Abstract
Advances in systematic computational biology and rapid elucidation of synergistic interplay between cis and trans factors governing transcriptional control have facilitated functional annotation of gene networks. The generation of data through deconstructive, reconstructive and database assisted promoter studies, and its integration to principles of synthetic engineering has started an era of designer promoters. Exploration of natural promoter architecture and the concept of cis engineering have not only enabled fine tuning of single or multiple transgene expression in response to perturbations in the chemical, physiological and environmental stimuli but also provided researchers with a unique answer to various problems in crop improvement in the form of bidirectional promoters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
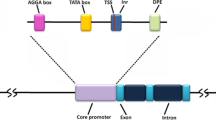
Revolution in sequencing technologies during the post genomic era spawned a huge amount of whole genome data that facilitated functional annotation of genes, proposing their evolution and expression in specific type of cells under certain environmental conditions. Expression of genes is regulated by a number of factors like promoter strength, cis- and trans-acting factors, cell growth stage, the expression level of RNA polymerase associated factors and other gene-level regulation. Efforts to characterize various promoters provided insights into control and modulation of gene expression, identifying cis elements as the major regulatory factors. The proximal and distal promoter regions include cis-regulatory elements that contain binding sites for trans-acting regulatory proteins known as transcription factors (TF) (reviewed in Novina and Roy 1996). In order to regulate the expression of gene under conditions like abiotic stress, biotic stress etc., we need to decipher the meaning of cis regulatory elements within the promoter region. Advances in promoter technology provide a framework for designing an expression cassette that could not only provide precise control of transgene activity but also modulate expression of a transgene in various contexts. A synthetic promoter is constructed using an array of cis-acting element from various sources as building blocks for the promoter engineering strategy. The concept may allow the researcher to reuse selectable markers after each gene modulation step, improve the expression characteristics, reduce unwanted background expressions and delimitate the number of genes that can be modulated simultaneously.
Naturally occurring promoters have long been available. They can be isolated and used to control the gene of interest put under their control. These constructs can then be inserted into appropriate host and used for analysing expression profile. But, however diverse the strengths of these promoters might be, they cannot provide a wide range of promoter strengths in a continuous manner. Constitutive promoter is a matter of choice in cases where steady state expression of gene is needed. A number of strong constitutive promoters have been derived from caulimoviruses, particularly cauliflower mosaic virus (CaMV). Some of the examples of plant promoters are CaMV 35S and 19S (Odell et al. 1985), the figwort mosaic virus (FMV) for full length transcript (Gowda et al. 1989), the nopaline synthase promoter (An et al. 1990), the octopine synthase promoter (Ellis et al. 1987) etc. CaMV 35S promoter is used extensively for the expression of plant genes as it is well characterised and is active in diverse variety of plants (Odell et al. 1985). The CaMV 35S promoter contains three domains: (1) Minimal core promoter containing TATA box from −46 to +8 bp with respect to transcription initiator site; (2) Domain A (also designated as sub-domain A1) expanding from −90 to −46 bp region that performs accessory role in increasing the transcriptional activity of upstream enhancers; (3) Domain B region (−343 to −90 bp) is further divided into five sub-domains (designated as B1–B5) that are recognised based on their interactions with various transcription factors. This region of upstream cis-regulatory sequence can function as transcription enhancer and also perform accessory role in increasing the transcriptional activity of enhancers (Benfey and Chua 1990; Fang et al. 1989). According to Benfey and Chua (1990) domains A and B regulate the tissue-specific gene expression patterns and their function were studied through combinatorial gain-of-function studies. However, domain A fused to minimal promoter alone doesn’t express gene. But domain A combined with domain B allows expression of transgene in plant. This indicates that complete domain B contains important sequences at the interface of sub-domains and the additive effect of sub-domains of domain B along-with domain A and minimal core promoter allows regulated expression of transgene. Recently, three studies conducted by Bhullar et al. (2003, 2007, 2010) have revealed that domain swapping or rearrangement of cis-elements in CaMV 35S promoter region creates synthetic CaMV 35S promoters with minimum sequence homology whose transgene activity is equivalent to that of the wild type CaMV 35S promoter. Hence, CaMV 35S promoter is a significant promoter model for studying combinatorial cis-engineering in synthetic plant promoters.
More recently many tissue specific promoters have been characterised that specifically direct gene expression in certain tissues only. These promoters are defined by specific cis-elements that allow expression of a gene. Zavallo et al. (2010) have isolated and functionally characterised two novel seed specific promoters namely HaFAD2-1 and HaAP10, from sunflower. These promoters contained seed specific motifs like AACA motif, ACGT element, E-Boxes, SEF binding sites and GCN4 motif. They further proved that promoter HaFAD2-1 has GUS activity in transformed seeds equivalent to constitutive CaMV 35S promoter in Arabidopsis transgenic plant. Hence this promoter has expression level same as CaMV 35S promoter.
Rediscovering the natural models
There are two approaches to study promoter structure and function in conjugation with different cis-elements and transgenes:
-
(1)
Deconstructive approach includes 5′ and 3′ deletion, analysis of activity of gene with respect to addition as well as removal of module or regulatory elements and specific mutations in nucleotide sequence of cis-elements. For example, certain auxin induced cis-regulatory elements like AUXRE (Auxin-Responsive Element) and ABRE (Abscisic Acid-Responsive Element) were characterised using above-mentioned as well as gain-of-function strategies (Liu et al. 1994). Kamisugi and Cuming (2005) used site directed mutagenesis to characterize a promoter in Physcomitrella patens and studied the evolution of the Abscisic acid-response in land plants.
-
(2)
Reconstructive approach allows addition of specific cis-regulatory elements individually or in combination, to the minimal strong constitutive promoter, thereby creating a synthetic promoter (Venter 2007; Venter and Botha 2010). It also involves generation of synthetic bidirectional promoter expressing more than one transgene under the regulation of specific and identified cis-elements. For example, Rushton et al. (2002), designed pathogen-inducible plant promoters by placing multiple cis-regulatory elements (that can mediate gene expression under pathogen attack) upstream to strong wild type CaMV 35S promoter. The study also revealed that pathogen induced gene transcription can also activate local wound-induced gene expression in plant. Cazzonelli and Velten (2008) also designed synthetic promoter carrying short direct repeated enhancer elements from Geminivirus, Nanoviruses, Badnaviruses and Caulimoviruses. These enhancer elements or short regulatory cassettes individually or in combination were placed upstream to minimal CaMV 35S promoter carrying reporter gene. This strategy resulted in higher expression of gene when direct repeat cassettes were placed in combination to the minimal promoter. Reconstructive approach also includes study of cis-elements in conjugation with minimal promoter. Mehrotra et al. (2005), Mehrotra and Panwar (2009) and Mehrotra and Mehrotra (2010) have studied two cis-motifs i.e. ACGT and GT which regulate pathogen defence and their respective functions when they are placed 50 nucleotides or 100 nucleotides upstream to TATA box. It was interpreted that single ACGT motif enhanced promoter expression by 2.39-fold when placed 100 nucleotides upstream to TATA box while two ACGT motifs separated by 5 nucleotides enhanced promoter expression by sixfold when placed 50 nucleotides upstream to minimal promoter.
A recent study by Mehrotra and Mehrotra (2010) indicated that two copies of ACGT when separated by 5 nucleotides allowed promoter activation by salicylic acid and when separated by 25 nucleotides, promoter was induced by abscisic acid but not salicylic acid. The differential induction is expected to involve the recruitment of different bZIP transcription factors (Lam and Lam 1995) and this change in spacing between two copies of a given motif can alter the signal pathway to which a promoter responds. Furthermore, single GT elements enhanced expression by nearly twofold when placed 50 nucleotides or 100 nucleotides upstream to promoter, but when two GT elements separated by 5, 10 or 25 nucleotides were placed, their expression plummeted significantly. Hence, it was proved that second copy of GT negatively regulated the expression of promoter when placed in close proximity to minimal promoter (Mehrotra and Panwar 2009).
-
(3)
Database Assisted Promoter Analysis: Merely the identification and conjugation of cis-elements to one type of promoter is not sufficient to predict transcriptional product of a transgene. That is because a cis-element active under the control of one promoter may not transcribe desired protein with another promoter (Tiwari et al. 2003). Hence, it is of utmost importance to annotate all the sequences in a genome and represent introns, exons, trans-acting factors, cis-regulatory elements in promoters with the help of regulatory networks and logic functions. Ettwiller et al. (2003) had predicted cis-elements in Sacchromyces cerevisiae genome using functional networks. They discovered cis-elements using information from protein–protein interactions and metabolic networks. They presented them as patterns using pattern discovery tool such as Teiresias and represented them as motifs. Furthermore, the motifs from metabolic networks were compared to motifs from protein–protein interaction information and gave an ‘overlap score’. Based upon this score and standard deviation of patterns from start codon, they were able to find 42 degenerate motifs from 647 original patterns in S. cerevisiae. They also verified that cis-regulatory sequences were composed of Adenine–Thymine rich patterns that offer transcription factor sites in the genome.
Certain regulatory sequence analysis tools are also freely available and allow retrieval of sequences in genomes, pattern discovery using oligo-analysis and dyad-analysis tools, pattern matching with putative transcription factor sites in sequenced set of genes and their upstream regions, Transcription Factor Binding Sites using Strings or Position-Specific Scoring Matches (PSSM) etc. (Helden 2003). Some of the scientists have linked transcription factor binding sites to defined set of basic functions. They have related the cis-regulatory module target sites individually or in combinations as logic operators like AND-OR-NOR. As per Istrail and Davidson (2004), each cis-regulatory sequence is occupied by a particular transcription factor and this site is given a particular ‘occupancy score’ that specifies equilibrium constants for the interaction of transcription factors and target sites. The scientist have represented interaction of transcription factors and target sites into four categories based upon the operation that they carry out, for example: (1) D, transcriptional activation operators, (2) F, basal transcription apparatus control operators, (3) G, combinatorial logic operators and (4) E, external control operators. For example, G operator can be represented as AND operator under the condition when a gene is expressed as a combined effect of two or more transcription factors. If any one of the transcription factor is not active, the gene would not be transcribed. All the operators for the cis-regulatory modules are further compiled and functionally interpreted.
Till date there are three main databases that identify transcription factor binding sites in upstream cis-regulatory elements in plant promoters. First is PLACE (Plant cis-Acting Regulatory DNA Elements) that compiles more than 469 cis-acting regulatory DNA motifs as reported on January 8, 2007. The database compiles all the motifs in plants that have been reported (Higo et al. 1999). Another database is PlantCARE (Plant cis-Acting Regulatory Elements) that compiles information about transcription factor sites, motif sequence, function, species, cell type, genes and represent cis-elements by positional matrices, consensus sequences and individual sites on particular sequences. The database also allows identification of new cis-elements from in silico data from transcriptome (Lescot et al. 2002). Third database TRANSFAC (Transcriptional Regulation from Patterns to Profiles) compiles eukaryotic transcription factors, their target genes and regulatory binding sites. It is the only database that provides structural and functional information of transcription factors (Hehl and Wingender 2001; Matys et al. 2003).
Fauteux and Strömvik (2009) generated a seeding DNA motif discovery algorithm and analyzed 54 seed storage protein (SSP) gene promoters from three plant families, namely Brassicaceae (mustards), Fabaceae (legumes) and Poaceae (grasses) with respect to certain representative species in Arabidopsis thaliana, soybean and rice, respectively. They could identify three conserved motifs, two RY-like and one ACGT-like. In comparison to other approaches that use a position weight matrix motif model, the seeder DNA motif effectively utilizes a string-based approach that identifies motifs that are statistically significant compared to a background set of sequences.
Studies characterizing different natural promoters (Kim et al. 2010; Saha et al. 2010; Xu et al. 2010b) are continuously increasing and provide the researchers with more and more cis-motifs which control gene expression. Saha et al. (2010) lately identified and characterized the cis-regulatory motifs responsible for tissue-specific expression in the −673 and +90 bases upstream of the LOJ (Lateral organ Junction) gene recognized as LOJ promoter. They found enhancer-like element in the distal region which stimulates a minimal promoter. Such examples where novel promoter elements are identified promises potential applications in genetic engineering.
Promoter studies to cis engineering…a designer shift
Synthetic promoters were initially created by three strategies: (1) By combining defined cis-regulatory element with strong constitutive promoter (Ito et al. 1998; Rushton et al. 2002; Gurr and Rushton 2005) or by duplicating the upstream enhancer domains in conjunction with strong promoter (Maiti et al. 1997); (2) By combining cis-regulatory elements from different promoters (Sawant et al. 2001); (3) By fusing two strong constitutive characterised promoters to develop hybrids that allow both the promoters to be active in either direction or by developing bidirectional promoter (Comai et al. 1990; Chaturvedi et al. 2006).
Sawant et al. (1999) carried out a computational analysis of conserved nucleotide sequences observed in promoter of highly expressing genes in plants and designed a ‘minimal expression casette’, Pmec and ‘Transcription activation module’, TAM (Sawant et al. 2001). TAM contains many cis-acting motifs responsive to salicylic acid, auxins, salt, abscisic acid, ethylene, gibberellic acid, jasmonate, a variety of abiotic as well as biotic stresses, development and nutrition related factors. Various cis-acting DNA motif could function as an activator by itself as well as a synergizing activator in the presence of other neighbouring homologous as well as heterologous motifs. This synergestic effect is well illustrated in the work of Sawant et al. (2005) where the effect of eight cis-acting motifs on transcription from the basal promoter (Pmec) was studied by placing these upstream of the TATA-box at the −38 position as in plant genes. Multimers of the eight different sequence elements were inserted, taking one at a time such that each of these caused 2–8-fold activation of the basal transcription. The complete module brought enhancement of 110-fold in transcription levels. This showed that all such factors have to be arranged in an orderly manner to give a fully functional enhanceosome-like complex. Chaturvedi et al. (2006) constructed a synthetic bidirectional expression module by placing Pmec on 5′ and 3′ side of TAM. They analyzed the expression in both directions by transient and stable transformation in tobacco. As a result, it was found that TAM worked as an activating enhancer functionally compatible to minimal promoter. Schlabach et al. (2010) used synthetic design to develop strong promoter where 10 mer DNA sequences in tandem were placed on microarray as 100 mers which yieled them an oligo nucleotide library of 52,429 unique sequence and were screened for the ability to activate GFP transcription from a CMV promoter.
An important approach to tune gene expression was developed by Jensen and Hammer (1998), Mijakovic et al. (2005) and Hammer et al. (2006). They attempted to control gene expression through construction of synthetic promoter libraries by introducing changes in the sequences flanking the −35 and −10 consensus sequences of bacterial promoters. Alper et al. (2005) devised a methodology for quantitative evaluation of gene expression by constructing library of synthetic promoters of varying strength. These promoters were constructed via directed evolution through mutagenesis of a constitutive promoter. The further analysis was made with various metrics and a study through integration of these constructs into genome to find out the dependency of phenotype on gene expression, was conducted. In this case, a derivative of the constitutive bacteriophage PL-lambda promoter was mutated through error-prone PCR, cloned into a reporter plasmid upstream of a low-stability GFP gene, and screened in E. coli. Based on GFP fluorescence a functional library of 22 mutants was obtained. The instabilities and inherent mutation rates associated with the over expression of endogenous genes by earlier used plasmid based systems were avoided with the use of this system. This and other promoter libraries have a broad host range, perhaps due to involvement of general polymerase machinery in the cell and a heterologous constitutive promoter.
Designer promoter…the result of cis engineering
Recent advances in synthetic promoter technology are enabling the production of novel promoters optimised to suit the requirements of a particular transgene and exercise tighter control. Inducible promoters are activated by one or more stimuli such as hormones (for example gibberellin, abscisic acid, jasmonic acid, salicylic acid, ethylene, auxin), environmental conditions (light, temperature), abiotic stress (water stress, salt stress, wounding) and biotic stress (microbes, insects, nematodes). Although inducible, these may sometimes direct expression in the absence of the stimulus.
Pathogen inducible promoters
Rushton et al. (2002) found that defence signaling could be well conserved across species boundaries at the promoter element level. An array of cis-acting elements (boxes W1, W2, GCC, JERE, S, Gst1, and D) recognised by specific transcription factors (WRKYs, ERFs, bZIPs, Mybs, Dofs and bHLHs) can mediate local gene expression in plants after pathogen attack. Hence, defined synthetic promoters were constructed. These contain tetramers of only a single type of element and monitored expression during interactions with a number of pathogens, including compatible, incompatible, and non host interactions. As it is known that spacing between individual cis-acting elements and/or between these elements is difficult to predict (Wray 1998), and needs to be determined experimentally, there were major differences in the inducibility of the various promoters for pathogens tested, the speed of induction and the basal expression levels. Although all of the promoter elements originated from other plants, they worked in Arabidopsis thaliana retaining their function in this heterologous background. Thereby, these elements can be used to make promoters for use in different plants and against different pathogens.
Heise et al. (2002) studied pathogen inducible Arabidopsis CMPG1 gene promoter and found that it’s also responsive to wounding. They found another pathogen-responsive element, F involved in the process. Hence; they constructed a synthetic promoter containing only F and were able to separate pathogen inducibility from wound inducibility. Rushton et al. (2002) also developed improved second-generation promoters by varying several parameters like the number of copies of an individual element in a promoter and hence, varied the strength and inducibility of a promoter. Importantly, this also can have the effect of reducing/eliminating some background expression because pathogen inducibility appears stronger than basal or wound-induced expression. What remains ahead is development of ‘lifestyle-specific’ promoters that are selectively inducible by biotrophic, hemibiotrophic or necrotrophic pathogens.
Römer et al. (2009) studied transcription- activator like (TAL) effector proteins that manipulate the hosts’ transcriptome to promote disease. Certain resistance (R) genes may be specifically activated by the respective TAL effectors by interaction with the corresponding UPT (UPregulated by TAL effectors). They showed that UPT boxes from different plant promoter can be assembled in vitro into one complex promoter in which each UPT box retains its TAL effector specificity and results in a single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens.
Recently, Kovalchuk et al. (2010) cloned the genomic sequences of seven γ-thionin (defensin) genes that are mainly expressed in developing grain from wheat and rice. While they studied the spatial and temporal activities of four defensin promoters in wheat and rice stable transformants, a strong basal activity was detected in the tissues which are most vulnerable to fungal and bacterial infections. All wheat and rice promoters were strongly induced by wounding in transgenic rice plants. Their work suggest that these PRPI (Pathogen Responsive and Pathogen Inducible) promoters will be useful for specific targeting and accumulation of pathogen resistance proteins in young, vulnerable tissues of developing and germinating grain.
‘Chemical switches’ that regulate transcription
A flexible system has evolved in the form of chemical-inducible systems for regulating gene expression. These chemical switches remains quiescent in the presence or absence of inducers, and therefore, does not inhibit physiological activities. Chemicals that can regulate transgene expression include the antibiotic tetracycline (tc), the steroids dexamethasone (dex), estradiol, copper, ethanol, the inducer of pathogen-related proteins benzothiadiazol, herbicide safeners, and the insecticide methoxyfenozide. When used in combination with tissue-specific promoters, the gene expression can be restricted to a given tissue at a specific time. To constitutively or conditionally express an inactive chimeric transcription activator, promoter activity system based on transcriptional activation, containing a heterologous DNA-binding domain (DBD), an activation domain (AD), a nuclear localization signal (NLS) or a regulatory domain of an animal steroid nuclear receptor, can be used. Steroids such as the glucocorticoid dexamethasone exhibit high specificity for the transcriptional activator, the glucocorticoid receptor (GR). Using domains from different proteins, Unger et al. (2002) established a steroid-inducible promoter for transgenic maize. In this system, the DNA-binding domain and the ligand-binding domain were taken from the estrogen receptor, whereas the activation domain was taken from the maize transcription factor C1. Male sterility could thus, be chemically controlled by conditionally expressing the male fertility gene MS45 in a male-sterile background.
Another strategy to regulate gene expression through inducers includes promoter inactivation system that utilizes the repression principle based on sterical interference of a repressor protein with proteins important for transcription. For example, in tetracycline inactivation system, the construction of fusion proteins between transcriptional transactivation domains and bacterial repressor proteins such as the Lac repressor or TetR (Tetracycline repressor) are undertaken. TetR binds to the tetracycline operator, Tet, in the absence of tetracycline. But when associated with tetracycline, dimeric TetR is converted to the monomeric form and released from its operator.
In contrast to above strategy, Gatz et al. (1992) developed a de-repression system in plants where a mutant TetR showing a “reverse phenotype” was employed. This reverse repressor binds DNA only in the presence of tetracycline. The target promoter, a modified 35S promoter, consisted of one or two copies of the tet operator placed upstream and downstream from the TATA-box, respectively. In the absence of tetracycline, over-expressed TetR binds to the Tet operator, and thereby prevents target gene expression but upon tetracycline binding; TetR is released from the operator, relieving the repression.
Ordiz et al. (2002) designed an ethanol inducible system based on the regulatory elements of the Aspergillus nidulans alcA promoter and adapted for use in plants. The target promoter contains the TATA box as well as upstream sequences of the alcA promoter fused to position 223 of the CaMV 35S promoter. A DNA binding protein of C6 zinc binuclear cluster family, AlcR binds to its target sequences within this promoter in the presence of ethanol or other inducers such as ethyl methyl ketone. When stably transformed into tobacco, these constructs mediate ethanol-dependent expression of transgenes. While using such systems, a risk of false expression exists because oxygen limitation in case of submerged environment of suspension culture may induce alcA in the absence of exogenously applied inducer. This has been overcome by development of an alc-GR system. Here, the rat glucocorticoid receptor (GR) domain has been fused to the AlcR transcription factor; conferring steroid-inducible control over alc-mediated gene expression (Roberts et al. 2005). Moore et al. (2006) discussed about various technologies for chemically inducible gene expression in plants.
Synthetic light switches
Gilmartin et al. (1990) reported about the molecular light switches for plant genes. However, Puente et al. (1996) studied the minimal promoter elements sufficient to mediate responses to light and developmental signals, and systematically analyzed the ability of four well characterized Light responsive Elements (LREs), individually or in selected combinations, to confer light responsiveness to non-light-regulated basal promoters. Their study revealed that individual motifs cannot confer any light responsiveness. However, if three distinct pair wise combinations of these motifs were made considering appropriate cell type, and developmental context, light responsiveness was observed. This was independent of the basal promoters used. Although the role of individual LREs can be quite different depending on the promoter context in which they are located but when in pairs these LREs tend to behave as units and signal integration points mediating both light and developmental control of gene expression.
Logemann and Hahlbrock (2002) while studying the crosstalk among stress responses in plants, found that ACE/ACE (ACGTcontaining element) functions as a Light-Regulatory as well as Elicitor-Responsive Unit. Their studies on Petroselinum crispum, have shown that one of the involved genes, encoding acyl-CoA oxidase (AOC), responds positively to UV light and negatively to a pathogen-derived elicitor through a promoter unit consisting of two almost identical ACEs. When introduced into an unrelated promoter or subjected to mutation, have shown the same type of response pattern. Such studies have generated considerable interest in elucidating the hierarchy of networks of transcription factors, and in identifying the key regulatory elements in different light-responsive developmental processes. A range of LREs have been studied in different promoters, many of which positively or negatively mediate gene expression in response to light. No single element is found in all light-regulated promoters, suggesting that a complex light-regulation network exists (Jiao et al. 2007).
Similarly, Evrard et al. (2009) proposed that FORCA, the regulatory cis- element found in a large number of Arabidopsis promoters, can serve as integration point of light and defense-related signals and thereby bring about changes at the transcriptome level in response to surrounding environmental conditions. FORCA is a frequently occuring hexameric promoter motif in case of many Arabidopsis genes that occur in clusters and are coexpressed in response to fungal or oomycete pathogens as well as defined light treatments. This element interacts with nuclear Arabidopsis proteins. It has been reported that such interactions are suppressed by defense-related stimuli and enhanced by prolonged exposure to constant light. Hence, FORCA like light-responsive promoter provides the building blocks of the strategy that can be used to develop synthetic promoters capable of coping up with two or more environmental conditions simultaneously.
Other types
Tissue-specific promoters confine transgene expression to a single plant part, tissue or cell-type. Ni et al. (1995) studied the strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase (mas) genes. It was found that a chimeric promoter, Mac, incorporating the mas region from +65 to −301 and 35S enhancer region from −90 to −941 expressed GUS activity at a level that was several times of double 35S promoter in transgenic tobacco plants. The changes in strength of expression were investigated for different combinations. For example if ocs activator was added to the mas promoter and activator or mas activator in combination with ocs promoter and activator. These combinations resulted in observance of GUS activity in a larger number of cell types, including xylem vessels and leaf non-specialized epidermal cells. The strongest promoter that they constructed consisted of a trimer of the ocs activator added to the mas activator and promoter ((ocs) 3mas).
An improved promoter for gene expression in cereal cells was developed by Last et al. (1991). It is based on truncated maize Adh1 promoter, with multiple copies of the Anaerobic Responsive Element from the maize Adh1 gene and ocs-elements from the octopine synthase gene of Agrobacterium tumefaciens. Promoter activity when measured in five different monocot species [wheat, maize, rice, einkorn (Triticum monococcum), and Lolium multiflorum] and one dicot (Nicotiana plumbaginifolia), the most highly expressing construct (pEmuGN) gave 10- to 50-folds higher expression than the CaMV 35S promoter in all the monocot species. The pEmu promoter should be valuable where a high level of gene expression is required in monocots.
With the work done by Pietrzak et al. (1989), it was found that transcriptional activity of a strong constitutive plant promoter, CaMV 35S promoter, could be modulated by addition of multiple synthetic oligonucleotides, carrying the heat shock promoter consensus sequence (CTXGAAXXTTCXAG, Pelham 1982). Under heat shock conditions such a promoter shows a threefold increase in activity. Similarly when an enhancerless 35S promoter was engineered, it gave increased activity under both normal and heat conditions, without a significant induction by the heat shock.
Coutu et al. (2007) developed 14 binary vectors pORE, suitable for Agrobacterium-mediated transformation of dicotyledonous plants and adaptable for biolistic transformation of monocotyledonous plants by using a combination of promoters (PHPL Arabidopsis thaliana hydroperoxide lyase promoter, PENTCUP2 Triticum aestivum lipid transfer protein promoter and PTAPADH Triticum aestivum lipid transfer protein promoter fused to an alcohol dehydrogenase intron). In 2008 Chung and Parish reported combinatorial interactions of multiple cis elements regulates the induction of the Arabidopsis XERO2 dehydrin gene by ABA and cold, Liu et al. (1994) constructed tuber-specific and cold-inducible chimeric promoters (CIPP) in potato. Deletion analysis of the CIPP and A. thaliana, cold inducible (cor15a) promoter showed the presence of a cis-element for tuber-specific and sucrose-responsive activity named TSSR and LTRE (Low Temperature Responsive element), respectively. So, they constructed two chimeric promoters using TSSR containing sequence and LTRE and measured the activity of a GUS reporter gene with and without cold-inducible conditions in transgenic potatoes. Results indicated that this chimeric promoter possessed a substantial cold-inducibility. So far, there are few promoters that can be used to control the expression of transgenes in potato in a tuber-specific manner combined with cold inducibility and this chimeric promoter may provide valuable tool for minimizing the accumulation of reducing sugars in cold-stored tubers. Recently, Tittarelli et al. (2009) have digitally identified various cold regulated promoters from peach using EST dataset.
Xu et al. (2010a) isolated and studied sequence for upstream region of endosperm-specific LPAAT (Lysophosphatidyl acytransferase) gene from coconut (Cocos nucifera L.) and searched for the promoter related elements. These elements were functionally analyzed in transgenic rice plants and were found to respond to fatty acid metabolism in endosperm cells. The special function and subcellular location of this LPAAT promoter make it a unique promoter candidate in crop endosperm modification in future. Sun et al. (2010) identified a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila. The promoter activity in case of leaves, roots, stems and flowers was as strong as CaMV 35S promoter but no promoter activity was detected in the seeds. This is beneficial for its application in crop engineering. Novel motifs responding to salt stress may exist in this 130 bp region.
Discussion
Critical aspects in cis engineering…how long can the magic of synthetic promoters last?
The synthetic promoter approach might be easy to implement because a collection of building block elements could be built up from a variety of plant species but it increases the possibility of Homology Based Gene Silencing (HBGS) due to sequence homology in transgene or promoter or cis-elements. HBGS is the inactivation of transgene either at transcription level or at post-transcription level caused due to presence of duplicated sequence in transgene and promoter, repeated sequence in transgene and native gene or presence of multiple copies of transgene in genome. It can lead to RNA-directed DNA methylation and inactivation of promoters (Wassenegger et al. 1994; Vaucheret et al. 2001) as well as co-suppression of another closely linked gene (Brusslan et al. 1993). So the challenge exists to develop synthetic promoters having minimum sequence homology or in other words to develop minimal core promoter with specific cis-regulatory element for specific expression. Bhullar et al. (2003) developed novel synthetic 35S promoter by two approaches: firstly, by swapping of domain A of CaMV 35S promoter in three ways: one by introduction of direct repeats of as-1 element (TGACG) in the synthetic DNA context with different flanking and intervening nucleotides, second by keeping the flanking and spacer nucleotides conserved as 7- and 5-bp, respectively, and third by replacement of subdomain A1 with octopine synthase element (ocs) from ocs promoter of Arabidopsis sp.; secondly, by replacing native cis-elements with cis-elements having same organization but no intervening homologous region to existing promoters. Results proved that novel promoters comprising new cis-elements expressed GUS gene levels comparable to wild-type 35S promoter but synthetic promoter with domain swapping revealed lower GUS gene activity.
More recently Bhullar et al. (2010) designed an approach to circumvent HBGS. They conjugated the silencing loci like 271 loci from tobacco to CaMV 35S promoter (carrying GUS reporter gene) that have reduced similarity to target promoter sequence. As a result, nearly 67% of transgenic lines escaped trans-inactivation of CaMV 35S promoter and there was delay in induction of silencing in synthetic promoter construct. Another approach to reduce HBGS is to develop gene cassettes having very less sequence homology to the endogenous gene or to one another if two or more transgenes are introduced. However, it is believed that artificial gene cassettes in the genome deplete basal transcription factors (TF) levels in genome. But in contrast, Sawant et al. (2005) revealed that many cis-elements together may provide additional TF-binding sites and contribute to the stability of Pre Initiation Complex (PIC) at TATA-box. Thus all the interactions mediated by upstream cis-regulatory modules allow higher expression of gene in plants. However, some other driving forces that can cause inactivation of transgene in plants, such as paramutation, position-effect variegation and chromatin mediated inactivation (Leeuwen et al. 2001; Meyer and Saedler 1996) may still change the picture.
Synthesizing bidirectional promoter: as new toolkits to manipulate plant genomes
Expression of more than one transgene in plants covers an important aspect of plant genetic engineering and genetic improvement of crop plants. However, regulation of multiple genes by single promoter can induce homology-based gene silencing of the transgene or native gene. Hence, gene stacking in plants can be achieved by the development of bidirectional promoter that allows simultaneous expression of two transgene. A bidirectional promoter can be constructed by the fusion of two unidirectional minimal promoters in opposite direction while they are regulated by common set of cis-motifs. Several scientists have developed bidirectional promoter using this strategy facilitating the expression of multiple transgene (Chaturvedi et al. 2006; Frey et al. 2001; Li et al. 2004). Bidirectional promoters not only solve the problem of gene silencing in plants but also minimize the amount of foreign DNA in transgenic plants.
Previously, artificial construction of bidirectional promoter using constitutive CaMV 35S promoter had lead to gene silencing of transgene in few cases (Xie et al. 2001). Now many reports have cited that bidirectional promoters exist naturally in plants. Hence, the naturally occurring bidirectional promoters in plants are now used to prevent homology based gene silencing and allow stacking of various genes. Most recently, Mitra et al. (2009) have proved that naturally occurring bidirectional promoter in Arabidopsis could be used to express two genes simultaneously. 2 Kb intergenic region of Arabidopsis chlorophyll a/b-binding protein cab1, cab2 genes was reported to be regulated by bidirectional promoter. This group checked the expression of GUS and GFP proteins fused to Arabidopsis cab1 and cab2 genes and found that both the genes were expressed in tissue specific and light induced manner, respectively.
Now that whole genome sequence of many plants have been deciphered, it is possible to use bioinformatic tools to predict putative bidirectional promoters in plants. Dhadi et al. (2009) have used PLACE and PlantCARE databases to identify cis-regulatory elements from three different sequenced plant genomes i.e. rice (Oryza sativa), Arabidopsis thaliana, and Populus trichocarpa. Based upon their knowledge on over-represented cis-elements in bidirectional promoters in respective genomes, they defined the functions of various cis-elements. Furthermore, based upon intergenic distances between promoters in bidirectional promoter and their co-expression patterns they predicted bidirectional promoters and also compared their expression with bidirectional promoters in humans.
References
Alper H, Fischer C, Nevoigt E, Stephanopoulos G (2005) Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA 102:12678–12683
An G, Costa MA, Ha S-B (1990) Nopaline synthase promoter 1S wound inducible and auxin inducible. Plant Cell 2:25–33
Benfey PN, Chua N-H (1990) The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250:959–966
Bhullar S, Chakravarthy S, Advani S, Datta S, Pental D, Burma PK (2003) Strategies for development of functionally equivalent promoters with minimum sequence homology for transgene expression in plants: cis-elements in a novel DNA context versus domain swapping. Plant Physiol 132:988–998
Bhullar S, Datta S, Advani S, Chakravarthy S, Gautam T, Pental D, Burma PK (2007) Functional analysis of the cauliflower mosaic virus 35S promoter: re-evaluation of the role of subdomains B5, B4, and B2 in promoter activity. Plant Biotechnol J 5:696–708
Bhullar S, Datta S, Burma PK (2010) Delayed trans-inactivation of synthetic domain A 35S promoters by “Tobacco 271 Locus” due to reduced sequence homology. Plant Mol Biol Rep 29:1–11
Brusslan JA, Karlin-Neumann GA, Huang L, Tobin EM (1993) An Arabidopsis mutant with a reduced level of cab140 RNA is a result of cosuppression. Plant Cell 5:667–677
Cazzonelli CI, Velten EJ (2008) In vivo characterization of plant promoter element interaction using synthetic promoters. Trans Res 17:437–457
Chaturvedi CP, Sawant SV, Kiran K, Mehrotra R, Lodhi N, Ansari SA, Tuli R (2006) Analysis of polarity in the expression from a multifactorial bidirectional promoter designed for high-level expression of transgenes in plants. J Biotechnol 123:1–12
Chung S, Parish RW (2008) Combinatorial interactions of multiple cis-elements regulating the induction of the Arabidopsis XERO2 dehydrin gene by abscisic acid and cold. Plant J 54:15–29
Comai L, Moran P, Maslyar D (1990) Novel and useful properties of a chimeric plant promoter combining CaMV 35S and MAS elements. Plant Mol Biol 15:373–381
Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Trans Res 16:771–781
Dhadi SR, Krom N, Ramakrishna W (2009) Genome-wide comparative analysis of putative bidirectional promoters from rice, Arabidopsis and Populus. Gene 429:65–73
Ellis JG, Llewellyn DJ, Walker JC, Dennis ES, Peacock WJ (1987) The ocs element: a 16 base pair palindrome essential for activity of the octopine synthase enhancer. EMBO J 6:3203–3208
Ettwiller LM, Rung J, Birney E (2003) Discovering novel cis-regulatory motifs using functional networks. Genome Res 13:883–895
Evrard A, Ndatimana T, Eulgem T (2009) FORCA, a promoter element that responds to crosstalk between defense and light signaling. BMC Plant Biol 9:1–13
Fang RX, Nagy F, Subramaniam S, Chua NH (1989) Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell 1:141–150
Fauteux F, Strömvik MV (2009) Seed storage protein gene promoters contain conserved DNA motifs in Brassicaceae, Fabaceae and Poaceae. BMC Plant Biol 9:126–136
Frey PM, Scharer-Hernandez NG, Futterer J, Potrykus I, Puonti-Kaerlas J (2001) Simultaneous analysis of the bidirectional African cassava mosaic virus promoter activity using two different luciferase genes. Virus Genes 22:231–242
Gatz C, Frohberg C, Wendenberg R (1992) Stringent repression and homogeneous depression by tetracyclin of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J 2:397–404
Gilmartin PM, Sarokin L, Memelink J, Chua NH (1990) Molecular light switches for plant genes. Plant Cell 2:369–378
Gowda S, Wu FC, Herman HB, Shepherd RJ (1989) Gene VI of figwort mosaic virus (caulimovirus group) functions in posttranscriptional expression of genes on the full-length RNA transcript. Proc Natl Acad Sci USA 86:9203–9207
Gurr SJ, Rushton PJ (2005) Engineering plants with increased disease resistance: how are we going to express it? Trends Biotechnol 23:283–290
Hammer K, Mijakovic I, Jensen PR (2006) Synthetic promoter libraries—tuning of gene expression. Trends Biotechnol 24:53–55
Hehl R, Wingender E (2001) Database-assisted promoter analysis. Trends Plant Sci 6:251–255
Heise A, Lippok B, Kirsch C, Hahlbrock K (2002) Two immediate-early pathogen-responsive members of the AtCMPG gene family in Arabidopsis thaliana and the W-box-containing elicitor-response element of AtCMPG1. Proc Natl Acad Sci USA 99:9049–9054
Helden JA (2003) Regulatory sequence analysis tools. Nucl Aci Res 31:3593–3596
Higo K, Ugawa W, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucl Aci Res 27:297–300
Istrail S, Davidson EH (2004) Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci USA 102:4954–4959
Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabea A (1998) A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase–specific transcription. Plant Cell 10:331–341
Jensen PR, Hammer K (1998) The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol 64:82–87
Jiao Y, Lau ON, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8:217–230
Kamisugi Y, Cuming AC (2005) The evolution of the abscisic acid-response in land plants: comparative analysis of group 1 LEA gene expression in moss and cereals. Plant Mol Biol 59:723–737
Kim Y-H, Bae JM, Huh G-H (2010) Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweet potato in response to plant developmental stage and environmental stress. Plant Cell Rep 29:779–791
Kovalchuk N, Li M, Wittek F, Reid N, Singh R, Shirley N, Ismagul A, Eliby S, Johnson A, Milligan AS, Hrmova M, Langridge P, Lopato S (2010) Defensin promoters as potential tools for engineering disease resistance in cereal grains. Plant Biotechnol J 8:47–64
Lam E, Lam YKP (1995) Binding site requirements and differential representation of TGA factors in nuclear ASF-1 activity. Nucl Acids Res 23:3778–3785
Last DI, Bretteli RIS, Chamberlain DA, Chaudhary AM, Larkin PJ, Marsh EL, Peacock WJ, Dennis ES (1991) pEmu: an improved promoter for gene expression in cereal cells. Theo App Genet 81:581–588
Leeuwen WV, RuttInk T, Borst-Vrenssen AWM, Plas LHWVD, Krol ARVD (2001) Characterization of position-induced spatial and temporal regulation of transgene promoter activity in plants. J Exp Bot 52:949–959
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Peer YVD, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl Acids Res 30:325–327
Li ZT, Jayasankar S, Gray DJ (2004) Bi-directional duplex promoters with duplicated enhancers significantly increase transgene expression in grape and tobacco. Transgenic Res 13:143–154
Liu Z-B, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6:645–657
Logemann E, Hahlbrock K (2002) Crosstalk among stress responses in plants: pathogen defense overrides UV protection through an inversely regulated ACE_ACE type of light-responsive gene promoter unit. Proc Natl Acad Sci USA 99:2428–2432
Maiti IB, Gowda S, Kiernan J, Ghosh SK (1997) Promoter/leader deletion analysis and plant expression vectors with the figwort mosaic virus (FMV) full length transcript (FLt) promoter containing single or double enhancer domains. Transgenic Res 6:143–156
Matys V, Fricke E, Geffers R, ßling EG, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos D-U, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucl Acids Res 31:374–378
Mehrotra R, Mehrotra S (2010) Promoter activation by ACGT in response to salicylic and abscisic acids is differentially regulated by the spacing between two copies of the motif. J Plant Physiol 167:1214–1218
Mehrotra R, Panwar J (2009) Dimerization of GT element interferes negatively with gene activation. J Genet 88:257–260
Mehrotra R, Kiran K, Chaturvedi CP, Anjuman S, Lodhi N, Sawant S, Tuli R (2005) Effect of copy number and spacing of the ACGT and GT cis elements on transient expression of minimal promoter in plants. J Genet 84:183–187
Meyer P, Saedler H (1996) Homology-dependent gene silencing in plants. Ann Rev Plant Physiol Plant Mol Biol 47:23–48
Mijakovic I, Petranovic D, Jensen PR (2005) Tunable promoters in systems biology. Curr Opin Biotechnol 16:329–335
Mitra A, Han J, Zhang ZJ, Mitra A (2009) The intergenic region of Arabidopsis thaliana cab1 and cab2 divergent genes functions as a bidirectional promoter. Planta 229:1015–1022
Moore I, Samalova M, Kurup S (2006) Transactivated and chemically inducible gene expression in plants. The Plant J 45:651–683
Ni M, Cui D, Einstein J, Narasimhulu S, Vergara CE, Gelvin SB (1995) Strength and tissue specificity of chimeric promoter derived from octopine and manopine synthase gene. Plant J 7:661–676
Novina CD, Roy AL (1996) Core promoters and transcriptional control. Trends Genet 12:351–355
Odell JT, Nagy F, Chua N-H (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313:810–812
Ordiz MI, Barbas CF-III, Beachy RN (2002) Regulation of transgene expression in plants with polydactyl zinc finger transcription factors. Proc Natl Acad Sci USA 99:13290–13295
Pelham HRB (1982) A regulatory upstream promoter element in the Drosophila Hsp 70 heat-shock gene. Cell 30:517–528
Pietrzak M, Burri M, Herrero J-J, Mosbach K (1989) Transcriptional activity is inducible in the cauliflower mosaic virus 35S promoter engineered with the heat shock consensus sequence. FEBS Lett 249:311–315
Puente P, Wei N, Deng XW (1996) Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J 15:3732–3743
Roberts GR, Garoosi GA, Koroleva O, Ito M, Laufs P, Leader DJ, Caddick MX, Doonan JH, Tomsett AB (2005) The alc-GR system. A modified alc gene switch designed for use in plant tissue culture. Plant Physiol 138:1259–1267
Römer P, Recht S, Lahaye T (2009) A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci USA 106:20526–20531
Rushton PJ, Reinstädler A, Lipka V, Lippok B, Somssich IE (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14:749–762
Saha D, Kumar V, Bhat SR, Srinivasan R (2010) Characterization of upstream sequences of the LOJ gene leads to identification of a novel enhancer element conferring lateral organ junction-specific expression in Arabidopsis thaliana. Plant Mol Biol Rep 28. doi:10.1007/s11105-010-0229-6
Sawant SV, Singh PK, Gupta SK, Madanala R, Tuli R (1999) Conserved nucleotide sequence in highly expressed genes in plants. J Genet 78:1–8
Sawant SV, Singh PK, Madanala R, Tuli R (2001) Designing of an artificial expression cassette for the high-level expression of transgenes in plants. Theor Appl Genet 102:635–644
Sawant S, Kiran K, Mehrotra R, Chaturvedi CP, Ansari SA, Singh P, Lodhi N, Tuli R (2005) A variety of synergistic and antagonistic interactions mediated by cis-acting DNA motifs regulate gene expression in plant cells and modulate stability of the transcription complex formed on a basal promoter. J Exp Bot 56:2345–2353
Schlabach MR, Hu JK, Li M, Elledge SJ (2010) Synthetic design of strong promoters. Proc Natl Acad Sci USA 107:2538–2543
Sun Q, Gao F, Zhao F, Li K, Zhang J (2010) Identification of a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila. BMC Plant Biol 10:90–102
Tittarelli A, Santiago M, Morales A, Meisel LA, Silva H (2009) Isolation and functional characterization of cold-regulated promoters, by digitally identifying peach fruit cold-induced genes from a large EST dataset. BMC Plant Biol 9:121–136
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543
Unger E, Cigan AM, Trimnell M, Xu RJ, Kendall T, Roth B, Albertsen M (2002) A chimeric ecdysone receptor facilitates methoxyfenozide-dependent restoration of male fertility in ms45 maize. Transgenic Res 11:455–465
Vaucheret H, Béclin C, Fagard M (2001) Post-transcriptional gene silencing in plants. J Cell Sci 114:3083–3091
Venter M (2007) Synthetic promoters: genetic control through cis engineering. Trends Plant Sci 12:118–124
Venter M, Botha FC (2010) Synthetic promoter engineering. Plant Develop Biol Biotechnol Persp 2(4):393–414
Wassenegger M, Heimes S, Riedel L, Sänger HL (1994) RNA-directed de novo methylation of genomic sequences in plants. Cell 76:567–576
Wray GA (1998) Promoter logic. Science 279:1871–1872
Xie M, He Y, Gan S (2001) Bidirectionalization of polar promoters in plants. Nat Biotechnol 19:677–679
Xu L, Ye R, Zheng Y, Wang Z, Zhou P, Lin Y, Li D (2010a) Isolation of the endosperm-specific LPAAT gene promoter from coconut (Cocos nucifera L.) and its functional analysis in transgenic rice plants. Plant Cell Rep 29:1061–1068
Xu W, Yu Y, Ding J, Hua Z, Wang Y (2010b) Characterization of a novel stilbene synthase promoter involved in pathogen- and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta 231:475–487
Zavallo D, Bilbao ML, Hopp HE, Heinz R (2010) Isolation and functional characterization of two novel seed-specific promoters from sunflower (Helianthus annuus L.). Plant Cell Rep 29:239–248
Acknowledgments
RM is thankful to Department of science and Technolgy,Government of India for financial support under fast track scheme for young scientists.We are also thankful to the facilities provided by BITS, Pilani.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mehrotra, R., Gupta, G., Sethi, R. et al. Designer promoter: an artwork of cis engineering. Plant Mol Biol 75, 527–536 (2011). https://doi.org/10.1007/s11103-011-9755-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9755-3