Abstract
The endosperm of cereal grains is an important resource for both food and feed. It contains three major types of tissue: starchy endosperm, the aleurone layer, and transfer cells. To improve grain quality and quantity using molecular methods, control of transgene expression directed by distinct temporal and spatial promoter activity is necessary. To identify aleurone layer-specific and/or transfer cell-specific promoters in rice, microarray analyses were performed, comparing the aleurone layer containing transfer cells and the other reproductive and vegetative tissues. After confirmation by RT–PCR analysis, we identified two putative aleurone layer and/or transfer cell-specific genes, AL1 and AL2. The promoter regions of these genes and β-glucuronidase (GUS) fusion constructs were stably transformed into rice. The GUS expression patterns indicated that the AL1 promoter was active exclusively in the dorsal aleurone layer adjacent to the main vascular bundle. In rice, transfer cells are differentiated in this region. Therefore, the promoter of the AL1 gene exhibits transfer cell-containing region-specific activity. The AL1 gene encodes a putative anthranilate N-hydroxycinnamoyl/benzoyltransferase. The promoter of this gene will be useful for enhancing uptake of nutrients from the mother cells and protecting filial seeds from pathogen attack.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The endosperm is the major storage organ in cereal grains. Molecular breeding of endosperm is necessary to further improve the quality and yield of grains, and therefore it is necessary to identify distinct tissue- and developmental stage-specific promoters of seeds. After anthesis, the endosperm of cereals develops three major types of tissue: the starchy endosperm, the aleurone layer, and transfer cells (Becraft 2001). After completion of cellularization in the endosperm, the aleurone layer, which is the storage site of proteins, lipids, phosphorus, and minerals, is first differentiated. The aleurone layer also plays a role in advancement of the seed germination process by providing enzymes, which hydrolyze the storage reserves in the endosperm and aleurone layer cells. The aleurone layer adjacent to the vascular bundle develops further into transfer cells with extensive networks of cell wall ingrowths to increase the surface area of the cellular membrane up to 22-fold (Thompson et al. 2001) and to facilitate the uptake of nutrients from the adjacent maternal tissues into the endosperm (Wang et al. 1994). The remaining cells develop into starchy endosperm, which accumulates starches and storage proteins.
In rice, aleurone layers are composed of one to two cell layers on the ventral side, one cell layer on the lateral side, and six to eight cell layers on the dorsal side adjacent to the major vascular bundle in the pericarp. Some cells in the dorsal aleurone layers sporadically form a wall ingrowth structure that is typical of transfer cells (Hoshikawa 1990). That is, not all of the cells adjacent to the main vascular bundle develop into transfer cells in rice. Therefore, in rice, the dorsal aleurone layer adjacent to the vascular bundle is designated the “transfer cell-containing region” in this study.
In contrast, all of the cells in the basal two or three cell layers adjacent to the pedicel develop into transfer cells in maize (Becraft 2001). Many endosperm transfer cell-specific genes have been reported in maize (Hueros et al. 1995, 1999; Gutierrez-Marcos et al. 2004), and they typically encode low molecular weight cysteine-rich proteins with an N-terminal hydrophobic signal peptide. Four types of such protein have been described in maize and named BETL-1 to -4 (Hueros et al. 1995, 1999). However, few studies have analyzed their promoter activity. The activity of BETL-1 promoter was confirmed by β-glucuronidase (GUS) expression analysis restricted to transfer cells of maize. Recently, genes encoding the low molecular weight cysteine-rich proteins OsPR9a and OsPR602 were identified in rice (Li et al. 2008). GUS expression analysis revealed that the promoters of these genes were highly active in the transfer cell-containing region. However, GUS activity was also detected in both maternal tissues and starchy endosperm cells adjacent to transfer cell-containing region and also in several floral tissues shortly before anthesis.
Although many genes are expressed only in the starchy endosperm of rice (Kawakatsu et al. 2008; Qu and Takaiwa 2004; Wu et al. 1998), endosperm transfer cell-specific genes and aleurone layer-specific genes have not yet been reported from rice. Some genes expressed in both the embryo and aleurone layer (Miyoshi et al. 1999; Yoshida et al. 1999) and some expressed in both the starchy endosperm and aleurone layer (Kawakatsu et al. 2008; Kuwano et al. 2009) have been reported previously. Aleurone layer-specific promoters as well as transfer cell-specific promoters have also been reported for other cereals. The promoter of the barley lipid transfer protein gene HvLTP2 induces aleurone layer-specific expression during barley seed development (Kalla et al. 1994). However, it directs expression in the embryo as well as the aleurone layer of transgenic rice (Opsahl-Sorteberg et al. 2004). These observations suggest that the activity of the promoter is not similar even among cereal species. Therefore, it is necessary to determine the promoter specificity for each plant species.
Here, we attempted to isolate aleurone layer-specific and/or transfer cell-containing region-specific promoters in rice. cDNA microarray technology has made it possible to monitor the expression of specific genes in a whole genome, analyzing thousands of genes at once. To detect differential gene expression among the developing embryo, endosperm, and aleurone layer containing transfer cells, we collected samples separately from rice caryopses 10 days after flowering (DAF). Then microarray analysis was performed to identify genes expressed in the aleurone layer containing the transfer cells, but not in any other parts of the seed or in other vegetative tissues.
Materials and methods
Plant material
Rice (Oryza sativa L. var. japonica cv. Nipponbare) plants were grown in pots in a greenhouse under natural light. Before the heading stage, the plants were transferred outside. Then, caryopses 10 DAF were collected. We only used superior caryopses that exhibited a high grain filling rate, because many genes show differences in expression between superior and inferior caryopses (Ishimaru et al. 2005; Kuwano et al. 2009). To collect the aleurone layers containing transfer cells, the lemmas, paleae, pericarps, and seed coats were removed from the caryopses, and then the aleurone layers were peeled off using forceps and a razor (Fig. 1). The samples were immediately frozen in liquid nitrogen. At the same time, the inner starchy endosperm and the embryo were colleted. Flowers and pistils were collected 1 day before flowering (DBF), caryopses were collected 7, 10, 14, 21, and 28 DAF, and leaves and roots of seedlings 7 days after germination (DAG) were collected. The seedlings were grown in a growth chamber at 28°C with 14 h/day illumination at an intensity of approximately 240 μmol m−2 s−1.
RNA isolation
Total RNA from rice tissues was extracted from the plant materials as described previously (Suzuki et al. 2007) using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). To eliminate residual genomic DNA, the RNA was treated with DNase I using an RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. To check for the absence of contaminating PCR bands derived from genomic DNA amplification, we used a pair of forward and reverse primers amplifying a product spanning one intron. For microarray and RT–PCR analyses, we used separate RNA preparations from independent tissue samples.
Microarray analysis
The quality of all RNA samples was examined by capillary electrophoresis with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Fluorescently labeled cDNAs were generated from 400 ng total RNA in each reaction using an Agilent Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies) and 10 mM Cy3- or Cy5-labeled dCTP (PerkinElmer, Boston, MA). Hybridization was performed according to the oligonucleotide microarray hybridization user’s manual with an Agilent in situ Hybridization Kit Plus (Agilent Technologies). We used a rice 44K oligo-microarray (Agilent Technologies) containing approximately 42,000 oligonucleotides synthesized based on the nucleotide sequence and full-length cDNA data of the Rice Annotation Project (RAP; http://www.rapdb.dna.affrc.go.jp/) to compare samples of the aleurone layer containing transfer cells, embryo, and endosperm. We used a rice 22K oligomicroarray (Agilent Technologies) containing approximately 22,000 oligonucleotides synthesized based on sequence data from the database of full-length cDNA clones from japonica rice (KOME; http://www.cdna01.dna.affrc.go.jp/cDNA/; Kikuchi et al. 2003) to compare samples of leaf, root, flower, pistil, and embryo. The arrays were scanned using a dual-laser DNA microarray scanner (Agilent Technologies).
RT–PCR analysis
The gene-specific primers used for RT–PCR are shown in Table 1. RT–PCR amplification was performed using a OneStep RT–PCR Kit (Qiagen). The actin gene was used as a control because its expression levels are approximately constant in various developmental stages (Yoshida et al. 2005). The experiment was repeated three times with three independent RNA samples.
Plasmid construction and transformation procedures
Regions of 1,728 base pairs (bp) of the AL1 promoter region and 2259 bp of the AL2 promoter region were amplified by PCR using specific primer pairs: 5′-CTCACGAGACCAATTATGAAAAGCGAGA-3′ and 5′-TGTCAAGGGTGCCGGTGAAGACGTAGAT-3′ for AL1, and 5′-TGGTTCGTGTCCTGAGGGCCACGCCT-3′ and 5′-TGGCGACCCGCTTGCTGAGGGTGCTG-3′ for AL2. The promoter regions were determined according to the sequence data in the database KOME. The following plasmids were constructed using the Invitrogen Gateway System (Invitrogen, Carlsbad, CA). The promoter regions were inserted into the pGWB3 vector (Nakagawa et al. 2007), which contains the GUS reporter gene and the Agrobacterium tumefaciens nopaline synthase (Nos) terminator. The resulting plasmids, AL1pro::GUS and AL2pro::GUS, were transferred into A. tumefaciens strain EHA105 by electroporation. Transgenic rice plants (cv. Kitaake) were produced as described previously (Toki et al. 2006).
Analysis of GUS gene expression
To examine the AL1 and AL2 promoter activities, leaves, secondary roots, and flowers 1 DBF, and immature seeds 7, 10, 14, 21, and 28 DAF were collected from the transgenic plants. Nineteen AL1pro::GUS and eighteen AL2pro::GUS transgenic plants were analyzed. All of the samples were subjected to histochemical analyses as described previously (Qu and Takaiwa 2004). The leaves were cut into 5-mm-thick sections, and seeds were cut into median transversal or longitudinal sections with a razor blade. The median transversal sections of seeds were embedded in 5% agar and cut into 100-μm-thick sections with a microslicer prior to microscopic observation.
Results
Microarray analyses
To identify aleurone layer-specific and/or transfer cell-specific genes, total RNAs from 10-day-old aleurone layers containing transfer cells, starchy endosperm, and embryos were compared by rice 44K oligomicroarray analysis. Clones with an average fluorescent intensity of >3,000 in the aleurone layers and <500 in both the embryos and the endosperm were selected as candidates for aleurone layer-specific and/or transfer cell-specific genes. This is because a fluorescence intensity of >3,000 is generally regarded as indicating abundant expression, while <500 is regarded as scarce expression. Only 56 of the approximately 42,000 clones were selected for further analysis.
To eliminate the possibility that the 56 candidate genes show expression in vegetative tissues, the Knowledge-based Oryza Molecular Biological Encyclopedia (KOME) cDNA database (Kikuchi et al. 2003) was searched to obtain information regarding the cDNA library from which the clones were derived. Forty-two clones were from cDNA libraries derived from vegetative tissues. The remaining 14 clones were from cDNA libraries of the panicles after heading or from cDNA libraries of the callus. These were included in the subsequent analyses.
Gene expression patterns of the 14 candidate clones were further monitored by rice 22K microarray with total RNA from leaves and roots 7 DAG, from flowers and pistils 1 DBF, and from embryos 10 DAF. Clones that had a fluorescence intensity of >500 in the leaves, roots, flowers, pistils, or embryos were excluded. Eight clones were used in subsequent analysis as candidates for aleurone layer-specific and/or transfer cell-specific genes. The Rice Annotation Project Database (RAP) locus IDs of these eight clones are Os01g0382200, Os02g0170300, Os02g0721100, Os02g0775300, Os03g0410000, Os07g0529000, Os10g0466800, and Os10g0467000.
RT–PCR analysis
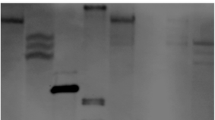
To confirm the aleurone layer and/or transfer cell-specific expression patterns of the eight selected candidate genes, RT–PCR analysis was performed using total RNA extracted from leaves and roots of 7 DAG flowers and pistils 1 DBF, and from immature embryos, the endosperm, and aleurone layers containing transfer cells 10 DAF. The transcripts corresponding to Os01g0382200 and Os03g0410000 were abundant only in aleurone layers containing transfer cells, and were undetectable in leaves, roots, flowers, pistils, embryos, and endosperm (Fig. 2). Therefore, RT–PCR analysis confirmed the results of microarray analyses for Os01g0382200 and Os03g0410000. The Os01g0382200 and Os03g0410000 genes were designated AL1 and AL2, respectively. However, the other genes did not show aleurone layer-specific expression in the RT–PCR analysis (Fig. 2).
RT–PCR analysis of the eight candidate aleurone layer-specific and/or transfer cell-specific genes. cDNA templates were prepared from leaves (L) and roots (R) of 7 DAG flowers (F) and pistils (P) 1 DBF, and immature embryos (Em), endosperm (En), and aleurone layers containing transfer cells (Al) 10 DAF. Lanes 1–8 indicate Os01g0382200, Os02g0170300, Os02g0721100, Os02g0775300, Os03g0410000, Os07g0529000, Os10g0466800, and Os10g0467000, respectively
Further RT–PCR analyses for AL1 and AL2 were also performed using total RNA extracted from developing seeds 7, 10, 14, 21, and 28 DAF. The AL1 transcripts were only detected 7 and 10 DAF, not 14, 21, or 28 DAF, suggesting that this gene is expressed in the early stages of seed development (Fig. 3). The AL2 transcripts were detected 7, 10, and 14 DAF, not 21 or 28 DAF.
Gene identification
BLASTX searches against the non-redundant protein database at NCBI (http://www.ncbi.nlm.nih.gov/) indicated that the AL1 gene encodes a hypothetical member of the transferase family, which includes anthranilate N-hydroxycinnamoyl/benzoyltransferase. The function of this gene in plants is unknown. Secondary structure prediction performed for the AL1 protein using the SOSUI program (available at http://www.bp.nuap.nagoya-u.ac.jp/sosui/) indicated that AL1 is a soluble protein with no transmembrane helices. Prediction of protein-sorting signals and localization sites in the amino acid sequences of AL1 was performed using the PSORT program (available at http://www.psort.hgc.jp/), and the protein was predicted to be located in the cytoplasm. On the other hand, the AL2 gene encodes the myb-related protein OsMYB3 (Suzuki et al. 1997), which is thought to be a transcription factor.
GUS expression analyses
To determine the promoter activities of the AL1 and AL2 genes, the AL1 and AL2 promoter regions were fused to the GUS reporter gene, and stably introduced into rice plants. Eighteen and nineteen fertile transformant lines were obtained by transformation with the constructs AL1pro::GUS and AL2pro::GUS, respectively. The GUS activities in leaves, roots, and flowers 1 DBF, and immature seeds 7, 10, 14, 21, and 28 DAF were examined. No GUS expression was detected in any of the tissues examined among the 19 lines of AL2pro::GUS transgenic plants (data not shown).
All GUS-expressing lines of AL1pro::GUS transgenic plants showed the same spatial and temporal patterns of GUS expression. However, the levels of expression differed between lines. GUS activity was seen only in the immature seeds (Fig. 4). In contrast, no GUS expression was detected in any of the samples of leaves, roots, and flowers of AL1pro::GUS transgenic plants (Fig. 4). This agreed well with the microarray and RT–PCR data for the AL1 gene. Longitudinal sections of seeds revealed that only the dorsal side, that is, the paleae side of the aleurone layer from top to bottom, showed GUS activity (Fig. 4). A high level of GUS expression was observed throughout seed development from 7 to 28 DAF. GUS activity was not detected on the ventral side of the aleurone layer, the embryo, or the starchy endosperm throughout seed development (Fig. 4). Observations of median cross-sections revealed that GUS expression was restricted to the aleurone layers adjacent to the dorsal main vascular bundle (Fig. 5). In rice, this region contains transfer cells (Hoshikawa 1990). That is, the AL1 promoter activity was only detected in the transfer cell-containing region.
GUS staining analysis of various organs in AL1pro::GUS plants. a, b, and c Leaf, root, and flower 1 DBF, respectively. d, e, f, g, and h Longitudinal sections of seeds 7, 10, 14, 21, and 28 DAF, respectively. i, j, and k Higher magnifications of (e). i, j, and k Embryo region, ventral side, and dorsal side of the seed, respectively. Bars indicate 1 mm in (a–h) and 0.5 mm in (i–k)
Analysis of promoter sequences
To elucidate the possible cis-acting elements responsible for gene expression in developing seeds, 2.0 kb of the AL1 promoter region was analyzed using the PLACE database (available at http://www.dna.affrc.go.jp/htdocs/PLACE; Higo et al. 1999). Although the endosperm box (TGATAAAG), which is conserved in many cereal genes and is involved in the expression of seed storage proteins, was not found in the AL1 promoter, other motifs responsible for seed-specific expression of storage proteins were found repeatedly (Table 2). In the AL1 promoter, the RY-repeat element (CATGCATG) responsible for seed-specific regulation was found at position −261. The AACAAAC motif was found at position −1761, the CTTTCGTGTAC motif was found at position −525, the ATATTTAWW motif was found at positions −995 and −1117, the RTTTTTR motif was found at positions −1202, −1516, −1528, and −1941, and the TACACAT motif was found at positions −539 and −747.
The plant hormone abscisic acid (ABA) plays a critical role in seed development (Marion-Poll 1997; Leung and Giraudat 1998), including induction and maintenance of seed dormancy, accumulation of storage products, and acquisition of desiccation tolerance. It is possible that the genes expressed in developing seeds contain ABA-responsive cis-acting elements in their promoter regions. Although a strong ABA-responsive element, (C/T)ACGTGGC, was not found in the AL1 promoter, the MYC element (CACATG) responsible for the ABA desiccation response was found at positions −263, −1653, and −1790 (Table 2).
The maize transfer cell-specific transcriptional activator ZmMRP-1 induces expression of transfer cell-specific genes, including Meg1 (Gutierrez-Marcos et al. 2004), BETL-1, and BETL-2 (Gomez et al. 2002). Barrero et al. (2006) investigated the interaction between ZmMRP-1 and the promoter of BETL-1. They found that a 12-bp motif, TATCTCTATCTC, is the strongest target motif of ZmMRP-1. In the AL1 promoter region, the same motif, GAGATAGAGATA, which is complementary to TATCTCTATCTC, was found at position −887 (Table 2). In addition, there are several repeats of the weaker target sequence motif, TAGATATAGATA, which is complementary to TATCTATATCTA, around the above strongest motif (Table 2).
Two of three rice seed-specific elements, CATGCATG and CTTTCGTGTAC, found in the AL1 promoter were not seen in the AL2 promoter (Table 2). The AL2 promoter also lacks transfer cell-specific elements. Furthermore, the AL2 promoter has a strong ABA responsive element, ACGTGKC, which is not present in the AL1 promoter.
Discussion
Rice 44K and 22K microarrays and RT–PCR analyses were used to identify genes that were expressed exclusively in the aleurone layers and/or in the transfer cells of developing seeds. One gene, Os01g0382200 (AL1), was designated as a transfer cell-containing region-specific gene. To our knowledge, this is the first report of a transfer cell-containing region-specific promoter activity in rice, as well as the first report of a putative anthranilate N-hydroxycinnamoyl/benzoyltransferase gene specifically expressed in a transfer cell-containing region.
Many transfer cell-specific genes have been reported in other cereals, and they usually encode low molecular weight cysteine-rich proteins with N-terminal hydrophobic signal peptides. In contrast, the AL1 protein is not a low molecular weight protein (406 amino acids), has no signal peptide, and is a soluble protein, and therefore it is a novel transfer cell-containing region-specific protein. The basal endosperm transfer layer-specific (BETL) proteins BETL-1 to -4 have been identified in maize. BETL-1 and BETL-3 are similar to defensin-like proteins, suggesting possible antimicrobial functions (Hueros et al. 1999). BETL-4 shows some identity to the Bowman-Birk family of α-amylase/trypsin inhibitors (Hueros et al. 1999). BETL-2 is a member of a small family of related peptides that possess antifungal activity (Serna et al. 2001). Another low molecular weight cysteine-rich protein, END1, was identified in barley transfer cells (Doan et al. 1996). The protein sequence of END1 provided no insight into its function. Royo et al. (2006) reported the identification of a receptor-like kinase, ZmLrk-1, which was induced by fungal infection in germinated seeds. In developing seeds, ZmLrk-1 transcripts can be detected exclusively at the basal endosperm transfer cell layer. These findings suggest that transfer cells play a role in defense of filial seeds from attack by pathogens as well as in uptake of nutrients from the mother cells. The AL1 gene encodes a putative transferase protein, which catalyses the first committed reaction of phenylpropanoid phytoalexin biosynthesis, that is, the formation of N-benzylanthranilate from benzoyl-CoA and anthranilate (Yang et al. 1997). Phytoalexins are produced in response to infection by parasites, and are essential for the expression of disease resistance. The AL1 may possibly constitute such a defense system.
In rice, not all of the cells adjacent to the dorsal main vascular bundle differentiate into transfer cells (Hoshikawa 1990). On the dorsal side of the endosperm, 6–8 cell layers develop into aleurone layer cells, and typical transfer cells with a wall ingrowth structure are scattered throughout this region. In GUS expression analysis, GUS activity was detected uniformly in the dorsal aleurone layer cells adjacent to the vascular bundle (Fig. 5), but was not detected in a scattered manner indicating the positions of transfer cells. This suggests that the function of AL1 is necessary throughout the whole transfer cell-containing region but not only in transfer cells. Further investigations are required to determine the precise role of AL1.
Recently, Li et al. (2008) identified two putative transfer cell-specific rice genes: OsPR602, which shows protein sequence identity with barley END1 (Doan et al. 1996), and OsPR9a, which contains a cysteine in a similar position to that in the maize defensin-like protein BETL-3 (Hueros et al. 1995). The results of GUS expression analyses indicated that these promoters were active not only in the transfer cells but also in several floral tissues shortly before pollination. The maize transfer cell-specific trans-acting element, ZmMRP-1, interacts with the cis-element, TATCTCTATCTC motif, in the BETL-1 promoter (Barrero et al. 2006). However, this 12-bp motif is not found in either the OsPR602 or OsPR9a promoter (Li et al. 2008). On the other hand, this motif is present in the AL1 promoter (Table 2), and is active only in the transfer cell-containing region. These results suggest that a common cis-element acts in maize and rice and ensures exclusive expression in the transfer cell-containing region. The introduction of a deletion and substitution mutation within the AL1 promoter would be helpful in determining the transfer cell-specific cis-elements in rice.
The promoter activity of TdPR60, the Triticum durum homolog of the barley END1 gene, was studied in stable transformed wheat, barley, and rice containing the promoter::GUS construct (Kovalchuk et al. 2009). The promoter activity in wheat and barley was found only in the endosperm transfer cells, but in rice was active within the starchy endosperm. These observations suggest diversity of cis-elements and transcription factors among cereal species, although the presence of cis-acting elements responsible for seed development and the ABA response in the AL1 promoter indicate that some elements are common among plant species (Table 2).
In this study, the GUS expression results were not entirely consistent with the results of the RT–PCR analysis. AL1 mRNA was detected in seeds only at 7 and 10 DAF, and disappeared after 10 DAF (Fig. 3), whereas AL1 promoter activity was detected throughout seed development (Fig. 4). One reason for this inconsistency is that because β-glucuronidase is comparatively stable, protein that was synthesized by 10 DAF might have still been present after 10 DAF.
We could not identify an aleurone layer-specific promoter in rice, although RT–PCR analysis indicated that the AL2 gene was specifically expressed in the aleurone layer (Fig. 2). The AL2 gene encodes the R2R3 MYB-domain protein with high similarity to the maize COLORLESS1 (C1) factor and Arabidopsis TRANSPARENT TESTA2 (TT2). The C1 gene plays a regulatory role in the production of anthocyanin pigments in the aleurone layer of the endosperm (Cone et al. 1986; Paz-Ares et al. 1987). The TT2 gene was expressed specifically in the seeds in early developmental stages and TT2 is involved in the regulation of flavonoid metabolism (Nesi et al. 2001). These results support the suggestion that the AL2 gene is expressed specifically in the aleurone layers and plays a role in the regulation of genes related to anthocyanin biosynthesis or other metabolism in this region. For the AL2pro::GUS chimeric gene, the genomic AL2 sequence from −2259 to −1 was used in this study. The ORF of AL2 was determined according to the sequence data in the KOME database. This 2259-bp fragment of the AL2 promoter region contains typical TATA and CCAAT boxes, located 144 and 181 bases upstream from the putative transcription initiation site, respectively. However, GUS analysis showed no AL2 promoter activity in the aleurone layer, or in any other plant tissue. According to a promoter sequence analysis, the AL2 promoter has fewer seed-specific cis-elements than the AL1 promoter (Table 2). These observations suggest that the AL2 promoter fragment was insufficient to induce gene expression in seeds and that cis-acting elements must be present in other regions of this gene. As the AL2 gene has no intron, the 5′ sequences further upstream of the promoter in this study and/or the 3′ flanking region of the AL2 gene may contain some regulatory cis-acting elements necessary for aleurone layer-specific expression (Hyder et al. 1992).
Several genes encoding proteins belonging to the lipid transfer protein family are expressed specifically in aleurone layer cells of cereals. The barley HvLTP2 gene was identified as an aleurone layer-specific gene and the promoter of HvLTP2 induced aleurone cell-specific expression in barley (Opsahl-Sorteberg et al. 2004). However, the HvLTP2 promoter is active in both the embryo and aleurone layer in transgenic rice. In maize, an aleurone-specific gene encoding a putative lipid transfer protein, AL9, was isolated (Gomez et al. 2009). The promoter activity of AL9 has not been tested in other plant species.
The AL1 promoter is suitable for transfer cell-containing region-specific expression of genes of interest in rice. This promoter offers the potential to improve both nutrient uptake from the maternal tissue and defense against pathogen invasion through transfer cells.
References
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell 9:1859–1868
Allen RD, Bernier F, Lessard PA, Beachy RN (1989) Nuclear factors interact with soybean β-conglycinin enhancer. Plant Cell 1:623–631
Barrero C, Muniz LM, Gomez E, Hueros G, Royo J (2006) Molecular dissection of the interactional activator ZmMRP-1 and the promoter of BETL-1. Plant Mol Biol 62:655–668
Becraft PW (2001) Cell fate specification in the cereal endosperm. Cell Dev Biol 12:387–394
Busk PK, Jensen AB, Pages M (1997) Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J 11:1285–1295
Cone KC, Burr FA, Burr B (1986) Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA 83:9631–9635
Doan DNP, Linnestad C, Olsen O-A (1996) Isolation of molecular markers from the barley endosperm coenocyto and surrounding nucellus cell layers. Plant Mol Biol 31:877–886
Ericson ML, Muren E, Gustavsson H-O, Josefsson L-G, Rask L (1991) Analysis of the promoter region of napin genes from Brassica napus demonstrates binding of nuclear proteins in vitro to a conserved sequence motif. Eur J Biochem 197:741–746
Gomez E, Royo J, Guo Y, Thompson R, Hueros G (2002) Establishment of cereal endosperm expression domains: identification and properties of maize transfer cell-specific transcription factor, ZmMRP-1. Plant Cell 14:599–610
Gomez E, Royo J, Muniz LM, Sellam O, Paul W, Gerentes D, Barrero C, Lopez M, Perez P, Hueros G (2009) The maize transcription factor myb-related protein-1 is a kea regulator of the differentiation of transfer cells. Plant Cell 21:2022–2035
Gutierrez-Marcos JF, Costa LM, Biderre-Petit C, Khbaya B, O’Sullivan DM, Wormald M, Perez P, Dickinson HG (2004) Maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16:1288–1301
Hattori T, Terada T, Hamasuna S (1995) Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J 7:913–925
Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43:136–140
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucl Acids Res 27:297–300
Hoshikawa K (1990) Anthesis, fertilization and development of caryopsis. In: Matsuo T, Shimizu S, Hoshikawa K, Maeda E, Yamazaki K (eds) Science of the Rice Plant, vol. 1 morphology. Nobunkyo, Tokyo, pp 273–306
Hueros G, Varotto S, Salamini F, Thompson D (1995) Molecular characterization of BET1, a gene expressed in the endosperm transfer cells of maize. Plant Cell 7:747–757
Hueros G, Royo J, Maitz M, Salamini F, Thompson D (1999) Evidence for factors regulating transfer cell-specific expression in maize endosperm. Plant Mol Biol 41:403–414
Hyder SM, Stancel GM, Nawaz Z, McDonnell DP, Loose-Mitchell DS (1992) Identification of an estrogen response in the 3′-flanking region of the murine c-fos protooncogene. J Biol Chem 267:18047–18054
Ishimaru T, Hirose T, Matsuda T, Goto A, Takahashi K, Sasaki H, Terao T, Ishii R, Ohsugi R, Yamagishi T (2005) Expression patterns of genes encoding carbohydrate-metabolizing enzymes and their relationship to grain filling in rice (Oryza sativa L.): comparison of caryopses located at different positions in a panicle. Plant Cell Physiol 46:620–628
Kalla R, Shimamoto K, Potter R, Nielsen PS, Linnestad C, Olsen OA (1994) The promoter of the barley aleurone-specific gene encoding a putative 7 kDa lipid transfer protein confers aleurone cell-specific expression in transgenic rice. Plant J 6:849–860
Kawakatsu T, Yamamoto MP, Hirose S, Yano M, Takaiwa F (2008) Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. J Exp Bot 59:4233–4245
Kikuchi S et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301:376–379
Kovalchuk N, Smith J, Pallotta M, Singh R, Ismagul A, Eliby S, Bazanova N, Milligan AS, Hrmova M, Langridge P, Lopato S (2009) Characterization of the wheat endosperm transfer cell-specific protein TaPR60. Plant Mol Biol 71:81–98
Kuwano M, Takaiwa F, Yoshida KT (2009) Differential effects of a transgene to confer low phytic acid in caryopses located at different positions in rice panicles. Plant Cell Physiol 50:1387–1392
Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49:199–222
Li M, Singh R, Bazanova N, Milligan AS, Shirley N, Langridge P, Lopato S (2008) Spatial and temporal expression of endosperm transfer cell-specific promoters in transgenic rice and barley. Plant Biotechnol J 6:465–476
Marion-Poll A (1997) ABA and seed development. Trends Plant Sci 2:447–448
Miyoshi K, Nakata E, Nagato Y, Hattori T (1999) Differential in situ expression of three ABA-regulated genes of rice, RAB16A, REG2 and OSBZ8, during seed development. Plant Cell Physiol 40:443–447
Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kitamura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2114
Opsahl-Sorteberg H-G, Divon HH, Nielsen PS, Kalla R, Hammond-Kosack M, Shimamoto K, Kohli A (2004) Identification of a 49-bp fragment of HvLTP2 promoter directing aleurone cell specific expression. Gene 341:49–58
Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6:3553–3558
Qu LQ, Takaiwa F (2004) Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol J 2:113–125
Royo J, Gomez E, Balandin M, Muniz LM, Hueros G (2006) ZmLrk-1, a receptor-like kinase induced by fungal infection in germinating seeds. Planta 223:1303–1314
Serna A, Maitz M, O’Connell T, Santandrea G, Thevissen K, Tienens K, Hueros G, Faleri C, Cai G, Lottspeich F, Thompson RD (2001) Maize endosperm secrets a novel antifungal protein into adjacent maternal tissue. Plant J 25:687–698
Suzuki A, Suzuki T, Tanabe F, Toki S, Washida H, Wu C-Y, Takaiwa F (1997) Cloning and expression of five myb-related genes from rice seed. Gene 198:393–398
Suzuki M, Tanaka K, Kuwano M, Yoshida KT (2007) Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L.): implications for the phytic acid biosynthetic pathway. Gene 405:55–64
Takaiwa F, Oono K (1990) Interaction of an immature seed-specific trans-acting factor with the 5′ upstream region of a rice glutelin gene. Mol Gen Genet 224:289–293
Thompson RD, Hueros G, Becker H-A, Maitz M (2001) Development and functions of seed transfer cells. Plant Sci 160:775–783
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976
Wang HL, Offler CE, Patrick JW, Uglade TD (1994) The cellular pathway of photosynthate transfer in the developing wheat grain. 1. Delineation of a potential transfer pathway using fluorescent dyes. Plant Cell Environ 17:257–266
Wu LS, Wang LD, Chen PW, Chen LJ, Tzen JT (1998) Genomic cloning of 18 kDa oleosin and detection of triacylglycerols and oleosin isoforms in maturing rice and postgerminative seedlings. J Biochem 123:386–391
Wu C, Washida H, Onodera Y, Harada K, Takaiwa F (2000) Quantitative nature of the prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. Plant J 23:415–421
Yang Q, Reinhard K, Schiltz E, Matern U (1997) Characterization and heterologous expression of hydroxycinnamoyl/bezoyl-CoA: anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol 35:777–789
Yoshida KT, Wada T, Koyama H, Mizobuchi-Fukuoka R, Naito S (1999) Temporal and spatial patterns of accumulation of transcript of myo-inositol-1-phosphate synthase and phytin-containing particles during seed development in rice. Plant Physiol 119:65–72
Yoshida KT, Endo M, Nakazono M, Fukuda H, Demura T, Tsuchiya T, Watanabe M (2005) cDNA microarray analysis of gene expression changes during pollination, pollen-tube elongation, fertilization, and early embryogenesis in rice pistils. Sex Plant Reprod 17:269–275
Acknowledgments
The authors would like to acknowledge Dr. T. Nakagawa for kindly providing the pGWB3 vector. The authors are grateful to Dr. J. Ito for tips on microscopic observations. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan to K. T. Y. (15380004) and by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project) to K. T. Y.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kuwano, M., Masumura, T. & Yoshida, K.T. A novel endosperm transfer cell-containing region-specific gene and its promoter in rice. Plant Mol Biol 76, 47–56 (2011). https://doi.org/10.1007/s11103-011-9765-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9765-1