assay buffer (final volume of 200 μl) consisted of 50 mM Bis Tris propane buffer (pH 7) containing 10% glycerol, 0.5 mM dithiothreitol, 20 mM MgCl2 and 0.5 mM MnCl2, 5 mM DTT. The putative synthase was incubated with 109.4 μM GPP (Echelon Biosciences) at a temperature of 30–32°C. To check the specific bivalent metal ion preferences, the enzyme was incubated with various concentration of MgCl2 (0–90 mM) and MnCl2 (0–10 mM). For testing the optimal pH value, the enzyme was incubated with Bis Tris buffer with a pH ranging from 5.5 to 6.5 and Bis Tris propane buffer from 6.5 to 9.0 at intervals of 0.5 units. The optimal temperature of the synthase was investigated by incubating the assay at different temperature (15–45°C). The enzyme assays were overlaid with 300 μl hexane. After 3 h, the reaction was stopped by shaking the tubes for 2.5 min, followed by a 2-min centrifugation. The hexane phase was analysed with GC-MS as described above. Addition of a defined concentration of an internal standard (cis nerolidol) into the hexane phase allowed quantification.
Determination of the Km value: 1 μg purified enzyme was incubated with GPP concentrations ranging from 10 to 60 mM under optimal linear assay conditions (50 mM Bis Tris propane buffer (pH 7.8), 50 mM MgCl2, 3 mM MnCl2, 10% glycerol, 5 mM DTT) at 35°C. The assays were overlaid with 180 μl hexane and the reaction was stopped by shaking for 1 min and a 2-min centrifugation. Immediately afterwards, 50 μl of the hexane phase was added to 2 ml liquid scintillation solution. Radioactivity was measured in a liquid scintillation counter (TriCarb, Packard BioScience). As a control, the assay was performed without enzyme.
Enzyme assay for crude protein extract
Aliquots (100 μl) of crude protein extract were incubated with 100 μl 50 mM Hepes KOH buffer (pH 8) containing 10% glycerol, 5 mM dithiothreitol (DTT), 20 mM MgCl2, 0.5 mM MnCl2. The putative synthase were incubated with 109.4 μl GPP (Echelon Biosciences) at 30–32°C for 3 h. The enzyme assays were overlaid with 200 μl hexane. After 3 h, the reactions were stopped by shaking the tubes for 2.5 min and a 2-min centrifugation. The hexane phase was analysed with GC-MS as described above. Cis nerolidol was used as an internal standard.
Immunolocalization of CIN
Flowers of N. suaveolens of various ages were harvested at the time of maximum enzyme activity (6 am, 1 day post-anthesis). Tissue pieces (2 × 3 mm) from defined areas of the petals were cut and immediately submersed in 4% (w/v) paraformaldehyde supplemented with 0.1% (v/v) Triton-X 100 in phosphate-buffered saline (PBS, 540 mM NaCl, 12 mM KCl, 6 mM KH2PO4, 32 mM Na2HPO4; pH 7.0–7.5). In order to remove all air from the parenchyme, samples were vacuum-infiltrated and final fixation was then performed at room temperature for 2 h. After being washed with PBS, the tissue pieces were stepwise dehydrated to 100% ethanol, passed through an ascending ethanol/polyethylene glycol (25, 50, 75% PEG) series and finally embedded in a mixture of PEG 1500 and PEG 4000 (2:1, v/v). Sections of 4 μm thickness were prepared on a sliding microtome (Jung, Heidelberg, Germany) and collected on poly-l-lysine-coated slides. After being rinsed with PBS, the sections were treated with NH4Cl (0.1 M) to block aldehydes, rinsed again with PBS and incubated for 30 min with 5% (w/v) bovine serum albumin (BSA) in PBS to reduce unspecific binding. Incubation with the specific antibody (antiserum against Nicotiana suaveolens terpene synthase raised in rabbit, diluted 1:500 in 5% (w/v) BSA in PBS) was performed overnight at 4°C. After being washed with 0.1% (w/v) and 1.0% (w/v) BSA in PBS, the sections were incubated with the secondary antibody, viz. anti-rabbit IgG labelled with alkaline phosphatase (diluted 1:500 in PBS; Sigma, Karlsruhe, Germany), for 90 min at 37°C. Sections were rinsed for 5 min with detection buffer (see Western blotting), incubated with 250 μl detection buffer and NBT/BCIP (4.5 μl NBT, nitroblue tetrazolium chloride per ml; 3.5 μl BCIP, 5-bromo-4-chloro-3-indolyl phosphate per ml) for 30 min. The reaction was stopped with 0.01 M Tris/HCl and 1 mM EDTA. Sections were covered with coverslips and sealed with nail varnish. Control sections were treated the same way but the primary antibody was replaced by preimmune serum (Biotrend Chemikalien GmbH). Micrographs were taken via a Carl Zeiss microscope (Axiostar plus) and a Canon powershot G5 camera.
Preparation of crude protein extract
Three petals were harvested every 3 h and placed in an ice-cold mortar. Samples of 0.2 g of the petals were extracted with 1 ml buffer containing 0.1 M sodium phosphate, 0.25 mM saccharose, 5 mM MgCl2, 1 mM CaCl2, 25 mM Na2S205, 2 mM DTT, 5 mM ascorbate, 2 μl mercaptoethanol, 0.1 g PVPP (poly vinyl polypyrolidon) and protease inhibitor cocktail tablets (Roche). The crude extracts were centrifuged for 15 min at 13,000 rpm (4°C). The supernatant was removed. Following addition of 10% glycerol, the crude extract was at −20°C. The crude extract was used for Western blots and enzyme activity tests.
SDS-polyacrylamide gel electrophoresis and Western blot analysis
Samples (15 μg) of the crude protein extract were loaded onto polyacrylamide gels (12.5%) and electrophoretically separated (Miniprotean, Bio-Rad Laboratories, Hercules, CA). Proteins were transferred to a PVDF (poly vinyl difluoride) membrane (Roth. Karlsruhe, Germany) by using a mini-tank blotting gel cassette (XCell II, Novex, San Diego, CA). The membranes were placed overnight in a Tris-buffered blocking solution (TBS; 20 mM) with 0.05% (v/v) Tween 20, 4% (w/v) skim milk, 1% (w/v) BSA. After incubation with the specific antibody (diluted 1:3,000 in blocking solution) for 1.5 h and repeated washes with TBS containing 0.05% (v/v) Tween 20 (TBS-Tween), the membrane was incubated with the secondary antibody (anti-rabbit alkaline phosphatase conjugate, diluted 1:20,000 in TBS-Tween; Sigma-Aldrich, St. Louis, MS) and washed again with TBS-Tween and TBS. The membrane was equilibrated in detection buffer (100 mM Tris–HCl, 150 mM NaCl, 50 mM MgCl2), incubated with CDP-Star (Roche, Mannheim, Germany; 0.25 μM in detection buffer) under darkness and analysed in a luminescent image analyser (LAS-1000, Fujifilm, Japan). Luminescence (17 min) was quantified with Fujifilm Image Gauge software. After the measurement of luminescence, the proteins were additionally stained with NBT/BCIP (50 mg/ml in dimethylformamide; Roche, Mannheim, Germany; 2:1).
Results
Terpenoid synthesis in N. suaveolens petal extracts at various time points during the day
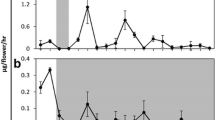
Higher nocturnal than diurnal monoterpene emission from Nicotiana flowers was previously published, and sabinene was shown to be rhythmically emitted with a reduced amplitude under constant illumination (Loughrin et al. 1993 and Raguso et al. 2003; Loughrin et al. 1991, respectively). The rhythmic emission of monoterpenes might be a consequence of fluctuating enzyme activities during the day and night, and during development, and/or a result of altered emission processes. Crude protein extracts of petals from various time points during the day and development were prepared and geranyl pyrophosphate (GPP) was added as the substrate. Terpene synthase(s) in the extracts converted the substrate into several products that were analysed via GC-MS. A typical product pattern obtained with a protein extract isolated from petals at 3 am on the 2nd day of flower opening is shown in Fig. 1A. The following monoterpenes were detected: α-pinene, sabinene, β-myrcene, limonene, 1,8-cineole, (+) α-terpineole and cis-nerolidol (the last-mentioned was added as an internal standard). Petals were then harvested at 3-h intervals from anthesis till the 5th day of flower development. Immediately after harvest, protein extracts were prepared and enzyme assays performed. The amounts of the five major monoterpenes were used to determine enzyme activities at eight time points during the day and at 5 consecutive days during flower development (Fig. 1B). The graphic presentation of the results showed that oscillations occurred in enzyme activity from anthesis till senescence in N. suaveolens flowers. These oscillations appeared synchronously for all five monoterpenes, although this pattern could best be observed for the major product 1,8-cineole. Generally speaking, low enzyme activites were present from 6 am till noon, whereas in the afternoon, activities increased, with maximum of enzyme activities being reached in darkness (100% = 17 pkat/mg total protein). The enzyme activity pattern (Fig. 1B) correlates well with the nocturnal emission pattern of the flowers. A more detailed inspection of the data revealed that (i) the oscillations in enzyme activity exhibited a shift of the enzyme activity maximum into the light phase with age, (ii) the enzyme activity maximum was split into two peaks, except at the day of anthesis and day 4. The biological relevance for this is presently unknown.
Terpene Synthase Enzyme Activity in N. suaveolens Petal Extracts. Plants were grown under 16 h light (L) and 8 h darkness (D; 10 pm to 6 am). (A) Petals of N. suaveolens were harvested from 2-day-opened flowers at 3 am and protein extracts were immediately prepared. The substrate geranyl pyrophosphate (GPP) was added and volatiles were analysed via GC-MS. Six monoterpene products were identified. Cis-nerolidol was added as an internal standard. (B) Petals were harvested at 3-h intervals from flowers at various developmental ages (day of anthesis until the age of 5 days), protein extracts were immediately prepared and enzyme assays were performed. Specific enzyme activities (pkat/mg) for five monoterpene products were determined for each time point during the day and during flower development. The highest specific activity was set at 100% (17.1 pkat/mg protein) and the relative specific activities were calculated respectively; mean values are shown (n = 2)
The oscillations of monoterpene emission from N. suaveolens flowers can be explained on the basis of rhythmic enzyme activities but two interesting questions can further be addressed: (i) is the set of emitted monoterpenes synthesized by one multi-product enzyme or individually by single product enzymes, and (ii) are these changes of enzyme activity attributable to regulation at the post-translational, translational and/or transcriptional level. To answer these questions, the isolation and characterization of a/the gene(s) encoding a/the respective terpene synthase/s are necessary.
Isolation of a CIN gene
A clone encoding a terpene synthase was isolated from a N. suaveolens cDNA library by using an internal sequence of a LIS from N. alata (Ganz and Piechulla, unpublished results). The first clone isolated was incompletely spliced and contained an intron of 148 nucleotides at nucleotide position 1,297 (C terminal part of the enzyme). A second isolated clone was incomplete at the 5′ end but, otherwise, the nucleotide sequence was identical to that of clone 1. Both clones were used as matrices to synthesize a complete clone. Specific primers were designed to amplify the N terminal and the C terminal part of clones 2 and 1, respectively. Both parts were used in another PCR to amplify the complete clone of 1,602 nucleotides (RR-motif with additional methionine to stop codon, accession No. EF175166). The sequence of this clone contained the full length of the coding sequence (without the transit peptide) of a terpene synthase (534 amino acids, theoretical molecular mass: 64.55 kD).
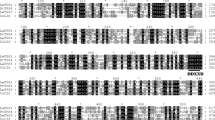
Two characteristic motifs of terpene synthases, the DDXXD motif (amino acids 359–363) and the RR(X)8W motif (amino acids 79–89), are present in the Nicotiana suaveolens terpene synthase (Fig. 2). Both motifs have a well-known function in the isomerization/cyclization process and binding of the divalent metal ion during the reaction mechanism (Asby and Edwards 1990, Facchini and Chapell 1992, Wise et al. 1998). Further motifs, which are also highly conserved in terpene synthases are the RRW (amino acids 307–309), the NMK (amino acids 392–394) and the CYMNE (amino acids 523–527) sequences. Finally, a 78 amino acid long sequence upstream of the RR(X)8W motif at the N terminal end of the enzyme is serine-rich (12×) and threonine-rich (3×) and therefore most likely a chloroplast target signal.
Sequence Alignment of Cineole Synthases (CINs). A CIN was isolated from a floral cDNA library of N. suaveolens (N.s.), sequenced and aligned with homologous genes from Citrus unshiu (C.u.), Arabidopsis thaliana (A.t.), Salvia officinalis (S.o.) (BLAST Search, Altschul et al. 1990, alignment with Clustal W). Filled square (1) RR-motif, filled square (2) RWW-motif, filled square (3) DDXXD-motif, filled square (4) CYMNE-motif
The amino acid sequence of the gene from N. suaveolens was aligned with 79 monoterpene synthases which were at least partly functionally characterized. The highest identity was found with a LIS from N. alata (Ganz and Piechulla, unpublished results). The function of the isolated terpene synthase from N. suaveolens could not be predicted because sequence identity or similarity in this context is not a distinctive criterion for functional analysis (Bohlmann et al. 1998).
The analysis of the phylogenetic relationship of the terpene synthase from N. suaveolens by using PAUP 4.0 and the neighbour-joining method has clustered this gene into the terpene synthase B subfamily, which combines monoterpene genes from a variety of angiosperm species (Fig. 3). This subfamily is distinct from other monoterpene synthase subfamilies, such as terpene synthase D (gymnosperms), terpene synthase G (e.g. OCI and MYR of Antirrhinum majus) and terpene synthase F (LIS of Clarkia breweri). The N. suaveolens terpene synthase clusters together with two sequences of Solanum lycopersicum in a monophyletic group together with the cluster of the Labiatae. The phylogenetic analysis of monoterpene synthases reveals that sequence identities or similarities are found between plant families but not between the functions of the enzymes, e.g. the N. suaveolens terpene synthase is less similar to the CINs from Salvia officinalis and Arabidopsis thaliana than to the LIS of N. alata.
Phylogenetic Relationship of CIN from N. suaveolens to other Monoterpene Synthases. Unrooted neighbour-joining phylogenetic tree based on amino acid sequence similarities. The tree was created with PAUP (version 4.0b10) and displayed by using TreeView. Gaps in the alignments (Clustal W) were removed for analysis. Bootstrap values below 50% were not included (1,000 replicates). Plant species and accession numbers used for tree constructions:Agastache rugosa LIM AY055214, Antirrhinum majus OCI AY195607, Antirrhinum majus MYR1 AY195608, Antirrhinum majus MYR2 AY195609,Arabidopsis thaliana MYR/OCI AF178535, Arabidopsis thaliana CIN AY691947, Arabidopsis thaliana LIM AB028607, Arabidopsis thaliana MTS AF497484, Arabidopsis thaliana LIM L161544, Arabidopsis thaliana OCI AY151086, Arabidopsis thaliana LIS AF497485, Artemisia annua LIS AF154124, Artemisia annua PIN AF276072, Cinnamonum tenuipile GER AJ457070, Citrus limon PIN AF514288, Citrus limon TER AF514286, Citrus limon LIMAF514287, Citrus limon LIM AF514289, Citrus unshiu OCI AB110642, Citrus unshiu MTS AB110638, Citrus unshiu PIN AB110641, Citrus unshiu TER AB110640, Citrus unshiu TER AB110639, Citrus unshiu LIM AB110636, Citrus unshiu LIMAB110637, Clarkia concinna LIS AF067602, Clarkia breweri LIS2 AF067603, Clarkia breweri LIS U58314, Eucalyptus globulus MTS2 AB266391, Eucalyptus globulus MTS1 AB266390, Lavandula angustifolia LIS DQ263741, Lavandula angustifolia LIM DQ263740, Lavandula latifolia LIS DQ421801, Lotus corniculatus var. japonicus OCI AY575970, Melaleuca alternifolia MTS AY279379, Mentha aquatica LIS AY083653, Mentha longifolia LIM AF175323, Mentha spicata LIM L13459, Nicotiana alata LIS -, Nicotiana suaveolens CIN EF175166, Ocimum basilicum TES AY693650, Ocimum basilicum MYR AY693649, Ocimum basilicum FEN AY693648, Ocimum basilicum LIS AY693647, Ocimum basilicum GER AY362553, Perilla citriodora LIS AY917193, Perilla citriodora GER DQ088667, Perilla frutescens var. crispa LIS AF444798, Perilla frutescens GER DQ234300, Perilla frutescens var. crispa GER DQ897973, Perilla frutescens MYR AF271259, Perilla frutescens LIM AF241793, Perilla frutescens LIMAB005744, Perilla frutescens var. acuta LIM D49368, Perilla frutescens LIM AF241792, Perilla frutescens var. frutescens LIM AF317695, Perilla citriodora LIM AF233894, Perilla citriodora LIM AF241790, Quercus ilex PIN AM283099, Quercus ilex MYR AJ304839, Rosmarinus officinalis CINDQ839411, Rosmarinus officinalis LIM DQ421800, Salvia officinalis CIN AF051899, Salvia officinalis SAB AF051901, Salvia officinalis BOR AF051900, Salvia stenophylla CAR AF527416, Schizonepeta tenuifolia LIM AF282875, Solanum lycopersicum MTS2 AY840092, Solanum lycopersicum MTS1 AY840091 BOR Bornyl diphosphate synthase, CAR Carene synthase, CIN Cineol synthase, FEN Fenchol synthase, GER Geraniol syntase, LIMLimonene synthase, LIS Linalool synthase, MTS Monoterpene synthase, MYR Myrcene synthase, OCI Ocimene synthase, PIN Pinene synthase, SAB Sabinene synthase, TER Terpinene synthase, TES Terpinolene synthase
Overexpression of the recombinant gene in E.coli and purification of the his-tagged enzyme with affinity chromatography revealed highly pure enzyme fractions as visualized by SDS gel electrophoresis and Coomassie blue staining (Fig. 4A). The purified enzyme was tested in an assay with tritium-labelled GPP or farnesyl pyrophosphate as substrate. H3-GPP was preferentially used as a substrate (data not shown). The products synthesized by the recombinant enzyme were analysed by using SPME and GC-MS. Seven products were detected (Fig. 4B): 1,8-cineole, limonene, sabinene, E-ß-ocimene, β-myrcene, α-pinene, and α-terpineole. Thus, the enzyme formed five cyclic (1,8-cineole, α-terpineole, α-pinene, sabinene and limonene) and two acyclic (β-myrcene, E-ß-ocimene) compound from the intermediate linalyl pyrophosphate (Fig. 4C). Since all compounds had previously been detected in the scent of N. suaveolens (Raguso et al. 2003), this result indicated that the majority of monoterpene products were synthesized by one enzyme. The major compound of this terpenoid mixture was 1,8-cineole, the reason for this multi-product enzyme originally being named CIN.
Determination of the Product Spectrum of N. suaveolens CIN. (A) The recombinant N. suaveolens terpene synthase was overproduced in E. coli and the his-tagged enzyme purified via Ni-NTA affinity chromatography. Protein samples of the chromatographic fractions were separated on a 12% SDS gel. M: protein marker, D: flow through, WF: wash fraction, E: eluted proteins with 200 mM imidazol. (B) Purified enzyme fraction E was incubated with GPP for 3 h at 32°C (see Material and Methods). SPME (30 min at 42°C) and GC-MS were used for product analysis. Product identification was performed via mass spectra and comparison of compounds in the NIST 147 library (National Insitute of Science and Technology). (C) Monoterpenes synthesized by CIN. GPP is converted to the enzyme-bound carbocation, which is the reactive intermediate for acyclic and cyclic monoterpene synthesis
The floral CIN from N. suaveolens has a molecular mass of 64.6 kD, with the highest enzyme activities (178 pkat/mg recombinant protein) being determined at 35°C and pH 8; Mg2+ ions are preferred over Mn2+. During incubation for 4 h on ice or 2 h at room temperature the enzyme loses approximately 20% of its activity. A data bank search with the CIN of N. suaveolens has revealed four CIN genes/enzymes from other plant species; one gene has been isolated from Salvia officinalis leaves (Wise et al. 1998, Peters and Croteau 2003), the second is expressed in root tissue of A. thaliana (Chen et al. 2004), and the third is found in flowers of Satsuma mandarines (Citrus unshiu Marc, Shimada et al. 2005) (Fig. 2). The CINs of N. suaveolens, A. thaliana, S. officinalis and C. unshiu vary in amino acid sequence identity from 41% to 50%, the molecular masses range from 60–70 kD and the length of transit peptides vary from 27 amino acids to 56 amino acids. The K m values for the substrate GPP are in the range of 0.2–1.1 μM. The root enzyme of A. thaliana exhibits a k cat of 0.001 s−1, whereas the N. suaveolens CIN synthase possesses a 300-fold higher turnover (0.33 s−1), revealing a catalytic efficiency of 2835 M−1 s−1. The product spectra of the A. thaliana root, S. officinalis leaf and N. suaveolens flower enzymes revealed 7–10 monoterpene compounds, composed of 52% to 79% of 1,8-cineole, and the floral CIN of C. unshiu almost exclusively synthesizes 97% 1,8-cineole.
Spatial expression of CIN
The origin of the N. suaveolens CIN was a flower-specific cDNA library. RT-PCR and Northern blots were performed to determine in which organs and tissues the N. suaveolens CIN was expressed. RNA was isolated from plant tissue harvested at 6 pm. This time point was chosen because nocturnal terpene synthase activities were known to be high (Fig. 1B). RT-PCR detected mRNA in buds and flowers but not in leaves, stems or roots (Fig. 5A). A Northern blot with RNA from several flower parts showed high expression levels in petals and stigma/style tissue, whereas in other floral tissues and other plant organs (leaves, shoots, roots, sepals), the levels were very low (Fig. 5B, C). This result indicated the existence of a CIN specifically expressed in petals and stigma/style in N. suaveolens flowers.
Expression of CIN in Various Organs and Tissues of N. suaveolens. (A) RT-PCR with NsTps1 forward and NsTps1reverse primers was performed on 1 μg RNA isolated from flowers, buds, stems, roots and leaves (harvest time: 6 pm). M: marker, +: positive control with cloned CIN in plasmid. (B) Plant tissues were harvested at 6 pm from 3-month-old plants. RNA was isolated and 5 μg total RNA was separated on denaturing agarose gels. RNA was blotted and hybridized with a gene-specific probe (379 nucleotides). A representative hybridization with the CIN probe revealed a transcript of ca. 1800 nucleotides in length. Tissue samples correspond to C. Blots were rehybridized with an 18S rDNA probe to allow normalization. (C) Two independent experiments and duplicated Northern blots were used for quantification of RNA levels (n = 4). Highest expression levels were set at 100% and relative transcript levels were calculated, ± SE is indicated
Thin sections of petals from N. suaveolens were prepared in order to localize the cells or cell layers harbouring the enzyme. Petals from flowers 1-day post-anthesis were incubated with the primary polyclonal antibody specifically detecting the CIN of N. suaveolens, followed by a goat anti-rabbit antibody labelled with alkaline phosphatase. The CIN enzyme was primarily found in the adaxial and abaxial epidermis cells but also to some extend in the mesophyll cells (Fig. 6A). Incubation with preimmune serum gave no antibody binding (Fig. 6B)
Localization of CIN in Petals of Nicotiana suaveolens. Petal lobes of N. suaveolens flowers were harvested at day 1 post-anthesis and embedded in PEG. Sections of 4 μm thickness were prepared and incubated with specific polyclonal antibodies (anti-CIN) (A) or preimmune serum (B), and goat anti-rabbit secondary antibodies labelled with alkaline phosphatase. Staining was performed with NBT/BCIP. Blue stain is present in adaxial and abaxial epidermis and, to a lower extent, in mesophyll cells
Nocturnal and circadian regulation of CIN
One of the goals of our investigations was the elucidation of at what molecular level the nocturnal terpene emission of N. suaveolens was regulated. The observed oscillations at the level of enzyme activity in petal extracts allowed three possible scenarios: the nocturnal activity changes were attributable to oscillations (i) at the protein level, (ii) at the transcript level or (iii) at both levels. Petals were harvested at 3-h intervals throughout the lifespan of the N. suaveolens flower (bud opening to 5-day-old flowers). Protein and RNA were separately extracted at each time point and Western and Northern blots were prepared. The experiment was repeated twice and two blots of each extraction series were made. Representative examples of both analysis are shown in Figs. 7A and 8A. To quantify the protein, a CDP-Star-labelled second antibody was used to allow detection with the LAS system (see Material and Methods; summarized data are plotted in Fig. 7B). The CIN protein levels oscillated from the 1st to the 6th day. The maximum of CIN protein accumulation was initially reached in the late afternoon at the transition from light to darkness, whereas in older flowers, the CIN protein maximum shifted to time points that lay in the middle of the light phase; therefore, the protein levels correlated with the oscillations observed for the enzyme activity.
Determination of CIN Protein Levels at Various Time Points of the Day and During N. suaveolens Flower Development. Plants were grown under 16 h light (L) and 8 h darkness (D; 10 pm to 6 am). (A) Petals of N. suaveolens were harvested at the indicated time points on 5 consecutive days after flower opening. Samples of 15 μg protein extract were separated by SDS-polyacrylamide gel electrophoresis and blotted onto PVDF membrane. Western blots were incubated with polyclonal antibodies against N. suaveolens CIN and the anti-rabbit antibody either labelled with alkaline phosphatase or CDP Star (*); representative blots show the CIN. (B) Chemiluminescence was quantified with the LAS system. The highest protein levels were set as 100%. D: darkness; min/max values are given for anthesis panels, 4th and 5th day post-anthesis; ± standard error is given for day 1 to day 3 post-anthesis based on three replicates of two independent experiments
RNA Expression Profile of CIN of N. suaveolens at Various Times During the Day. Plants were grown under 16 h light (L) and 8 h darkness (D; 10 pm to 6 am). (A) Petals of N. suaveolens were harvested at the indicated time points for 5 consecutive days after flower opening. RNA was isolated and 5 μg total RNA was separated on denaturing agarose gels. RNA was blotted and hybridized with a CIN-specific probe. Blots were rehybridized with an 18S rDNA probe for normalization. (B) For quantification, two independent experiments and duplicate Northern blots were used (n = 4); the highest expression levels were set to 100% and relative transcript levels were calculated; ± SE is indicated
The CIN RNA levels determined at various time points during the day and throughout flower development are shown in representative Northern blots (Fig. 8A). The RNA levels of several experiments were quantified and plotted (Fig. 8B). A characteristic oscillation pattern could be observed with low transcript levels during the day and high levels at the transition from light to darkness. Highest transcript levels were reached at 9 pm. This mRNA pattern was in accordance with the CIN emission and activities and protein levels in petals (Figs. 1B and 7B). The congruent oscillations at the transcript, protein and enzyme activity levels suggested that the nocturnal emission of monoterpenes from N. suaveolens flowers was a consequence of transcriptional control.
Since the transcription level turns out to be the key control point responsible for the evening and night emission of several monoterpenes from N. suaveolens flowers, the question remains as to how the characteristic 24-h emission pattern can be sustained throughout flower development. One possibility is that a circadian clock may be involved in the regulation of the floral CIN of N. suaveolens.
To address this problem, plants were transferred to constant conditions, either continuous illumination (LL) or darkness (DD), to determine mRNA expression patterns under free-running conditions (without the influence of environmental cues). A representative Northern blot is shown in Fig. 9A. Prior to the shift into LL conditions, plants were grown under a light/dark regime (darkness 10 pm till 6 am). Treatment of 3 days under LL revealed a rhythmic mRNA pattern with pronounced amplitudes (left part of Fig. 9B). The maxima of these mRNA accumulations occurred at the subjective L/D transition and tended to shift into the subjective night. This indicated that under free-running conditions, the endogenous clock had a period length longer than 24 h. When plants were transferred back into light/dark conditions (darkness 10 am till 6 pm), pronounced mRNA rhythms with high amplitudes were manifested. The maxima appeared in the middle of the light phase because of the shift of the light and dark regime (darkness from 10 pm till 6 am to 10 am till 6 pm) (middle part of Fig. 9B). Under continuous darkness (DD), a pronounced oscillation pattern was once again observed but with slightly reduced amplitudes. At the first day under DD, the maximum appeared in the second half of the subjective light phase, whereas during the second day under DD, the maximum further tended to be shifted into the subjective dark phase. The oscillations under both constant conditions (LL and DD) revealed period lengths longer than 24 h, reflecting the endogenous clock under free-running conditions in N. suaveolens flowers.
RNA Expression Profiles of CIN in N. suaveolens Petals Under Free-running Conditions. Plants were grown under 16 h light (L) and 8 h darkness (D, 10 pm to 6 am) before they were shifted to continuous light conditions (LL for 3 days). Subjective nights (s.n.) and subjective days (s.d.) are indicated. Thereafter, light conditions were altered to 16 h light and 8 h darkness (darkness from 10 am to 6 pm) for 3 days (DL). Light was then switched off and plants were kept in complete darkness for 3 days (DD). (A) Petals of N. suaveolens were harvested at the indicated time points. RNA was isolated and 5 μg total RNA was separated on denaturing agarose gels. RNA was blotted and hybridized with a CIN-specific probe. Blots were rehybridized with an 18S rDNA probe for normalization. (B) For quantification, duplicate Northern blots of one experiment were used; mean values are shown (n = 2). The highest expression levels were set to 100% and relative transcript levels were calculated
Discussion
In many plant species, the emission of floral scent varies as a result of environmental changes, after pollination, during flower development and during the day. Such alterations are a prerequisite for scent compounds to act as inter- and intra-organismic communication signals. For example, the variation of odour composition of Ophrys sphegodes flowers influences the behaviour of the male pollinator (Ayasse 2006). A decrease or alteration of floral bouquet after pollination changes the attractiveness of these flowers. The advantage of post-pollination changes in scent emission is most likely that pollinators are directed to the unpollinated flowers of the plant.
Another reason for the occurrence of variable scent emission is the high energy and carbon costs required for the synthesis and maintanance of scent volatiles. To limit such costs, plants have evolved mechanisms to optimize the emission of volatile organic compounds (VOCs). Although many plants continuously emit scent at constant levels, other flowers emit scent with increasing and decreasing VOC levels and/or vary the bouquet composition (Tollsten 1993; Pichersky et al. 1994; Schiestl et al. 1997; Wang et al. 1997; Pott et al. 2002). Some flowers emit volatiles periodically, e.g. at specific times during the day or night, and are referred to a diurnal or nocturnal emitters, respectively (Dudareva et al. 2000). This rhythmic scent emission correlates well with the appearance of the type of insect that pollinates the flower, e.g. the day-active insects (bees, bumble bees, butterflies) in contrast to the night-active moths. Although the emission of scent is, in many plant species, limited to certain times during development or day, the bouquet composition can be complex. As a consequence, the coordinated activation of at least three biochemical pathways, the terpenoid, phenylpropanoid and the fatty acid pathways, and their respective enzymes is necessary to allow the simultaneous synthesis and emission of VOCs.
The flowers of Nicotiana suaveolens emit high levels of benzenoid derivatives such as methyl benzoate, methyl salicylate, benzyl benzoate and benzyl salicylate, and eight cyclic and acyclic monoterpenes at night but lower levels during the day (Loughrin et al. 1993; Raguso et al. 2003). The rhythmic and coordinated pattern of emission continues throughout the lifespan of the flower (approximately 5–7 days). Both, methyl benzoate and methyl salicylate have been shown to be synthesized by one methyltransferase (the S-adenosyl methionine: benzoic acid/salicylic acid carboxyl methyltransferase, BSMT), which facilitates the simultaneous appearance of both compounds in the scent of N. suaveolens (Pott et al. 2004). The underlying mechanisms responsible for the coordinated increase and decrease of floral monoterpene volatiles in the scent is not known for N. suaveolens or for other plant species. To address this question, we have isolated and characterized a monoterpene synthase from N. suaveolens floral tissue. A single synthase (1,8-CIN) synthesizes at least six out of eight floral monoterpenes in N. suaveolens. From the highest to lowest levels, this enzyme produces 1,8-cineole > β-myrcene = limonene = sabinene > (E)-β- ocimene = α-terpineole = α-pinene, which approximately reflects the gradient of monoterpene emission from the flower (1,8-cineole > β-myrcene = limonene = sabinene = (E)-β-ocimene > α and β-pinene > α-terpineole). The isomer β-pinene has not been identified in the GC-MS profile of the overexpressed enzyme. Since β-pinene is only a minor scent product of N. suaveolens flowers, the amount of this compound synthesized by the isolated enzyme is probably under the detection limit in our system.
The simultaneous emission of seven monoterpenes from N. suaveolens flowers can now be explained by the presence of a single gene/enzyme, which is a multi-product monoterpene synthase. Interestingly, many other Nicotiana species also emit the monoterpenes of the “cineole cassette”, e.g. N. longiflora, N. plumbaginifolia, N. langsdorffi, N. boraniensis, N. forgetiana, N. alata, and N. mutabilis, whereas N. rustica, N. africana, N. cavicola and N. ingulba do not emit these monoterpenes, and N. sylvestris emits only β-myrcene and (E)-β-ocimene (Raguso et al. 2003, 2006). N. longiflora, N. plumbaginifolia, N. langsdorffi, N. boraniensis, N. forgetiana, N. sylvestris and N. alata are combined into the section Alatae, which is distinct from the section Suaveolentes (N. suaveolens, N. africana, N. cavicola and N. ingulba) (Aoki and Ito 2000; Chase et al. 2003). The taxa of the Alatae section emit the “cineole cassette of monoterpenes”, whereas the Suaveolentes do not emit these monoterpenes. An exception however is the namegiving species N. suaveolens. Apparently, many taxa of the Suaveolentes have either recently lost the CIN gene, never evolved it or do not express it, whereas active enzyme is present and expressed in N. suaveolens flowers. The subgenus Rustica is distinct from the Alatae and Suaveolentes. With regard to monoterpene emission, N. rustica flowers resemble many taxa of the Suaveolentes. The emission of the two monoterpenes β-myrcene and (E)-β-ocimene from N. sylvestris (Alatae) flowers is distinct from all other investigated Nicotiana species. Possible explainations might be: (i) the Alatae-specific CIN has undergone mutations that have altered the enzyme and its product spectrum or (ii) two new genes have been established after gene duplication and diversification. Two individual genes synthesizing the floral monoterpenes β-myrcene or (E)-β-ocimene have recently been isolated from Antirrhinum majus (Dudareva et al. 2003). Both genes are phylogentically distinct from other terpene synthases clustering in the terpene synthase G subfamily. The sequencing and functional analysis of the gene(s)/enzymes(s) might clarify the genetic situation in N. sylvestris.
The enzyme properties of the four CINs isolated and characterized so far are similar. Each of these enzymes is specifically expressed either in leaves, roots, or flower tissue and three are clearly multi-product enzymes synthesizing almost the same product spectrum, with a contingent of 50–80% of 1,8-cineole. The flower-specific CIN from Citrus however produces 97% cineole but no minor products have been identified, whereas flower-specific A. thaliana enzyme (At3g 25810) does not synthesize 1,8-cineole but the other members of the “cineole cassette”. All four CINs are phylogenetically related and cluster in the terpene synthase subfamily B, which comprises most of the angiosperm monoterpene synthases. The enzymes present in the floral tissue of A. thaliana and C. unshiu have lost the ability to synthesize the complete “cineole cassette of monoterpenes”; in citrus, this may be attributable to the specific breeding of the horticulture cultivar Satsuma mandarin Marc. Miyagawa-wase.
The regulation of the rhythmic emission of scent volatiles from N. suaveolens has also been addressed in this investigation. Three possible control points or levels, which may theoretically facilitate the oscillations of monoterpene emission have been examined: the transcriptional (mRNA, Fig. 8), translational (protein, Fig. 7) and post-translational (enzyme activity, Fig. 1B) levels. At all three levels, oscillations with a maximum at the transition from day to night have been observed. This leads to the conclusion that fluctuations at the transcript level are the critical regulatory check point and that the alterations in protein and enzyme activity are a consequence of transcript oscillations. However, regulation might additionally occur at the translational and post-translational level, or at the level of substrate supply. For the first time, we have now demonstrated that, in N. suaveolens petals, all three levels exhibit oscillations, unlike those of the carboxyl methyltransferases previously investigated, e.g. the BSMT of N. suaveolens (Pott et al. 2004; Effmert et al. 2005, Dreschler and Piechulla unpublished results), the benzoic acid carboxyl methyltransferase of Antirrhinum majus (Kolosova et al. 2001a) and the salicylic acid carboxyl methyltransferase of Stephanotis floribunda (Pott et al. 2003). A reason that might have contributed to the previous results lies in the detection method used for Western blots to determine protein levels. Our experience is that secondary antibodies conjugated with alkaline phosphatase do not adequately discriminate small quantitative differences of proteins. We have therefore used preferentially CDP-Star-labelled antibodies and determined the chemiluminescent signal with the LAS system. This allows better quantification and has enabled us to observe varied protein levels during day/night and throughout flower development.
The rhythmic appearence and disappearence of the 1,8-CIN transcript and of its protein and enzyme activity supports the observed nocturnal emission of the floral monoterpenes. The molecular mechanism that would best explain this observation is the “circadian clock”. Such endogenous clocks, which are molecular networks responsible for the precise timing of molecular and cellular processes have previously been extensively investigated. Many components of the plant circadian clock directly or indirectly influencing rhythmic gene expression have been determined, although the complete system is still far from being understood (Hoffrogge et al. 2003; McClung 2006). To establish whether monoterpene synthesis and emission in N. suaveolens is under the control of the circadian clock, we have placed plants under constant illumination or constant darkness to eliminate the action of the external “zeitgeber”. Oscillations of the 1,8-CIN transcripts under free-running conditions have revealed a period length of longer than 24 h, indicating a slower-running (than 24 h) endogenous clock. External light/dark signals synchronize the transcript oscillations and are the basis for the precise nocturnal emission of the monoterpenes from N. suaveolens flowers. Thus, circadian regulation may exist for monoterpene emission as has been determined for a diurnally leaf-expressed β-pinene synthase of Artemisia annua and a floral monoterpene synthase of A. majus (Lu et al. 2002; Dudareva et al. 2003). The rhythm of the A. annua pinene synthase persists for 7 days under LL or DD and, since the period length is slightly shortened under DD, a faster-running (than 24 h) endogenous clock seems to act in A. annua.
The precise diurnal and nocturnal timing of monoterpene emission from flowers appears to be the result of circadian clock control acting primarily at the level of the accumulation of the CIN transcript. This monoterpene synthase regulation is well coordinated with the expression of another scent synthesizing enzyme, the carboxyl methyltransferases of N. suaveolens , which catalyse the production of methyl benzoate and methyl salicylate (Effmert et al. 2005). These results clarify two established strategies for facilitating the simultaneous emission of scent compounds: (i) synthesis of VOCs by multi-product enzymes and (ii) the coordination of the biosynthetic pathways by a circadian clock.
Finally, the cellular localization of the 1,8-CIN has been addressed. Many terpene synthases that are present in vegetative tissue are primarily localized in specific glands, ducts or trichomes (Werker et al. 1985; Turner et al. 2000a, b). No glands and ducts have yet been found on petal epidermal surfaces but trichomes are present on the abaxial site of N. alata and N. suaveolens petals (Effmert et al. 2006). The adaxial and the abaxial epidermis cells of N. suaveolens, and not the trichomes, contain the methyltransferase BSMT (Rohrbeck et al. 2006) and the CIN (Fig. 6). Monoterpene synthesis in N. suaveolens and Clarkia breweri (LIS) occurs equally in the upper and lower epidermal cells of the petals, whereas in A. majus and S. floribunda, the synthesis of fragrance volatiles only takes place in the adaxial epidermis of the petals (Dudareva et al. 1996; Kolosova et al. 2001b; Effmert et al. 2006; Rohrbeck et al. 2006). This difference might be explained by the different inflorescences. Nicotiana and Clarkia present individual flowers, which can be freely reached from the pollinator regardless of whether the scent is emitted from the adaxial and/or abaxial epidermis, whereas Stephanotis and Antirrhinum flowers are arranged in complex inflorescences in which the adaxial epidermis presents the surface of the inflorescence, so that the emission of the scent components only occurs from this surface to the surrounding space.
Abbreviations
- LIS:
-
Linalool synthase
- CIN:
-
Cineole synthase
- GPP:
-
Geranyl pyrophosphate
- VOCs:
-
Volatile organic compounds
- LL:
-
Continuous illumination
- DD:
-
Continuous darkness
- LD :
-
Light/dark regime
- SPME:
-
Solid phase micro extraction
References
Altschul SF, Gish W, Miller W, Byers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:402–410
Andersson S (2006) Floral scent and butterfly pollinators. In: Dudareva N, Pichersky E (eds) Biology of floral scent. Taylor & Francis Group, Boca Raton, 199–217
Aoki S, Ito M (2000) Molecular phylogeny of Nicotiana (Solanaceae) based on the nucleotide sequence of the matK gene. Plant Biol 2:316–324
Asby M, Edwards PA (1990) Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J Bio Chem 265:13157–13164
Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Gen Genomics 267:730–745
Ayasse M (2006) Floral scent and pollinator attraction in sexually deceptive orchids. In: Dudareva N, Pichersky E (eds) Biology of floral scent. Taylor & Francis Group, Boca Raton, 219–241
Bohlmann J, Meyer-Gauen G, Croteau RB (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95:4126–4133
Bohlmann J, Gershenzon J, Aubourg S (2000) Biochemical, molecular genetic and evolutionary aspects of defense-related terpenoid metabolism in conifers. Recent Adv Phytochem 34:109–150
Chang S, Puryear J, Caiurney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko Y, Joseph J, Savolainen V, Parokonny AS (2003) Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann Bot 92:107–127
Chen F, Tholl D, D’Auria JC, Farooq A, Pichersky E, Gershenzon J (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15:481–494
Chen F, Ro D-K, Petri J, Gershenzon J, Bohlmann J, Pichersky E, Tholl D (2004) Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol 135:1956–1966
Cheng SH, Seemann JR (1998) Extraction and purification of RNA from plant tisue enriched in polysaccharides. Methods Mol Biol 68:27–32
Davis EM, Croteau R (2000) Cyclization enzymes in the biosynthesis of monoterpenes and diterpenes. Top Curr Chem 209:53–95
Dobson HEM (2006) Relationship between floral fragrance composition and type of pollinator. In: Dudareva N, Pichersky E (eds) Biology of floral scent. Taylor & Francis Group, Boca Raton, 147–197
Dudareva N, Cseke L, Blanc VM, Pichersky E (1996) Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8:1137–1148
Dudareva N, Piechulla B, Pichersky E (2000) Biogeneses of floral scents. In: Janik J (eds) Horticultural reviews. vol 24. John Wiley & Sons, Inc, New York, 31–54
Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Fäldt J (2003) (E)-β-ocimene and β-myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 15:1227–1241
Effmert U, Saschenbrecker S, Ross J, Negre F, Fraser CM, Noel JP, Dudareva N, Piechulla B (2005) Floral benzenoid carboxyl methyltransferases: from in vitro to in planta function. Phytochemistry 66:1211–1230
Effmert U, Buss D, Rohrbeck D, Piechulla B (2006) Localization of the synthesis and emission of scent compounds within the flower. In: Dudareva N, Pichersky E (eds) Biology of floral scent. New York, Tayler & Francis Group, Boca Raton, 105–123
Facchini PJ, Chappell J (1992) Gene familiy for an elicitor-induced sesquiterpene cyclase from tobacco. Proc Natl Acad Sci USA 89:11088–11092
Goodspeed TH (1954) The genus Nicotiana. Chronica Botanica Company, Walthman, Mass., USA
Gutermann I, Shalit M, Menda N, Piestun D, Dafny-Yellin M, Shalev G, Bar E, Davydov O, Ovadis M, Wang J, Adam Z, Pichersky E, Lewinsohn E, Zamir D, Vainstein A, Weiss D (2002) Rose scent: genomics approach to discovering novel floral fragrance-related genes. Plant Cell 14:2325–2338
Hoffrogge R, Mikschofski H, Piechulla B (2003) Suface plasmon resonance spectoscopy (SPR) interaction studies of the circadian-controlled tomato LHCa4*1 (CAB 11) protein with its promoter. Chronobiol Int 20:543–558
Knudsen JT, Gershenzon J (2006) The chemical diversity of floral scent. In: Dudareva N, Pichersky E (eds) Biology of floral scent. Taylor & Francis Group, Boca Raton, pp. 27–52
Knudsen JT, Eriksson R, Gershenzon J, Stahl B (2006) Diversity and distribution of floral scent. Bot Rev 72:1–120
Kolosova N, Gorenstein N, Kish CM, Dudareva N (2001a) Regulation of circadian methyl benzoate emission in diurnally and nocturnally emitting plants. Plant Cell 13:2333–2347
Kolosova N, Shermann D, Karlson D, Dudareva N (2001b) Cellular and subcellular localization of S-adenosyl-L-methionine: benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methylbenzoate in snapdragon flowers. Plant Physiol 126:956–964
Loughrin JH, Hamilton-Kemp TR, Anderson RA, Hildebrand DF (1990) Volatiles from flowers of Nicotiana sylvestris, N. otophora and N. malus x domestica: headspace components and day/night changes in their relative concentrations. Phytochemistry 29:2473–2477
Loughrin JH, Hamilton-Kemp TR, Anderson RA, Hildebrand DF (1991) Circadian rhythm of volatile emission from flowers of Nicotiana sylvestris and N. suaveolens. Physiol Plant 83:492–496
Loughrin JH, Hamilton-Kemp TR, Burton HR, Anderson RA (1993) Effect of diurnal sampling on the headspace composition of detached Nicotiana suaveolens flowers. Phytochemistry 32:1417–1419
Lu S, Xu R, Jia JW, Pang J, Matsuda SPT, Chen XY (2002) Cloning and functional characterization of a β-pinene synthase from Artemisia annua that shows circadian pattern of expression. Plant Physiol 130:477–486
McClung CR (2006) Plant circadian rhythms. Plant Cell 18:792–803
Peters RJ, Croteau RB (2003) Alternative termination chemistries utilized by monoterpene cyclases: chimeric analysis of bornyl diphosphate, 1,8-cineole, and sabinene synthase. Arch Biochem Biophys 417:203–211
Pichersky E, Raguso RA, Lewinsohn E, Croteau R (1994) Floral scent production in Clarkia (Onagraceae). I. Localization and developmental modulation of monoterpene emission and linalool synthase activity. Plant Physiol 106:1533–1540
Pichersky E, Lewinsohn E, Croteau R (1995) Purification and characterization of S-linalool synthase, an enzyme involved in the production of floral scent in Clarkia breweri. Archiv Biochem Biophys 316:803–807
Pott MB, Pichersky E, Piechulla B (2002) Evening-specific oscillations of scent emission, SAMT enzyme activity, and SAMT mRNA in flowers of Stephanotis floribunda. J Plant Physiol 159:925–934
Pott MB, Effmert U, Piechulla B (2003) Transcriptional and post-translational regulation of S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase (SAMT) during Stephanotis floribunda flower development. J Plant Physiol 160:635–643
Pott BM, Hippauf F, Saschenbrecker S, Chen F, Ross J, Kiefer I, Slusarenko A, Noel JP, Pichersky E, Effmert U, Piechulla B (2004) Biochemical and structural characterization of benzenoid carboxyl methyltransferases involved in floral scent production in Stephanotis floribunda and Nicotiana suaveolens. Plant Physiol 135:1946–1955
Raguso RA, Levin R.A, Foose SE, Homberg MW, McDade LA (2003) Fragrance chemistry, nocturnal rhythms and pollination “syndromes” in Nicotiana. Phytochemistry 63:265–284
Raguso RA, Schlumpberger BO, Kaczorowski RL, Holtsford TP (2006) Phylogenetic fragrance patterns in Nicotiana sections Alatae and Suaveolentes. Phytochemistry 67:1931–1942
Rohrbeck D, Buss D, Effmert U, Piechulla B (2006) Localization of methyl benzoate synthesis and emission in Stephanotis florbunda and Nicotiana suaveolens flowers. Plant Biol 8:615–626
Schiestl FP, Ayasse M, Paulus HF, Erdmann D, Franke W (1997) Variation of floral scent emission and postpollination changes in individual flowers of Ophrys sphegodes subsp. shegodes. J Chem Ecol 23:2881–2895
Shimada T, Endo T, Fujii H, Hara M, Omura M (2005) Isolation and characterization of (E)-β-ocimene and 1,8-cineole synthases in Citrus unshiu Marc. Plant Sci 168:987–995
Tollsten L (1993) A multivariante approach to post-pollination changes in the floral scent of Platanthera bifolia (Orchidaceae). Nord J Bot 13:495–499
Trapp SC, Croteau RB (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158:811–832
Turner GW, Gershenzon J, Croteau RB (2000a) Distributions of peltate glandular trichomes on developing leaves of peppermint. Plant Physiol 124:655–664
Turner GW, Gershenzon J, Croteau RB (2000b) Development of peltate glandular trichomes of peppermint. Plant Physiol 124:665–680
Wang J, Dudareva N, Bhakta S, Raguso RA, Pichersky E (1997) Floral scent production in Clarkia breweri (Onagraceae). II. Localization and developmental modulation of the enzyme S’adenosyl-L-methionine:(iso)eugenol O-methyltransferase and phenylpropanoid emission. Plant Physiol 114:213–221
Werker E, Ravid U, Putievsky E (1985) Glandular hairs and their secretions in the vegetative and reproductive organs of Salvia sclarea and S. dominica. Isr J Bot 34:239–252
Wise ML, Savage TJ, Katahira E, Croteau R (1998) Monoterpene synthases from common sage (Salvia officinales). J Biol Chem 273:14891–14899
Acknowledgements
The authors thank Rita Heese (University of Rostock) for her technical assistance, Diana Rohrbeck for her help with in situ hybridizations and Sandra Saschenbrecker for many RNA samples. We also thank Natalia Dudareva (University Purdue, Indiana, USA) and Eran Pichersky (University Ann Arbor, Michigan, USA) for their initial help in clone isolation and enzyme characterization. This project was financially supported by the DFG to BP.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roeder, S., Hartmann, AM., Effmert, U. et al. Regulation of simultaneous synthesis of floral scent terpenoids by the 1,8-cineole synthase of Nicotiana suaveolens . Plant Mol Biol 65, 107–124 (2007). https://doi.org/10.1007/s11103-007-9202-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9202-7