Abstract
Dehydration is the most crucial environmental constraint on plant growth and development, and agricultural productivity. To understand the underlying mechanism of stress tolerance, and to identify proteins for improving such important trait, we screened the dehydration-responsive proteome of chickpea and identified a tubby-like protein, referred to as CaTLP1. The CaTLP1 was found to predominantly bind to double-stranded DNA but incapable of transcriptional activation. We investigated the gene structure and organization and demonstrated, for the first time, that CaTLP1 may be involved in osmotic stress response in plants. The transcripts are strongly expressed in vegetative tissues but weakly in reproductive tissues. CaTLP1 is upregulated by dehydration and high salinity, and by treatment with abscisic acid (ABA), suggesting that its stress-responsive function might be associated with ABA-dependent network. Overexpression of CaTLP1 in transgenic tobacco plants conferred dehydration, salinity and oxidative stress tolerance along with improved shoot and root architecture. Molecular genetic analysis showed differential expression of CaTLP1 under normal and stress condition, and its preferential expression in the nucleus might be associated with enhanced stress tolerance. Our work suggests important roles of CaTLP1 in stress response as well as in the regulation of plant development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental stress limits growth, development and crop productivity (Bray 2004; Chaves and Oliveira 2004), thereby playing a major role in determining the geographic distribution of many plant species. Major environmental stresses are united by the fact that at least part of their detrimental effect on plant performance is caused by disruption of water status. Water-deficit or dehydration is the most common environmental stress to which plants are often exposed, and in many regions it is the bottleneck to agricultural development (Chaves et al. 2003). Transitory or constant dehydration adversely affects the morpho-anatomical, physiological and biochemical processes in plants, though the extent of damage varies considerably at different developmental stages. The perception and dehydration-induced signal transduction to switch on adaptive responses are critical steps in determining the survival of plants exposed to dehydration, and elucidation of the nature of these mechanisms has been an interesting area of research. However, very few plants have been subjected to biochemical and molecular studies to analyse the mechanisms of dehydration tolerance (Bartels et al. 1996; Pandey et al. 2010; Choudhary et al. 2009). The use of crop plants of agricultural importance seems to be a potential approach in investigating dehydration tolerance as different cultivars are available with differing degrees of tolerance. This provides correlative evidence for biomolecules putatively involved in dehydration response. Further, the transient and moderate dehydration treatments in stress genomic studies probably describes the most common form of dehydration that majority of plants are likely to encounter.
The plant cell wall or extracellular matrix (ECM) is a dynamic organization; essential not only for cell division, enlargement, and differentiation, but also acting in diverse environmental stress (Ellis et al. 2002; Vogel et al. 2004). Though the protein part accounts for only 10 % of cell wall mass, they contribute substantially to the cellular dynamics as they comprise several hundreds of different molecules with diverse functions (Carpita and Gibeaut 1993). Previously, we developed the differential ECM-specific proteome of a grain legume, chickpea, being subjected to progressive dehydration. The temporal changes in the proteome led to the identification of a tubby-like protein (TLP), designated CaTLP1 putatively involved in dehydration response (Bhushan et al. 2007). The tubby gene was first identified from obese mice via positional cloning (Kleyn et al. 1996; Noben-Trauth et al. 1996). Since then, a wide array of cellular functions of the gene have been postulated, which includes vesicular trafficking (Ikeda et al. 2002), mediation of insulin signalling (Kapeller et al. 1999), gene transcription (Boggon et al. 1999), G-protein signalling (Santagata et al. 2001), and ribosomal RNA synthesis (He et al. 2000), among others. The tubby-like protein genes are reported to be present in organisms ranging from single-celled to multicellular eukaryotes (Riechmann et al. 2000; Lai et al. 2004), suggesting that TLPs might carry out some fundamental role in cells in which they are expressed. This suggestion has been further strengthened by the occurrence of disease phenotypes in mutated tubby (Banerjee et al. 1998; Hagstrom et al. 1999), and also by the embryonic lethality of mice homozygous for a disrupted tubby allele (Ikeda et al. 2001). While four members of this family are currently known in mammals (Ikeda et al. 1999), plants harbour a large number of TLPs; for instance, 11 members in Arabidopsis (Lai et al. 2004), and 14 members in rice (Liu 2008; Kou et al. 2009). In addition to the typical 270 amino acids conserved C-terminal Tub domain, plant TLPs have evolved with F-box conserved at the N-terminal sequence, which is otherwise highly divergent in animal. Preliminary investigations on mammalian tubby protein, a resident of plasma membrane (Carroll et al. 2004), indicate that it interacts with G-protein and translocates to nucleus on perception of signal (Santagata et al. 2001). Also, it has been shown that these proteins may function as bipartite transcription regulators by binding to double-stranded DNA and activating transcription (Boggon et al. 1999). While a wide array of cellular functions of TLPs have been established in animals (Ikeda et al. 2002), their role in plants is still elusive. Nevertheless, the highly conserved evolution of tubby proteins and the existence of redundancy suggest their indispensable role in plants.
In recent years, this protein family has been shown to be involved in ABA-dependent signalling in Arabidopsis (Lai et al. 2004) and pathostress response in rice (Kou et al. 2009). The mode of action of tubby family proteins in plants is believed to be regulated by ABA (Lai et al. 2004), which is characteristically implicated in different stress responses. We report here the molecular cloning and characterization of CaTLP1 as a step toward elucidating its function in dehydration tolerance. The sequence of CaTLP1 displayed high similarity with the tubby-like protein genes earlier reported in Arabidopsis and rice (Lai et al. 2004; Kou et al. 2009). Overexpression of CaTLP1 in transgenic tobacco plants showed enhanced growth and development when compared to wild-type plants. Also, CaTLP1-overexpressing plants showed improved tolerance to dehydration, high salinity and oxidative stress, indicating its possible role in multivariate stress-responsive pathways. These results not only provide new insights into the underlying mechanism of action of plant TLPs, but may also facilitate the targeted genetic manipulation in crop plants to improve stress tolerance.
Materials and methods
Plant materials, growth conditions, and stress treatments
The chickpea seedlings were grown in pots containing a mixture of soil and soilrite (2:1, w/w) in an environmentally controlled growth room, and maintained at 25 ± 2 °C with 50 ± 5 % relative humidity under 16 h photoperiod (270 μmol m−2 s−1 light intensity). The 3-week-old seedlings were subjected to dehydration condition for a period of 7 days by withdrawing water, and tissues were harvested at different intervals. Furthermore, pots containing dehydrated seedlings were rewatered and allowed to recover for a period of 24 h, and the rehydrated tissues (R24) were harvested. In a separate set of experiment, the seedlings were supplemented with half-Hoagland’s medium until 3 weeks, followed by treatment with different concentrations (100, 250, and 500 mM) of NaCl in the same medium. The tissues were harvested 24 h after the treatment. The treatment of ABA was carried out by spraying three-week-old seedlings with 100 μM ABA, and tissues were harvested at targeted intervals. Methyl jasmonate (100 μM) treatment was given in a similar fashion and tissues were collected at 8, 16, and 24 h. For salicylic acid treatment, a 5 mM solution was prepared in ethanol and sprayed on the leaflets. The harvested tissues were instantly frozen in liquid nitrogen and stored at −80 °C, unless stated otherwise.
Expression of CaTLP1 in Escherichia coli, and mass spectrometric identification
The full-length CaTLP1 was inserted in-frame into pGEX4T-2 expression plasmid (Amersham Biosciences, UK) utilizing BamHI and NotI restriction sites engineered at 5′- and 3′-ends, respectively to yield the construct pGEXCaTLP1. The insert sequence and reading frame were verified by nucleotide sequencing, and the construct was transformed into E. coli BL21 (DE3). The GST-tagged protein was produced by inducing transformed cells with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The recombinant protein was purified from the bacterial lysates with glutathione-Sepharose beads (Amersham Biosciences, UK) and resolved by 12 % SDS-PAGE.
Mass spectrometric analysis of the protein was performed as described earlier (Bhushan et al. 2006). In brief, trypsin-digested peptides were analyzed by electrospray ionization time-of-flight mass spectrometry (LC/MS/TOF) using a Tempo nanoflow MDLC system coupled to Q-STAR Elite spectrometer (Applied Biosystems, USA). The spectra were analysed to identify the protein of interest using Mascot sequence matching software (Matrix Science, USA).
Nucleic acids analysis and construction of phylogenetic tree
The cDNA sequence of CaTLP1 was determined by ABI Prism 3700 DNA Analyzer (Applied Biosystems, USA). A phylogenetic tree was constructed from amino acid alignment by neighbour-joining method using the default parameters. Multiple alignment of proteins were performed with ClustalW program, and a phylogram was obtained by MEGA software version 5.05 (Tamura et al. 2011).
Genomic DNA was extracted using DNeasy kit (Qiagen, USA), and aliquots of 10 μg DNA were digested with different restriction enzymes. In a separate set of experiment, RNA was isolated using Tripure reagent (Roche Applied Science), as recommended by manufacturer. The electrophoresis of nucleic acids was performed as described (Sambrook and Russell 2001), blotted overnight onto Hybond-N membrane (Amersham Biosciences, UK), and fixed at 1,200 J/cm for 30 s using Stratalinker UV crosslinker (Stratagene, USA). Immobilized nucleic acids were hybridized with [32P]-labelled 314-bp CaTLP1 fragment (amplified with internal primers 5′-TGGATAGTCAACCTCCACATG-3′ and 5′-GGTTGCCAGAGTATGAATCTG-3′).
Subcellular localization
Subcellular localization of CaTLP1-EYFP fusion protein was examined in vivo in stably transformed Arabidopsis seedlings. The CaTLP1-EYFP construct in pGWB441 gateway vector was used to transform Agrobacterium cells. The bacteria were grown to stationary phase in liquid culture at 25–28 °C, 250 rpm. in sterilized YEP (10 g yeast extract, 10 g peptone, and 5 g NaCl per liter) supplemented with spectinomycin (100 mg ml−1). Cultures were initiated with 1:100 dilution of primary overnight cultures and grown for roughly 18–20 h. Cells were harvested by centrifugation (5,500g) for 20 min at room temperature. The harvested cells were resuspended in inoculation medium [1/2 strength MS Medium (M-5519; Sigma Chemicals), 5.0 % sucrose, 44 nM benzylamino purine (10 ml l−1 of a 1 mg ml−1 stock in DMSO) and 0.008 % Silwet L-77] pH adjusted to 5.7 to a final OD600 of approximately 2.0. Arabidopsis plants were transformed by floral-dip method (Clough and Bent 1998). The primary roots of transformants were examined under confocal microscope (Leica Microsystems). A cell wall specific dye, propidium iodide (PI), was used to stain the cell wall of the primary root cells.
Electrophoretic mobility shift assay and determination of transactivation
Nucleic acid binding experiments were performed using the recombinant protein as described earlier (Boggon et al. 1999). The recombinant protein (0.5 mg) was incubated in electrophoretic mobility shift assay (EMSA) buffer [25 mM MOPS (pH 8.0), 2 mM DTT, 120 mM KOAc and 2 mM EDTA] by mixing 0.5 pmol of either [32P]-labelled 20 bp dsDNA or 20 b ssDNA for 30 min at 30 °C. The reactions were resolved by electrophoresis on 6 % TBE-polyacrylamide gel at 80 V for 2 h. The protein-DNA complexes as well as free probes were visualized by autoradiography with exposure time of 30 min at 4 °C. To determine the activation domain, the coding sequence of CaTLP1 was cloned in yeast expression plasmid pGBKT7 (Clontech Laboratories, USA) at NdeI-BamHI restriction site to express CaTLP1 fused to GAL4 DNA-BD. The construct was transformed into yeast strain AH109 (Cagney et al. 2000) harboring MEL1 reporter gene. The cells were plated on synthetically defined medium deficient in tryptophan. The transformants were then analyzed for α-galactosidase expression. The GAL4 AD domain from the plasmid pGADT7 (Clontech Laboratories, USA) was cloned in-frame with the plasmid pGBKT7 and used as positive control, while pGBKT7 plasmid transformed in AH109 cells served as negative control.
Transformation and analysis of CaTLP1-overexpressing plants for stress tolerance
Full-length coding sequence of CaTLP1 was cloned into the polylinker site of the binary vector pBI121 (Clontech Laboratories, USA) under the control of CaMV 35S promoter. The construct was mobilized into Agrobacterium tumefaciens strain EHA105. Agrobacterium mediated transformation of tobacco (Nicotiana tabacum L. var. Petit Havana) was carried out using standard protocol (Hobbs et al. 1990). Regenerated shoots were rooted on phytohormone-free rooting medium supplemented with cefotaxime (250 mg l−1) and kanamycin (100 mg l−1), transferred to soil, and grown in standardized greenhouse conditions. Seventeen independent transgenic events harboring CaTLP1 were grown for further analyses. The wild-type and the transgenic plants were studied parallely in the same growth room and comparative morpho-anatomical, molecular, and physiological analyses were carried out. Morpho-anatomical studies were accomplished by scanning electron microscopy (EVO LS10, Carl Zeiss).
Leaf-disc bioassay was carried out to determine dehydration, salinity, and oxidative stress tolerance using 1 cm2 leaf-discs excised from 30-day-old, both wild-type and transgenic, tobacco plants. The discs were floated on different concentrations of sorbitol (200 mM), NaCl (250 and 500 mM), H2O2 (1 mM) and menadione (5 and 7 μM) for osmotic, salinity and oxidative stress, respectively. The chlorosis of the discs was examined after 72 h of incubation. Chlorophyll retention assay was done in leaf-discs subjected to salinity stress as described earlier (Porra 2002). The chlorophyll content was calculated using following formulas:
-
1.
\( \left[ {\text{Chl a}} \right] = \left( {12.00 \, 9 \, *{\text{A}}_{663.8} } \right) - \left( {3.11 \, 9 \, *{\text{A}}_{646.8} } \right); \)
-
2.
\( \left[ {\text{Chl b}} \right] = \left( {20.78 \, 9*{\text{ A}}_{646.8} } \right) - \left( {4.88 \, 9*{\text{A}}_{663.8} } \right); \)
-
3.
\( \left[ {{\text{Chl a}} + {\text{b}}} \right] = \left( {17.67 \, 9*{\text{A}}_{646.8} } \right) + \left( {7.12 \, 9*{\text{A}}_{663.8} } \right); \)
where A is the absorption measured at respective wavelengths.
Comparative analysis of net photosynthesis and transpiration rate
Net photosynthesis and rate of transpiration were measured using a gas exchange fluorescence system GFS3000 (Heinz Walz GmbH, Germany). The measurements were made under saturating irradiance (1,000 μmol photons m−2 s−1) and constant airflow of 750 μl s−1 at 25 °C. Five independent leaves of three randomly chosen plants each, from wild-type and transgenic plants, were placed directly into the measuring cell (3 cm2) for the measurements.
Immunoblot analysis
Immunoblot analysis was done by resolving protein extracts from wild-type and transgenic plants on a uniform 12.5 % SDS-PAGE, and electro transferring onto nitrocellulose membrane at 150 mA for 2 h. The membranes were blocked with 5 % (w/v) non-fat milk for 1 h and incubated with anti-CaTLP1 polyclonal antibody for 2 h. The antibody was raised in rabbit against an antigenic peptide (CVDPSHNVSSEEQER) (Sigma-Genosys, USA). The blot was eventually incubated with alkaline phosphatase conjugated secondary antibody for 1 h and the signals were detected using NBT/BCIP (nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate) method.
Statistical analyses
Each experiment was replicated at least three times and values are expressed as means ± SE. All mean comparisons were done using paired t test for independent samples. The measurements for determining net photosynthetic rate and leaf conductance were analyzed by one-way analysis of variance (ANOVA).
Results
Molecular cloning and structural organization of dehydration-responsive CaTLP1
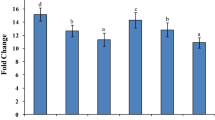
In a previous study, we had developed differential proteome of extracellular matrix of chickpea under dehydration (Bhushan et al. 2007). A critical analysis of the proteome revealed a spot representing tubby-like protein (TLP) that was found to be highly regulated under dehydration (Fig. 1a). A threshold level of expression of TLP was observed in unstressed tissues, indicating its possible role in plant growth and development. Under dehydration, expression of the protein reached maximal level at 72 h but decreased during 96–120 h, which might be due to feedback inhibition by the accumulated protein itself. The expression level was further elevated at later stages of dehydration and also at rehydrated stage (Fig. 1b). Here, we describe cloning of the candidate gene encoding TLP and its possible role in stress tolerance. The 3′-rapid amplification of cDNA using degenerate primers, designed from peptide sequence tags, yielded a partial fragment of 1.0 kb homologous to a chickpea EST sequence (gi-7635492). The coding region of the gene was isolated, cloned in pGEM-T vector and named as pCaTLP1. Further, the full-length ORF of CaTLP1 was cloned in-frame with GST cDNA in pGEX4T-2 and expressed in Escherichia coli, BL21 (DE3), which could encode the fusion protein with an estimated molecular mass of 72 kDa (Fig. S1). The expressed protein was excised from the gel, in-gel digested with trypsin, and confirmed by mass spectrometry. A representative mass spectrum is shown in Fig. S2. In silico analysis revealed that the actual transcript size of CaTLP1 is 1.722 kb with a coding region of 1.236 kb, and 215-bp 5′- and 271-bp 3′-UTRs, respectively.
Identification of dehydration-responsive tubby-like protein from differential cell wall proteome. a Cell wall proteins were electrofocused onto 13 cm IPG strip (pH 4–7), and resolved on 12.5 % (w/v) SDS-PAGE. Gel images were analyzed using PDQuest, version 7.2.0, and the TLP spot in normalized densities among different time intervals was identified. The gel section containing the candidate spot is magnified in the lower panel. Molecular weight marker in kDa is indicated on the right. b Temporal changes of the TLP under progressive dehydration and recovery stage at 24 h (R24). Boxed areas are the magnified gel sections and the TLP spots are encircled. The bar graphs cited below are indicative of the fold-expression in terms of band intensity of TLP plotted against the duration of dehydration. The error bars in the graph represent the standard deviation of the values taken from three replicate gels. *Significant difference (* P < 0.05, ** P < 0.005) between control and given time point. Asterisks indicate significant difference with day 0 (P < 0.05)
To study the genomic organization, we cloned the CaTLP1 genomic DNA spanning 2.299 kb fragment, indicating the presence of intron/s. The comparison of the full-length cDNA sequence with the corresponding genomic DNA sequence revealed that the coding region of the CaTLP1 is interrupted by three introns (Fig. 2a). The deduced protein sequence analysis and domain search (http://www.expasy.org/prosite) suggest that CaTLP1 comprises of 55 aa residues F-box domain (Pfam00646) and 294 residues Tub domain (PF01167) (Fig. S3).
Structural and phylogenetic analysis of CaTLP1. a Schematic representation of the CaTLP1 and exon–intron organization. b An unrooted phylogenetic tree showing evolutionary relationship of CaTLP1 with its orthologs. The phylogram was generated using the neighbor-joining algorithm of MEGA software, version 5.05. The numerical represents the bootstrap value. Scale bar indicates an evolutionary distance of 0.1 aa substitution per position in the sequence. E exon, I intron, UTR untranslated regions
In order to evaluate the phylogenetic and evolutionary relationship of CaTLP1 within the TLP protein family, amino acid sequence was aligned with fifteen orthologs from other plants as well as animals, and an unrooted phylogram was generated. At first glance, two major groups were apparent from the phylogram (Fig. 2b). Plant TLP family proteins clustered in a single group, suggesting that they might originate from a single ancestral gene. However, the organization of TLPs in different members within the plant group suggests an evolutionary divergence in this protein family (Yang et al. 2008). Interestingly, TLP1 of mouse, TLP3 of human and fruit flies clustered together forming a separate group. Furthermore, these members were found to be close neighbors of CaTLP1, showing the cross-kingdom similarity.
Transcriptional regulation by dehydration, salt and ABA treatments, and tissue expression pattern of CaTLP1
To investigate the dehydration response of CaTLP1 at the transcript level, we carried out Northern blot analysis. The CaTLP1 transcripts were induced within 5 h of dehydration showing maximum accumulation at 12 h and remained so up to 16 h. However, the transcripts showed a drastic decrease after 24 h and reached the background level at 48 h (Fig. 3a). Next, we examined whether CaTLP1 transcription is affected by ABA and other stress conditions. The CaTLP1 was induced by ABA after 30 min of treatment, quickly reaching to saturation at 1 h. The transcript level remained the same until 3 h, but dropped to the background level after 4 h of treatment (Fig. 3b). It is known that the dehydration-responsive pathway often overlaps with that of ABA and salt stress, and thus the response of CaTLP1 transcript under salt stress was investigated. In a dose-response experiment, CaTLP1 was induced by treatment with higher concentrations of NaCl. The transcripts were most strongly induced at 250 mM NaCl (Fig. 3c), while there was no induction at lower concentrations. These results altogether suggest that CaTLP1 may participate in general osmotic stress, and this participation may, in part, depend on ABA.
Expression profile of CaTLP1 in response to different stresses and phytohormone treatments. Three-week-old chickpea seedlings were subjected to a dehydration, b ABA (100 μM) treatment, c salt stress at different concentration of NaCl (100–500 mM), d methyl jasmonate (100 μM), and salicylic acid (5 mM). The lanes represent various time points during the treatments. e Northern blot showing differential expression of CaTLP1 in different organs with 10 μg total RNA in each lane. The RNA blot was hybridized with a [32P]-labelled 0.5 kb 5′-CaTLP1 fragment. The image of ethidium bromide-stained rRNA shows equivalent loading and RNA quality. R root, S stem, L leaf, F flower, SP seed pod
It is known that the genes involved in disease resistance are often expressed differentially during plant-pathogen interaction. Jasmonic acid (JA) and salicylic acid (SA) are essential compounds in the pathogen- and wound-signalling pathways and reported to mimic pathostress response in plants (Clarke et al. 2000). To examine whether CaTLP1 was responsive to pathogen infection, chickpea seedlings were treated with JA and SA. There was very little change in the accumulation of CaTLP1 transcripts in seedlings treated with JA and SA, suggesting that the CaTLP1 possibly has little or no role in pathostress signalling (Fig. 3d).
To determine the organ specificity of CaTLP1 expression, we compared transcript abundance in different organs (roots, stems, leaflets, flowers and pods) by Northern analysis. The expression of CaTLP1 was low but detectable in roots, though to a lesser extent than that in flowers and pods. However, the expression in vegetative organs such as stems and leaves were substantially higher (Fig. 3e), suggesting a possible synergistic quantitative relationship of CaTLP1 to seedling growth and development.
CaTLP1 is a low copy gene of a multiple gene family
To characterize the CaTLP1 gene in more detail, we performed Southern blotting, which could be used to obtain preliminary information on the size and organization of the corresponding genomic sequence and the number of gene copies. Genomic DNA of chickpea was digested with different restriction enzymes and blotted to nylon membrane. The blot was hybridized with [32P]-labelled 314-bp CaTLP1 fragment under high stringency conditions. The restriction enzymes SspI and SacI, having single site in the probe region, yielded two bands, whereas HindIII, EcoRI, and BamHI having no restriction sites yielded a single band, indicating the presence of single copy of the CaTLP1 (Fig. 4). However, Southern analysis when performed with 3′-conserved region of the CaTLP1 cDNA as probe, displayed multiple bands (Fig. S4). These results confirm that the genes encoding TLPs in the chickpea genome are organized in low copy multigene family.
In silico prediction of CaTLP1 localization
As CaTLP1 was initially identified as a cell wall resident, we thought it likely that the candidate protein would contain transport signal sequence. An examination of the amino acid sequence using subcellular localization (SCL) Predictor CELLO v.2.5 (Yu et al. 2006) revealed that CaTLP1 is duly localized to the extracellular space as well as nucleus (Fig. S3). The nuclear localization of the protein can be attributed to the presence of nuclear localization signals in the coding sequence. However, the cell wall localization is striking in that the protein does not contain any signature sequence for translocation to the extracellular space, as revealed by SecretomeP (http://www.cbs.dtu.dk/services/SecretomeP/). Since many proteins with no known signal peptide are reported to be secreted to the extracellular space (Slabas et al. 2004; Pandey et al. 2010), we reasoned that CaTLP1 may have unique characteristics to be efficiently targeted to the cell wall.
Ectopic expression, localization and stress-induced accumulation of CaTLP1
To verify that the observed dual localization of CaTLP1 occurs in vivo, we generated transgenic Arabidopsis plants expressing CaTLP1-YFP fusion protein. The transgenic plants showed predominant expression of the fusion protein in cell walls and to a lesser extent in nuclei (Fig. 5a). There is increasing number of proteins which are reported to reside in multiple compartments, having similar and/or distinct functions. Furthermore, many stress-responsive proteins are known to shuttle between different subcellular compartments, albeit their role in signal transduction mechanisms remains largely unexplored. To investigate whether translocation of CaTLP1 in the nucleus is influenced by stress condition, the transgenic plants were subjected to dehydration, and YFP signals were monitored by confocal microscopy. The intensity was determined as an average of the maximum intensities in different regions of the organelles studied. The YFP signals were relatively stronger in the nuclei as compared to the extracellular space (Fig. 5b, c), suggesting that the preferential accumulation of CaTLP1 might be influenced by stress condition.
Subcellular localization of CaTLP1-YFP fusion protein. The roots of stably transformed A. thaliana show the expression of CaTLP1-YFP. a Confocal images of epidermal cells were captured in 6-day-old seedlings. Upper left panel shows magnified cells with CaTLP1-YFP fluorescence, while right panel shows the brightfield image of cell patterns. Lower left panel shows fluorescence of PI-stained cell walls and nucleus, whereas right panel shows fluorescence of PI-stained cells overlaid onto brightfield images of cells expressing CaTLP1-YFP. b Localization of CaTLP1-YFP in Arabidopsis primary roots under dehydration. The seedlings were removed from MS plate and subjected to dehydration on glass slide for 15 min. Upper left panel shows yellow fluorescence of CaTLP1-YFP, while right panel shows brightfield image of roots. Lower left panel shows fluorescence of PI-stained cell walls and nucleus, whereas right panel shows fluorescence of PI-stained cells overlaid onto brightfield images of cells expressing CaTLP1-YFP. c Zoomed images of dehydrated cells expressing CaTLP1-YFP with overlaid images on brightfield (1st and 2nd sections) and PI-stained cells with overlaid images (3rd and 4th sections). The arrows indicate stress-induced preferential accumulation of CaTLP1-YFP. The bars indicate the extent of resolutions
To summarize, the expression analysis of CaTLP1-YFP fusion protein established fluorescence in both extracellular matrix and nucleus of transgenic Arabidopsis plants (Fig. 5a–c). The stress-induced preferential accumulation of CaTLP1 in the nucleus confirms that the localization of the protein under normal and stress condition varies considerably, suggesting its possible role in stress response.
CaTLP1 binds to double-stranded DNA but is incapable of transcriptional activation
The TLPs have been shown to act as transcription factor in mammalian cells (Boggon et al. 1999). To determine whether CaTLP1 has DNA-binding property, gel shift experiment was conducted using glutathione S-transferase (GST) tagged fusion protein. The GST-tagged CaTLP1 was purified as described. Since A-like structure of oligo d(A/T) is metastable and that of oligo d(G/C) is stable, which in turn stabilizes the protein-DNA interaction, we used d(T20)/d(A20) and d(G20)/d(C20) for DNA-binding assay. The [32P]dCTP-labelled double-stranded 20 mer DNA probe was prepared by annealing the oligonucleotides d(T20)/d(A20) and d(G20)/d(C20), while purified GST was used as a negative control. There was band shift with GST-CaTLP1 fusion protein in case of double-stranded DNA molecules, while no shift was observed with the purified GST (Fig. 6a). Further, the binding of radio-labelled probe was effectively competed out by addition of 100-fold excess of cold oligonucleotides. Similarly, gel shift assay was carried out using single-stranded d(C20) and d(T20) oligonucleotides as probe which yielded no or little band shift (Fig. 6b). These results suggest that the CaTLP1 binds avidly to double-stranded DNA, but poorly to single-stranded DNA. This specificity indicates that the binding is not the result of non-specific electrostatic interactions rather depend on specific determinants, characteristic of double-stranded DNA (Berg 1990).
a CaTLP1 demonstrates a preference for binding double-stranded DNA. Gel-shift assays were carried out using double-stranded 20 mer d(A/T) and d(G/C), and b single-stranded 20 mer d(C20) and d(T20) [32P]-labelled probes. The shift was competed out by 100-fold excess of cold oligos. The arrows represent the band-shifts. c Transactivation analysis of CaTLP1. The CaTLP1 coding sequence fused with GAL4-DB in the vector pGBKT7 was used for transformation into a yeast strain AH109. Yeast colonies harbouring no vector, empty vector (pGBKT7), and recombinant pGBKT7-CaTLP1 were grown on YPDA, synthetic medium lacking Trp, and on SD media lacking Trp but supplemented with X-α-gal. The GAL4AD domain from pGADT7 was cloned in the vector pGBKT7 to be used as positive control. FP free probe
Transcriptional activators are known to be modular proteins with distinct domains for DNA binding and transcriptional activation (Jhonson and McKnight 1989). It is possible that CaTLP1 might have the potential to activate a large number of stress-responsive genes, if it had transactivation capability. To investigate such a possibility, the ORF of CaTLP1 was introduced in-frame with the GAL4 DNA-binding domain in yeast expression vector pGBKT7. The pGBKT7-CaTLP1 construct was then transformed into yeast strain AH109, harboring MEL1 reporter gene. The colonies were observed for time-dependent colour development; but strangely, they remained white even after 3 days of incubation whereas the vector colonies turned blue (Fig. 6c). The inability of CaTLP1 to activate reporter gene may be due to the lack of activation domain, corroborating an earlier report by Lai et al. (2004).
Overexpression of CaTLP1 confers stress tolerance in transgenic plants
Since chickpea is highly recalcitrant to in vitro regeneration and genetic transformation (Sanyal et al. 2005), we developed transgenic tobacco plants constitutively expressing CaTLP1 under the control of CaMV 35S promoter to investigate its stress-responsive function. A total of 17 independent transgenic events were confirmed by PCR analysis; the transcripts, and protein expression were determined by Northern and Western blot analyses, respectively (Fig. 7a–c). None of the transgenic lines displayed any phenotypic abnormality throughout their life cycle. Two independent overexpressing lines from F2 generation, one with high (T1) and the other with moderate expression (T4) of the transgene, were selected for further characterization. Southern blot analysis revealed the integration of three and two copies of CaTLP1 in T1 and T4 lines, respectively (Fig. 7d). The CaTLP1-overexpressing seedlings were transplanted and grown alongside the wild-type plants under identical conditions. The transgenic plants displayed a higher growth rate, as evident by increased shoot height and biomass, when compared to wild-type plants (Fig. 8a). Average root length in 15-day-old transgenic plants was more than two-fold than that of wild-type plants (Fig. 8b). Similarly, the shoot length of the transgenic plants was significantly higher (Fig. S4). The average area of leaves on the transgenic plants produced during early- to mid-vegetative phase (15–45-day-old plants) was about 2.5-fold higher than those of wild-type plants (Fig. 8c). However, there was no appreciable change in the average area of leaves produced later in the vegetative phase and just prior to flowering (data not shown). Since the average leaf area and primary root length of the transgenic plants were higher than those of wild-type control plants, their anatomical features were examined and found to be significantly different. As shown in Fig. 8d, the average area of epidermal cells of the leaves and epiblema cells of the roots of transgenic plants were 1.5–1.8-fold higher than those of wild-type plants, which is likely to be the main reason for their rapid growth.
Molecular characterization of transgenic tobacco plants expressing CaTLP1. a PCR amplification of CaTLP1 using gene-specific primers. b Expression of CaTLP1 in transgenic plants as determined by RNA gel-blot analysis. Total RNA was extracted from wild-type and transgenic plants, and probed with [32P]-labelled 0.5 kb 5′-CaTLP1 fragment. Ethidium bromide-stained rRNA is shown as loading control. c Western blot analysis of independent transgenic lines. Total protein was extracted from both wild-type and transgenic plants, and probed with anti-CaTLP1 polyclonal antibody. The representative Coomassie-stained gel shows uniform protein loading. d Southern blot analysis of CaTLP1-overexpressing plants. Genomic DNA of two independent transgenic events (T1 and T4) was digested with EcoRI that has no restriction site in the probe. The blot was hybridized with [32P]-labelled GUS cDNA probe. Molecular weight marker in kb is indicated on the left
Phenotypic screening of wild-type and CaTLP1-overexpressing tobacco plants. a Morphology of 15- to 45-day-old wild-type and transgenic tobacco plants; b root phenotype of 15-day-old plants; and c leaf-area. d Observation under scanning electron microscope (SEM) shows size of primary root cells (upper panels) and the leaf epidermal cells (lower panels). The bar indicates the extent of resolution. The representative T1 and T4 transgenic plants expressing CaTLP1 are shown. Days were counted after seedlings had been transplanted to soil. The error bars in the graphs represent the standard deviation of the values taken from three plants for each of two independent transgenic lines. * Significant difference (* P < 0.05, ** P < 0.005) between control and given time point. Asterisks indicate significant difference with day 0 (P < 0.05)
To determine the stress tolerance of the transgenic plants, chlorophyll retention rate was monitored by leaf-disc assay. Both wild-type and CaTLP1-overexpressing plants were subjected to osmotic stress by incubating leaf-discs in different concentrations of sorbitol and NaCl. The leaf-discs of transgenic plants could retain higher percentage of chlorophyll when compared with that of wild-type plants (Fig. 9a, b). We also investigated whether the CaTLP1 overexpression affected oxidative stress tolerance in transgenic plants. Leaf-discs from wild-type and transgenic plants were subjected to different concentrations of ROS generating chemicals such as H2O2 and menadione. The leaf-discs from the wild-type plants bleached completely under higher concentrations of these chemicals, while the transgenic plants showed very little chlorosis. However, leaf-discs treated with distilled water remained green throughout the experiment (Fig. 9a). The higher retention rate of chlorophyll in the transgenic plants even after 72 h of incubation confirms the observed phenotypic differences (Fig. 9b). These results suggest that stress-induced loss of chlorophyll was lower in CaTLP1-overexpressing plants, reflecting their better ability to withstand such stress.
Differential stress-response of wild-type and CaTLP1-overexpressing plants. a Observation of chlorosis due to osmotic or oxidative stress on the leaf-discs subjected to sorbitol, NaCl, H2O2, and menadione treatments. Leaf-discs (1 cm2) of 30-day-old wild-type and transgenic tobacco plants were floated on different concentrations of the solutions as indicated. The discs floated on sterile distilled water served as experimental control. b Quantitation of chlorophyll content in leaf-discs of wild-type and transgenic plants under progressive osmotic stress of 250 mM NaCl. The experiments were done in triplicates (n = 3), and average mean values were plotted against duration of stress. c Phenotypic appearance of transgenic plants compared to wild-type plants treated with 1 μM ABA. d Fresh weight of 3-week-old seedlings grown in MS media supplemented with ABA compared to the fresh weight of the seedlings in unstressed condition expressed as percent relative fresh weight. The error bars in the graphs represent the standard deviation of the values taken from three plants for each of two independent transgenic lines. * Significant difference (* P < 0.05, ** P < 0.005) between control and given time point. Asterisks indicate significant difference with day 0 (P < 0.05)
To evaluate ABA-mediated stress response, 8-day-old wild-type and transgenic seedlings were transferred to medium supplemented with 1 μM ABA and allowed to grow for 3 weeks. The response was measured in terms of percent relative fresh weight, i.e., fresh weight of the seedlings grown in experimental conditions relative to the fresh weight of the same line grown in control condition. The relative fresh weight was 58 % in T1, followed by 50 % in T4 and 31 % in the wild-type plants (Fig. 9c, d). These results altogether confirm the involvement of CaTLP1 in dehydration and salt-stress responsive pathways, and the paramount role of ABA in such responses.
Since net photosynthetic rate is known to be affected by various stress conditions (Allen and Ort 2001; Zgallai et al. 2005), we investigated in planta photosynthetic rate which mirror the true impact of stress response on physiological characteristics of plants. Under progressive dehydration, the photosynthetic rate in transgenic plants remained unaltered in initial 24 h, decreased linearly up to 96 h, and sustained thereafter. In contrast, the decrease in photosynthetic rate in wild-type plants was found to be proportional to the duration of stress treatment. In wild-type plants, photosynthetic rate reduced to 12 % after 120 h of dehydration, while the rates were 60 and 50 % in transgenic T1 and T4, respectively (Fig. 10a, e). During the time-course of dehydration, the detached leaves of transgenic plants lost water more slowly as compared to wild-type plants (Fig. 10b, c). It is likely that expression of CaTLP1 might be associated with dehydration tolerance by maintaining turgidity and relative water content. The resistance to dehydration in transgenic plants could be attributed, at least in part, to their ability to efficiently close their stomata and reduce rate of transpiration. This possibility is exemplified by the observation of reduced transpiration rate in transgenic plants under dehydration when compared to wild-type plants (Fig. 10d, e). Although the reason for higher photosynthetic but lower transpiration rate in transgenic plants is presently unclear, it might be due to stress-induced changes in stomatal behavior (Araujo et al. 2011; Zhang et al. 2008; Roelfsema and Hedrich 2005).
Improved dehydration tolerance in transgenic tobacco plants expressing CaTLP1. a Dehydration-induced temporal changes of net photosynthesis in wild-type and transgenic plants. b Measurement of water loss in the excised leaves of wild-type and transgenic plants. The fresh weight (FW) was measured at different time intervals as indicated, and water loss was calculated from the decrease in FW compared with time zero. Mean values were normalised to the leaf-area of 10 leaves for each of three independent experiments. c Phenotypic appearance of wild-type and transgenic plants, wherein the rate of water-loss was compared. d Changes in transpiration rate of transgenic and wild-type plants in a time-dependent manner under dehydration. The dehydration treatment was administered as described in ‘Materials and methods’. Values are expressed as mean from five independent leaves of three randomly chosen plants. e Phenotypic appearance of wild-type and transgenic plants, wherein the rate of photosynthesis and transpiration were measured
Discussion
In plants, the extracellular matrix is the first compartment that senses the stress signals, transmits them to the cell interior, and eventually influences the cell fate decision (Clarke 2001; Bhushan et al. 2007). It is postulated that the stress-responsive proteins in this compartment might play a variety of functions during cellular adaptation. Therefore, the knowledge on stress adaptation would help in formulating mechanism for defense, and bridging the gap between molecular mechanisms by which plants sense and respond to such stress. In a search for novel dehydration-responsive proteins, we identified a tubby-like protein from the chickpea cell wall (Bhushan et al. 2007). During the past few years, there have been several reports on the tubby-like protein gene family in animals, including human, though there is far less information on their equivalance in plants. While AtTLP9 of Arabidopsis has been reported to be involved in ABA-depended signalling pathway (Lai et al. 2004), induced expression of OsTLPs of rice has been reported to be responsive to bacterial infection (Kou et al. 2009). The TLPs seem to constitute a large family in plants, and sequence comparison indicates that they share a close sequence similarity. The sequence analysis of CaTLP1 revealed a well-conserved Tub-domain (294 residues spanning 117–411 aa) at the C-terminal region and F-box (55 residues spanning 51–106 aa) at the N-terminal region (Fig. S3).
It is known that there is a close relation between physiological function of a protein and its intracellular location. In mammalian cells, tubby protein is reported to be localized in the nucleus as well as in plasma membrane (Santagata et al. 2001), and shown to have a direct link in G-protein signalling that regulate the gene expression. Mutation studies in the tubby-like protein family gene showed significant abnormalities and phenotypes that do not always overlap despite overlapping expression patterns (Ikeda et al. 2002). Therefore, it is obvious that TLPs are multifunctional proteins, and may play multiple independent functional roles. To our knowledge, there has been no attempt to demonstrate the localization of tubby protein in plants, and this is the first report documenting that CaTLP1 is localized predominantly in the cell wall and nucleus (Fig. 5). The multiorganellar localization of CaTLP1 can be attributed to its role in early stress response primarily for perceiving the signals at the cell wall, and then driving them by nuclear translocation. The regulation of CaTLP1 function, through control of dual localization, is of interest because this characteristic feature is well suited for its effective functioning under stress conditions.
Boggon et al. (1999) earlier demonstrated that the mouse Tubby protein is a bipartite transcriptional regulator. The characteristic properties of CaTLP1, particularly stress-induced preferential accumulation in the nucleus and DNA-binding activity support it, at least partly, to be a putative transcription factor (Figs. 5, 6). However, its inability to induce the transcription of reporter gene may account for the absence of transactivation domain in CaTLP1, which is in consistence with the earlier report by Lai et al. (2004). Unlike highly diverse N-terminal sequence of animal TLPs, CaTLP1 contains conserved F-box domain. The role of F-box is established as cell surface receptor and transcription modulator (Kipreos and Pagano 2000). These proteins use broad and different mechanisms for target recognition, the most common mechanism being the formation of a Skp1/Cullin/F-box (SCF) complex. Experimental data from work with yeast indicate that Skp1 interacts with the Cdc53/cullin and F-box proteins to form this characteristic complex (Krek 1998; Patton et al. 1998). Lai et al. (2004) has shown the physical interaction between AtTLP9 and ASK1 (Arabidopsis Skp1 like 1). The presence of the conserved F-box domain in CaTLP1 suggests that it might play key role in stress tolerance, possibly by protein–protein interaction.
Overexpression of CaTLP1 in transgenic tobacco plants rendered improved tolerance to dehydration, salinity and oxidative stresses. There were significant increases in root and leaf development, net photosynthesis, and plant biomass in CaTLP1-overexpressing lines (Figs. 8, 9). A related F-box containing protein, DOR has recently been reported to confer drought tolerance in plants by regulating ABA-induced stomatal closure (Zhang et al. 2008). It is possible that CaTLP1 might influence stomatal closure in a similar fashion in order to minimize water loss, which is corroborated by higher expression of CaTLP1 in leaves (Fig. 3e). Based on the findings of the present study and a comprehensive literature survey on TLPs, we propose that CaTLP1 may be associated with synaptic function in stress tolerance (Lai et al. 2004; Ikeda et al. 2002; Boggon et al. 1999). As described above, CaTLP1 comprises of at least two characteristic domains, Tub and F-box, and is localized in the cell wall, presumably serving as a molecular sensor. Following the possible signal transduction initiating events, it translocates to the nucleus to carry out targeted biological function. Perhaps CaTLP1 binds to DNA, and interacts with the regulator with the help of Tub and F-box domains, respectively. F-box domain may either regulate the activity or recruit other regulators at the promoter that can effectively switch on and off the downstream gene expression. Our future efforts will focus on further characterization of CaTLP1 in stress response, and to evaluate its use as modifier of plant development and stress tolerance.
References
Allen DJ, Ort DR (2001) Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci 6:36–42
Araujo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, Martinoia E, Jordana X, Damatta FM, Fernie AR (2011) Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23:600–627
Banerjee P, Kleyn PW, Knowls JA, Lewis CA, Ross BM, Parano E, Kovats SG, Lee JJ, Penchaszadeh GK, Ott J, Jacobson SG, Gilliam TC (1998) TULP1 mutation in two extended Dominican kindreds with autosomal recessive retinitis pigmentosa. Nat Genet 18:177–179
Bartels D, Furini A, Ingram J, Salamini F (1996) Responses of plants to dehydration stress: a molecular analysis. Plant Growth Regul 20:111–118
Berg OG (1990) In: Revzin A (ed) The biology of nonspecific DNA-protein interactions. CRC Press, Boca Raton, pp 71–85
Bhushan D, Pandey A, Chattopadhyay A, Choudhary MK, Chakraborty S, Datta A, Chakraborty N (2006) Extracellular matrix proteome of chickpea (Cicer arietinum L.) illustrates pathway abundance, novel protein functions and evolutionary perspect. J Proteome Res 5:1711–1720
Bhushan D, Pandey A, Choudhary MK, Datta A, Chakraborty S, Chakraborty N (2007) Comparative proteomic analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol Cell Proteomics 11:1868–1884
Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L (1999) Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286:2119–2125
Bray EA (2004) Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J Exp Bot 55:2331–2341
Cagney G, Uetz P, Fields S (2000) High-throughput screening for protein–protein interactions using two-hybrid assay. Methods Enzymol 328:3–14
Carpita NC, Gibeaut D (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3:1–30
Carroll K, Gomez C, Shapiro L (2004) Tubby proteins: the plot thickens. Nat Rev Mol Cell Biol 5:55–63
Chaves MM, Oliveira MM (2004) Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. J Exp Bot 55:2365–2384
Chaves MM, Maroco JP, Pereira JS (2003) Understanding plant response to drought—from genes to the whole plant. Funct Plant Biol 30:239–264
Choudhary MK, Basu D, Datta A, Chakraborty N, Chakraborty S (2009) Dehydration-responsive nuclear proteome of rice (Oryza sativa L.) illustrates protein network, novel regulators of cellular adaptation, and evolutionary perspective. Mol Cell Proteomics 8:1579–1598
Clarke SE (2001) Cell signalling at the shoot meristem. Nat Rev Mol Cell Biol 2:276–284
Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12:2175–2190
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signalling to jasmonate and ethylene responses. Plant Cell 14:1557–1566
Hagstrom SA, Duyao M, North MA, Li T (1999) Retinal degeneration in tulp1-/- mice: vesicular accumulation in the interphotoreceptor matrix. Invest Ophthalmol Vis Sci 40:2795–2802
He W, Ikeda S, Bronson RT, Yan G, Nishina PM, North MA, Naggert JK (2000) GFP-tagged expression and immunohistochemical studies to determine the subcellular localization of the tubby gene family members. Mol Brain Res 81:109–117
Hobbs SL, Kpodar P, DeLong CM (1990) The effect of T-DNA copy number, position and methylation on reporter gene expression in tobacco transformants. Plant Mol Biol 15:851–864
Ikeda S, He W, Ikeda A, Naggert JK, North MA, Nishina PM (1999) Cell-specific expression of tubby gene family members (tub, Tulp1, 2, and 3) in the retina. Invest Ophthalmol Vis Sci 40:2706–2712
Ikeda A, Ikeda S, Gridley T, Nishina PM, Naggert JK (2001) Neural tube defects and neuro epithelial cell death in Tulp3 knockout mice. Hum Mol Genet 10:1325–1334
Ikeda A, Nishina PM, Naggert JK (2002) The tubby-like proteins, a family with roles in neuronal development and function. J Cell Sci 115:9–14
Jhonson PF, McKnight SL (1989) Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem 58:799–839
Kapeller R, Moriarty A, Strauss A, Stubdal H, Theriault K, Siebert E, Chickering T, Morgenstern JP, Tartaglia LA, Lillie J (1999) Tyrosine phosphorylation of tub and its association with Src homology 2 domain-containing proteins implicate tub in intracellular signalling by insulin. J Biol Chem 274:24980–24986
Kipreos ET, Pagano M (2000) The F-box protein family. Genome Biol 1:3002.1–3002.7
Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L, Charlat O, Woolf EA, Tayber O, Brody T, Shu P, Hawkins F, Kennedy B, Baldini L, Ebeling C, Alperin GD, Deeds J, Lakey ND, Culpepper J, Chen H, Glücksmann-Kuis MA, Carlson GA, Duyk GM, Moore KJ (1996) Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell 85:281–290
Kou Y, Qiu D, Wang L, Li X, Wang S (2009) Molecular analyses of the rice tubby-like protein gene family and their response to bacterial infection. Plant Cell Rep 28:113–121
Krek W (1998) Proteolysis and the G1-S transition: the SCF connection. Curr Opin Genet Dev 8:36–42
Lai CP, Lee CL, Chen PH, Wu SH, Yang CC, Shaw JF (2004) Molecular analysis of the Arabidopsis TUBBY-like protein gene family. Plant Physiol 134:1586–1597
Liu Q (2008) Identification of rice TUBBY-like genes and their evolution. FEBS J 275:163–171
Noben-Trauth K, Naggert JK, North MA, Nishina PM (1996) A candidate gene for the mouse mutation tubby. Nature 380:534–538
Pandey A, Rajamani U, Verma J, Subba P, Chakraborty N, Datta A, Chakraborty S, Chakraborty N (2010) Identification of extracellular matrix proteins of rice (Oryza sativa L.) involved in dehydration-responsive network: a proteomic approach. J Proteome Res 6:3443–3464
Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M (1998) Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev 12:692–705
Porra RJ (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73:149–156
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome wide comparative analysis among eukaryotes. Science 290:2105–2110
Roelfsema MRG, Hedrich R (2005) In the light of stomatal opening: new insights into ‘the Watergate’. New Phytol 167:665–691
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L (2001) G-protein signalling through tubby proteins. Science 292:2041–2050
Sanyal I, Singh AK, Kaushik M, Amla DV (2005) Agrobacterium-mediated transformation of chickpea (Cicer arietinum L.) with Bacillus thuringiensis cry1Ac gene for resistance against pod borer insect Helicoverpa armigera. Plant Sci 168:1135–1146
Slabas AR, Ndimba B, Simon WJ, Chivasa S (2004) Proteomic analysis of the Arabidopsis cell wall reveals unexpected proteins with new cellular locations. Biochem Soc Trans 32:524–528
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vogel JP, Raab TK, Somerville CR, Somerville SC (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40:968–978
Yang Z, Zhou Y, Wang X, Gu S, Yu J, Liang G, Yan C, Xu C (2008) Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 92:246–253
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64:643–651
Zgallai H, Steppe K, Lemeur R (2005) Photosynthetic, physiological and biochemical responses of tomato plants to polyethylene glycol-induced water deficit. J Integrat Plant Biol 47:1470–1478
Zhang Y, Xu W, Li Z, Deng XW, Wu W, Xue Y (2008) F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis thaliana. Plant Physiol 148:2121–2133
Acknowledgments
This research was supported by grants from the Department of Biotechnology (DBT), Government of India. The authors thank Council of Scientific and Industrial Research, Government of India for providing pre-doctoral fellowship to VW, KJ and SG. The authors also thank Dr. Suchismita Dass for critical reading of the manuscript, and Mr. Jasbeer Singh for illustration and graphical representation in the manuscript
Conflict of interest
The authors don’t have conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
V. Wardhan and K. Jahan contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2012_9925_MOESM2_ESM.tiff
Fig. S2 ESI-MS analysis of CaTLP1 showing “multicharge envelope” of signals from differentially charged forms of the protein. (TIFF 2099 kb)
Fig. S4 Southern analysis of chickpea showing CaTLP1 is a member of multigene family. (TIFF 1360 kb)
11103_2012_9925_MOESM5_ESM.tiff
Fig. S5 Comparative analysis of CaTLP1-overexpressing tobacco plants and the wild-type counterparts in terms of biomass and shoot length. (TIFF 2076 kb)
Rights and permissions
About this article
Cite this article
Wardhan, V., Jahan, K., Gupta, S. et al. Overexpression of CaTLP1, a putative transcription factor in chickpea (Cicer arietinum L.), promotes stress tolerance. Plant Mol Biol 79, 479–493 (2012). https://doi.org/10.1007/s11103-012-9925-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-012-9925-y