Abstract
A number of basic leucine zipper (bZIP) transcription factors are known to function in stress signaling in plants but few have thus far been functionally characterized in rice. In our current study in rice, we have newly isolated and characterized the OsABF1 (Oryza sativa ABA responsive element binding factor 1) gene that encodes a bZIP transcription factor. Its expression in seedling shoots and roots was found to be induced by various abiotic stress treatments such as anoxia, salinity, drought, oxidative stress, cold and abscisic acid (ABA). Subcellular localization analysis in maize protoplasts using GFP fusion vectors indicated that OsABF1 is a nuclear protein. In a yeast experiment, OsABF1 was shown to bind to ABA responsive elements (ABREs) and its N-terminal region was necessary to transactivate the downstream reporter gene. The homozygous T-DNA insertional mutants Osabf1-1 and Osabf1-2 were more sensitive in response to drought and salinity treatments than wild type plants. Furthermore, the upregulated expression of some ABA/stress-regulated genes in response to ABA treatment was suppressed in these Osabf1 mutants. Our current results thus suggest that OsABF1 is involved in abiotic stress responses and ABA signaling in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant growth and productivity are greatly affected by environmental stresses such as drought, high salinity, and low temperature. Upon exposure to abiotic stress conditions, plants undergo a variety of changes from physiological adaptation to gene expression. Among the many stress-inducible genes are those that directly protect against environmental stress such as osmoprotectants, chaperones, and detoxification enzymes. Others include transcription factors and protein kinases that regulate gene expression and signal transduction during the stress responses (Seki et al. 2003). Thus, the timely expression of stress-responsive genes is crucial to the plant’s ability to survive under different environmental stress conditions (Chen and Zhu 2004; Yamaguchi-Shinozaki and Shinozaki 2006; Chinnusamy et al. 2007; Shinozaki and Yamaguchi-Shinozaki 2007).
A number of experiments, including transcriptome analyses, have identified many transcription factors that respond to abiotic stresses including bZIPs, zinc finger proteins, AP2/EREBPs, bHLHs, and NACs (Chen et al. 2002; Seki et al. 2002; Rabbani et al. 2003; Chen and Zhu 2004; Oh et al. 2005, 2009; Nijhawan et al. 2008). Various transcription factors bind to specific cis-acting elements in the promoters of stress-responsive genes and thereby function as major mediators of plant tolerance to abiotic stress (Meshi and Iwabuchi 1995; Yanagisawa 1998; Liu et al. 1999; Riechmann and Ratcliffe 2000; Chen and Zhu 2004; Chinnusamy et al. 2007; Shinozaki and Yamaguchi-Shinozaki 2007; Fode et al. 2008). These stress-inducible transcription factors thus form a complex signaling network in response to abiotic stresses.
The bZIP proteins compose a large family of transcription factors that harbor a bZIP domain composed of a basic region and a leucine zipper (Hurst 1994; Jakoby et al. 2002). The conserved basic region is responsible for sequence-specific DNA binding, whereas the less conserved leucine zipper region contains amphipathic sequences in a coiled-coil form and confers dimerization specificity. The leucine zipper sequence thus mediates homo- and/or hetero-dimerization of the bZIP transcription factors. In plants, the bZIP transcription factor genes are expressed in a variety of ways such as organ-specific, stimulus-responsive, development-dependent, and cell cycle-specific (Schindler et al. 1992; Minami et al. 1993; de Vetten and Ferl 1995; Chern et al. 1996; Uno et al. 2000; Jakoby et al. 2002; Rodriguez-Uribe and O’Connell 2006; Nijhawan et al. 2008).
The phytohormone abscisic acid (ABA) plays an essential role in the adaptive response of plants to abiotic stresses including droughts, cold, and high salinity. ABA is also involved in various aspects of plant growth and development such as seed maturation, dormancy, inhibition of cell division, and germination (Leung and Giraudat 1998; Yamaguchi-Shinozaki and Shinozaki 2005; Christmann et al. 2006; Kim 2007). A number of bZIP transcription factors are known to participate in ABA and/or stress signaling (de Vetten and Ferl 1995; Jakoby et al. 2002; Lopez-Molina et al. 2002; Fujita et al. 2005). The bZIP transcription factors interact with specific ABA-responsive elements (ABRE), which harbor the conserved motif PyACGTGGC, in the promoter regions of many ABA-inducible genes and thereby transactivate downstream gene expression (Niu et al. 1999; Kim et al. 1997; Yamaguchi-Shinozaki and Shinozaki 2005). The bZIP transcription factors can thus be designated as ABA responsive element (ABRE)-binding factors (ABFs) or ABRE-binding proteins (AREBs). In Arabidopsis, 13 of 75 bZIP transcription factors belong to the A group that contains ABF genes among 10 subfamilies (Jakoby et al. 2002).
Expression of the Arabidopsis bZIP transcription factor family genes, ABF2/AREB1, ABF4/AREB2, and ABF3, is upregulated by ABA, dehydration, and salinity stress in vegetative tissues (Uno et al. 2000). In a transient experiment using protoplasts, the transcription of a reporter gene driven by ABRE is also activated by these bZIP transcription factors (Uno et al. 2000; Nakashima et al. 2006). The constitutive overexpression of ABF3 in Arabidopsis and rice exhibits enhanced drought tolerance (Kang et al. 2002; Oh et al. 2005). In addition, the overexpression of rice OsbZIP23 and OsbZIP72, which are positive regulators of ABA signaling, enhances abiotic stress tolerance (Lu et al. 2008; Xiang et al. 2008). ABF proteins also function in plant growth and development. For example, the Arabidopsis abi5 mutant shows decreased sensitivity to ABA by disrupting ABA signal transduction during seed germination, indicating that AtABI5 links ABA signal transduction with gene expressions in seeds (Finkelstein and Lynch 2000). In monocot plants, TRAB1 and HvABI5, rice and barley homologs of AtABI5, physically interact with their corresponding AtABI3 homologs, OsVP1 and HvVP1, and regulate ABA-inducible gene expression (Hobo et al. 1999; Nakamura et al. 2001; Casaretto and Ho 2003). Moreover, a bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance (Zou et al. 2008). These previous data suggest that ABF proteins function in a conserved ABA signal transduction pathway in both dicot and monocot plant species.
Rice is one of the most significant staple crops and is a model monocot species. However, whilst a number of bZIP transcription factors involved in stress signaling have now been identified in Arabidopsis, few of the 89 known bZIP transcription factors have been functionally characterized in rice (Nijhawan et al. 2008). In the present study, we have identified an abiotic stress-inducible bZIP transcriptional factor from rice, OsABF1. The expression pattern of OsABF1 was investigated in rice shoots and roots exposed to various stress conditions. The ABRE binding activity and transactivation ability of OsABF1 were evaluated using a yeast one-hybrid system. To investigate the in vivo functions of OsABF1, T-DNA insertional mutants of OsABF1 were analyzed under salinity and drought treatments. The expression of known ABA responsive genes was also examined in these mutants. The role of OsABF1 in ABA-dependent abiotic stress signaling in rice is discussed.
Materials and methods
Plant materials, growth conditions and stress treatments
Rice (Oryza sativa L. cultivars Dongjin and Hwayoung) seeds were sterilized with 70% ethanol, immersed in distilled water for 1 day and then grown for 14 days in a plant growth chamber (28 ± 1°C, 80% relative humidity and 14/10 h day/night photoperiod). Fourteen-day-old seedlings were placed under zero oxygen conditions for 0, 2, 4, 8, 12, and 24 h followed by 4 and 12 h recovery periods from this stress. Oxygen deprivation was carried out by using pure nitrogen gas in AtmosBag™-inflatable polyethylene isolation chambers (Sigma–Aldrich, Milwaukee, WI) (Kato-Noguchi and Morokuma 2007). For other stress treatments, seed surfaces were sterilized and immersed in distilled water, transferred to a growth chamber (28 ± 1°C, 80% relative humidity and 14/10 h day/night photoperiod) and grown in distilled water for 14 days. The seedlings then underwent different treatments including drought (10% polyethylene glycol), salinity (250 mM NaCl), oxidative stress (10 mM H2O2), cold (4°C), and ABA (100 μM) for 0, 2, 4, 8, 12, and 24 h.
The T-DNA insertional mutant lines of OsABF1, Osabf1-1 and Osabf1-2 were identified from the rice T-DNA Insertion Sequence Database (Jeong et al. 2006; http://www.postech.ac.kr/life/pfg/risd/index.html). Homozygous lines of Osabf1-1 and Osabf1-2 were isolated by PCR screening using OsABF1 gene-specific and T-DNA specific primers.
cDNA cloning
The OsABF1 full length cDNA clone was isolated by RT-PCR using total RNA extracts from drought-treated shoots of rice (O. sativa L. cv. Dongjin) seedlings with the primer pairs 5′-AAGCTTATGATGGCGTCGAGGGTG-3′ (F) and 5′-GGTACCCTACCACTCCATCGAGTT-3′ (R). The PCR products were inserted into pLUG-TA vector (iNTRON Biotechnology, Seoul, Korea) and the cDNA sequence was deposited into GenBank under the accession number GQ904238.
RT-PCR
Total RNA from stress-treated seedlings was extracted using Trizol reagent (Gibco-BRL, Grand island, NY). First strand cDNA was synthesized with 2 μg of purified total RNA using PrimeScritpt™ Reverse Transcriptase (Takara Bio Inc., Shiga, Japan). Oligo (dT) was used as a primer and the RT reaction was incubated at 42°C for 1 h in a total volume of 25 μl. Gene specific primers were used to examine the expression patterns of OsABF1 including other transcripts in rice seedlings under different stress treatments. OsDEG10 was used as a positive PCR marker to confirm the effects of the different abiotic stress treatments (Park et al. 2009).
Total RNA was also isolated from ABA-treated seedling shoots of the T-DNA mutant lines and their respective wild type plants for gene expression analysis. The rice actin gene was amplified as an internal control to quantify the relative amounts of cDNA (McElroy et al. 1990). Primer sequences used for the RT-PCR analysis of the OsABF1 gene, regulatory genes and rice actin are shown in Table 1.
Phylogenetic analysis
The Clustal W, Pole BioInformatique Lyonnasis (PBIL) program (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html) was used to perform multiple sequence alignments. A phylogenetic tree was constructed using MEGA software version 4.0 via the neighbor-joining method (Tamura et al. 2007). Bootstrap analysis was performed with 1,000 replicates and bootstrap values are shown as percentages.
Subcellular localization of GFP fusion proteins
The full-length cDNA of the OsABF1 gene was amplified by PCR using the primer pairs 5′-CACCATGATGGCGTCGAGGGTGATGGCG-3′ (F) and 5′-CTACCACTCCATCGAGTTTGTTCT-3′ (R, N-terminal GFP fusion) or 5′-CCACTCCATCGAGTTTGTTCTTCT-3′ (R, C-terminal GFP fusion). PCR products were inserted into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA). Validated cDNA inserts were then subcloned into p2FGW7 for N-terminal GFP fusion or p2GWF7 for C-terminal GFP fusion (Karimi et al. 2002), using LR clonase (Invitrogen). The resulting GFP-OsABF1 and OsABF1-GFP fusion constructs driven by the CaMV35S promoter were delivered into maize mesophyll protoplasts using a polyethylene glycol (PEG)-calcium mediated method (Hwang and Sheen 2001). This was followed by 12–24 h incubation to enable transient expression. Chlorophyll autofluorescence and OsHXK5NLS-RFP were used as chloroplast and nuclear markers, respectively (Cho et al. 2009). Expression of fusion constructs was monitored using a confocal microscope (LSM 510 META, Carl Zeiss, Jena, Germany).
Yeast one-hybrid experiment
The cDNA sequences of the full OsABF1 open reading frame (ORF) excluding the stop codon, the C-terminal bZIP region excluding the stop codon (OsABF1ΔN), and the N-terminal region excluding the bZIP domain (OsABF1ΔC), respectively, were amplified by PCR using appropriate primer pairs. These PCR products were cloned using the pLUG-TA cloning vector systems, sequenced and then fused in frame with the pYESTrp2 vector containing the GAL4 DNA binding domain to construct pYESTrp2/OsABF1, pYESTrp2/OsABF1ΔN, and pYESTrp2/OsABF1ΔC. The yeast strain carrying the pYC7-Int plasmid was used as a lacZ reporter system. The pYC7-Int/ABRE construct was prepared by inserting a trimer of the Em1a element (GGACACGTGGCG) into the SmaI site of pYC7-Int. pYESTrp2/OsABF1, pYESTrp2/OsABF1ΔN and pYESTrp2/OsABF1ΔC were transformed into yeast cells carrying pYC7-Int or pYC7-Int/ABRE. The pYESTrp2 empty vector and pYESTrp2/AtABF3 (Choi et al. 2000) were also transformed into the yeast cells as negative and positive controls, respectively. The transformants were incubated on SD-Ura-Trp plates at 30°C until positive clones were observed. The identified positive clones were streaked on fresh SD-Ura-Trp plates to purify the colonies.
Transactivation assay in yeast
Four to five yeast colonies were incubated in liquid SD medium at 30°C overnight. The cultures were then diluted several times with fresh yeast extract-peptone-dextrose (YPD) medium and incubated at 30°C for 3–5 h. The A600 was then measured at the end of the growth period and 1.5 ml of each culture was harvested by brief centrifugation and suspended in 1.5 ml of Z buffer (60 mM Na2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4·7H2O, pH 7.0). The cultures were again harvested by centrifugation and resuspended in 0.3 ml of Z buffer and separated into 0.1 ml aliquots which were freeze thawed three times using liquid nitrogen to lyse the cells. An aliquot of 0.7 ml of Z buffer supplemented with β-mercaptoethanol (100 ml Z buffer and 0.27 ml of β-mercaptoethanol) was then added to these preparations. The reaction was initiated by the addition of 0.16 ml of a 4 mg/ml stock solution of O-nitrophenyl β-d galactopyranoside (ONPG) at 30°C. After a color change to yellow, the reaction was stopped by adding 0.4 ml 1 M Na2CO3. The β-galactosidase activity at A420 was expressed in Miller units.
Stress tolerance in rice mutants
For salt treatment, rice plants were grown hydroponically in a growth chamber (28 ± 1°C, 80% relative humidity and 14/10 h day/night photoperiod) for 14 days and then transferred into 250 mM NaCl solution for 3 days. These plants were then transferred to normal growth conditions for 12 days. For dehydration treatment, rice plants grown for 14 days were transferred to a dish for 12 h, then rehydrated and grown for 14 days. The number of plants that continued to grow was counted.
Results
Identification of the OsABF1 transcription factor
We performed microarray experiments to monitor rice genes that undergo altered expression under conditions of abiotic stress (data not shown). From these analyses we selected and further characterized a bZIP transcription factor, OsABF1, which was found to be induced by various types of abiotic stress. An 801 bp cDNA clone of OsABF1 was isolated from rice seedlings using gene specific primers based on the TIGR Rice Genome Annotation Database (http://blast.jcvi.org/euk-blast/index.cgi?project=osa1; LOC_Os01g64730). Our sequence analysis indicated that OsABF1 is identical to the predicted gene OsZIP12 from the OsZIP family list (Nijhawan et al. 2008). A comparison of OsABF1 to other coned bZIP proteins revealed that it shows the highest homology to OsbZIP40, GBF4, and AtbZIP13, transcription factors that were previously classified as group A bZIPs (Supplementary Fig. S1) (Lu et al. 2008). The basic region of the OsABF1 bZIP domain exhibits high sequence similarity to the basic domain of these three bZIP proteins. The leucine zipper region of OsABF1 contains five heptad repeats. Thus, OsABF1 can be classified as one of the group A bZIPs that comprise the ABF family.
To evaluate the divergence of OsABF1 from other bZIPs, a phylogenetic tree was constructed for the group A bZIPs of Arabidopsis and rice. This analysis indicated that all of the group A bZIPs can be classified into three subgroups (Fig. 1), each containing both rice and Arabidopsis bZIPs, thus suggesting that the divergence of these subgroups predated the divergence of dicots and monocots.
OsABF1 expression under abiotic stress treatments
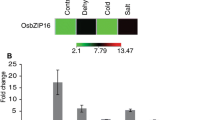
The expression patterns of the OsABF1 gene were analyzed in the shoots and roots of rice seedlings under various treatments such as anoxia, drought, salinity, cold, osmotic and oxidative stresses, and ABA by RT-PCR (Fig. 2). OsABF1 transcripts were not detectable in untreated control samples (Fig. 2a). In contrast, OsABF1 was upregulated more strongly in shoots than in roots after 4 h in response to anoxia treatment (Fig. 2b). In drought and salinity treated seedlings, OsABF1 expression was highly induced after 2 h (Fig. 2c, d). Upon oxidative stress (H2O2) and cold (4°C) treatments, the OsABF1 gene was similarly induced within 2 h, reached a maximum level after 8–12 h and then gradually decreased (Fig. 2e, f). These results demonstrate that OsABF1 is rapidly upregulated under all of the abiotic stress conditions tested. In addition, we further examined whether OsABF1 responds to treatment with the phytohormone ABA and found this gene to be highly induced within 2 h in ABA-treated seedlings (Fig. 2g). A well-known abiotic stress inducible gene, OsDEG10 was used as a positive control in these experiments as the transcript levels of this gene was previously found to be increased under conditions of anoxia, salinity, drought, NaCl, methyl viologen and cold (Park et al. 2009). Our current analyses confirmed that OsDEG10 is expressed within 12 h in all of the stress-treated rice seedlings (Fig. 2h).
Expression patterns of the OsABF1 gene in the shoots and roots of rice seedlings. a Expression of the OsABF1 gene in rice seedlings under normal (distilled water) conditions without stress treatment at 0, 2, 4, 8, 12 and 24 h. b Expression of the OsABF1 gene in the shoots and roots of rice seedlings at 0, 2, 4, 8, 12 and 24 h after anoxia treatment, and after 4 h (R4) and 12 h (R12) of recovery from this stress. c–g Expression pattern of the OsABF1 gene during drought (10% PEG), salinity (250 mM NaCl), oxidative stress (10 mM H2O2), cold (4°C) conditions, and ABA (100 μM) treatments in the shoots and roots of rice seedlings at 0, 2, 4, 8, 12 and 24 h after stress onset. h Expression pattern of OsDEG10 in the shoots of rice seedlings after 12 h of anoxia, drought (10% PEG), salinity (250 mM NaCl), ABA (100 μM), oxidative (10 mM H2O2) or cold (4°C) stress conditions. The rice actin1 gene was used as an internal control
Subcellular localization of OsABF1
To determine the subcellular localization of the OsABF1 protein, we generated GFP-OsABF1 and OsABF1-GFP fusion constructs under the control of CaMV35S promoter. These constructs were then expressed in the mesophyll protoplasts of maize. The results of this experiment revealed that both the OsABF1-GFP and GFP-OsABF1 fusion proteins are expressed in the nuclei of maize protoplasts (Fig. 3a, b), as confirmed by their colocalization with the nuclear marker, OsHXK5NLS-RFP (Cho et al. 2009). These data thus indicate that OsABF1 is a nuclear protein that may act as a transcription factor to regulate the expression of downstream genes.
Subcellular localization of GFP-OsABF1 and OsABF1-GFP fusion proteins in transfected mesophyll protoplasts of maize. a GFP-OsABF1; b OsABF1-GFP. Chlorophyll autofluorescence and NLS-RFP were used as chloroplast and nuclear markers, respectively. A false color (blue) was used for chlorophyll autofluorescence to distinguish it from GFP (green) and RFP (red) fluorescence
Transactivation ability of OsABF1
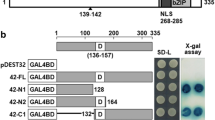
A yeast one-hybrid experiment was performed to test the transactivation ability of OsABF1. OsABF1, OsABF1ΔC, and OsABF1ΔN sequences were cloned into a yeast expression vector pYESTrp2 (Fig. 4a) which contains the B42 domain under the control of the yeast GAL1 promoter. Thus, the expression of cDNA inserts fused to the B42 activation domain is inducible by glucose. This construct was used to transform the yeast strain pYC7-Int that harbors a trimer of the Em1a element, a conserved ABRE domain. In the yeast strain transformed with pYESTrp2/OsABF1 grown on SD-Ura-Trp, β-galactosidase activity was found to be three-fold higher than compared with the control construct (Fig. 4b). This transactivation ability remained for OsABF1ΔC but was not observed in the yeast strain containing OsABF1ΔN indicating a requirement for the N-terminal region. Hence, our results suggest that OsABF1 binds to cis-acting elements containing the ABRE core sequence and thereby drives the transcription of downstream genes. It is noteworthy in this regard that no enzymatic activity was detectable in our analysis of the pYC7-Int strain that lacks the ABRE promoter (data not shown).
Transactivation assay of the OsABF1 protein. a Vectors used in a yeast one-hybrid assay. b Transactivation activity in yeast. β-Galactosidase activity is expressed in Miller units. pYESTrp2/AtABF3 and pYESTrp2 empty vector were used as a positive and a negative control, respectively. The bar indicates SD values
Analysis of OsABF1 mutants
To examine the function of OsABF1 during conditions of abiotic stress, we isolated two independent T-DNA mutant alleles of OsABF1, Osabf1-1 and Osabf1-2, generated from Hwayoung (HY) and Dongjin (DJ) wild type plants, respectively. Osabf1-1 contains a T-DNA insertion in the second exon whereas Osabf1-2 harbors this insertion in the first intron (Fig. 5a). The absence of OsABF1 transcripts in the homozygous mutants was confirmed by RT-PCR and these plants were then subjected to stress tolerance testing (Fig. 5b). In these experiments, the mutant and corresponding wild type plants were grown hydroponically for 14 days and transferred into a 250 mM NaCl solution for a further 3 days. After the salt treatment, the plants were allowed to recover in hydroponic solution for 12 days. No mutant plants survived this treatment, whereas 22–29% of the wild type plants were alive at the end of the experimental period (Fig. 5c, d). These results demonstrate that Osabf1 mutants are more susceptible to high salinity than wild type plants.
Survival rate of OsABF1 mutant plants under abiotic stress. a Schematic diagram of the OsABF1 genomic structure and T-DNA insertion sites in the mutant alleles Osabf1-1 and Osabf1-2. The box and solid lines indicate exons and introns, respectively. The position of the T-DNA is indicated by a triangle. b Expression analysis by RT-PCR of OsABF1 in the wild type rice plants, Hwayoung (HY) and Dongjin (DJ), and in corresponding T-DNA mutants at 12 h after ABA (100 μM) treatment. c Survival rate of Osabf1 mutants when subjected to high salinity (250 mM NaCl). d Representative phenotype of wild type and T-DNA mutants treated with high salinity. Plants on the left and right, respectively, are surviving and dead wild type plants. e Survival rate of Osabf1 mutants subjected to dehydration. f Representative phenotype of wild type and T-DNA mutants treated with dehydration. Plants on the left and right, respectively, are representative surviving and dead plants from each line
We conducted a parallel stress tolerance assay for dehydration in which 14-day-old seedlings were exposed to dry conditions for 12 h and later placed in a hydroponic solution to recover for 14 days. The results showed that 23–42% of the mutant plants and 54–65% of the wild type plants survived (Fig. 5e, f). Wild type plants are thus more tolerant to dehydration than Osabf1 mutants. These results again suggest that the OsABF1 plays an important role in enhancing the tolerance of rice plants to abiotic stress conditions including high salinity and dehydration.
Expression of ABA/stress-regulated genes in Osabf1 mutants
Abscisic acid-dependent and independent regulatory systems are known to be involved in the control of stress-responsive gene expression (Yamaguchi-Shinozaki and Shinozaki 2005, 2006). To evaluate the possible regulatory role of OsABF1, we compared the expression patterns of ABA/stress-regulated genes in both Osabf1 mutant and wild type rice seedlings treated with ABA. Six abiotic stress-inducible genes were selected to investigate the regulatory function of OsABF1: OsABA45 (LOC_Os12g29400), Asr1 (LOC_Os02g33820), SalT (LOC_Os01g24710) and OsNAC (LOC_Os01g66120) that are induced by cold, drought, high salinity, and ABA in rice seedlings (Rabbani et al. 2003); OsLEA3 (LOC_Os05g46480) encoding a late embryogenesis abundant protein induced by ABA; and SKC1 (LOC_Os01g20160) encoding a cation transporter involving salt response in rice seedlings (Zou et al. 2008). Interestingly, the upregulated expression of the OsNAC, OsLEA3, and OsABA45 genes in wild type plants exposed to ABA was significantly suppressed in the Osabf1 mutants (Fig. 6). No significant differences in the expression profile were found for the other genes. These data indicate that OsABF1 plays a regulatory role during the expression of specific ABA/stress-inducible genes.
RT-PCR analysis of ABA/stress regulated genes in Osabf1 mutant and wild type rice plants. a Hwayoung and Osabf1-1; b Dongjin and Osabf1-2. Expression of the marker genes was analyzed in the mutant and control plants at 0, 2, 4, 8, 12 and 24 h after ABA (100 μM) treatment. The rice actin1 gene was used as an internal control
Discussion
The bZIP transcription factor family plays an important role in abiotic stress tolerance in plants. Of the 10 identified groups (A, B, C, D, E, F, G, H, I and S) of the Arabidopsis bZIP family, group A includes the ABF genes (Jakoby et al. 2002). In rice, 14 out of 89 members of the bZIP family belong to group A. This is based on the highly conserved motif in the basic and hinge region and the phosphorylation site for Ca2+-dependent protein kinase (R/KxxS/T) in two residues that are C-terminal to a conserved Leu within this motif (Nijhawan et al. 2008). Consistently, amino acid sequence alignments demonstrate that OsABF1 is similar to other bZIP proteins including OsbZIP40, GBF4, and AtbZIP13, also classified as group A bZIP proteins. Together with our phylogenetic analysis, these findings suggest the possibility of functional conservation among these transcription factors.
It has been demonstrated previously that the expression of many ABA/stress-regulated genes in plants is mediated by cis elements sharing the ACGT sequence. Cis-acting regulatory elements are important molecular switches involved in the transcriptional regulation of a dynamic network of genes (Yamaguchi-Shinozaki and Shinozaki 2005, 2006). The result of subcellular localization experiments suggested that OsABF1 functions as a transcriptional regulator in the nucleus (Fig. 3). Consistently, other bZIP proteins are also present in the nucleus during embryo maturation (Zou et al. 2008). Our current yeast one-hybrid analysis indicates that OsABF1 can bind to the ABRE element and retain transactivation ability (Fig. 4) indicating that OsABF1 binds to ABRE and activates the expression of downstream genes. Similar observations of OsABI5, OsbZIP23, and OsbZIP72 have been reported in yeast, i.e., that these proteins bind to motifs harboring the ABRE core sequence (Zou et al. 2008; Xiang et al. 2008; Lu et al. 2008).
Several bZIP ABA responsive transcription factors have been isolated in different tissues in rice such as RITA1 in rice seeds (Izawa et al. 1994), OsZIP1a and OsZIP2a in the vegetative parts of rice (Nantel and Quatrano 1996), OsbZ8 in developing embryos (Nakagawa et al. 1996) and OsABI5 in young rice seedlings (Zou et al. 2008). In our present study, OsABF1 expression was found to be significantly induced in the shoots and roots of rice seedlings by various abiotic stresses such as anoxia, drought, salinity, oxidative stress and ABA (Fig. 2). Our data thus indicate that OsABF1 plays an important role in response to abiotic stresses. To address the functions of OsABF1 during ABA signaling, we characterized its null mutants. Seedlings of the Osabf1 mutants were found to be more sensitive to high salinity and dehydration treatments compared with wild type plants (Fig. 5). Similarly, the OsbZIP23 null mutant was more sensitive to salinity and drought and the overexpression of this gene enhanced tolerance to drought and salinity (Xiang et al. 2008). Our results are consistent with the finding that abf3 and abf4 mutants display defects in response to ABA, salinity and dehydration in Arabidopsis (Kim et al. 2004). Hence, OsABF1 likely plays a positive role as an ABA responsive transcription factor in abiotic stress signaling.
The upregulated expression of the ABA-regulated genes, OsNAC, OsLEA3, and OsABA45, was suppressed in each case in Osabf1 mutants (Fig. 6). The expression of OsLEA3 and OsABA45 was previously found to be increased in OsbZIP23 and OsbZIP72 overexpressing plants, whereas OsABA45 expression was shown to be decreased in an OsbZIP23 mutant line (Xiang et al. 2008; Lu et al. 2008). Thus, both OsbZIP23 and OsbZIP72 appear to play positive regulatory roles in ABA signaling and their overexpressing lines are more tolerant to salinity and drought. OsNAC has been found to be induced by ABA, and OsNAC-overexpressing plants show improved tolerance to dehydration and salt stresses (Nakashima et al. 2007; Zheng et al. 2009). This suggests the involvement of OsABF1 in the ABA signal transduction pathway. Hence, we propose OsABF1 as a positive regulator in the ABA-dependent abiotic signaling pathway. Additional studies will be required to obtain further insight into the mechanism by which OsABF1 regulates the expression of OsNAC, OsLEA3, and OsABA45 in response to ABA. The expression of SKC1, Asr1 and SalT was found not to be significantly altered in the Osabf1-1 and Osabf1-2 mutants. This suggests that the regulation of these genes may follow a distinct ABA-dependent abiotic pathway.
In summary, we report herein the ABA/stress-inducible expression of OsABF1 and its function in stress tolerance in rice seedlings. The role of OsABF1 in the ABRE-mediated expression of downstream target genes and thus in stress tolerance was demonstrated by a yeast one-hybrid assay and a loss of function approach in our experiments. Thus, our present results provide evidence for the involvement of OsABF1 in stress tolerance and ABA signaling in rice.
References
Casaretto J, Ho TD (2003) The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 15:271–284
Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgern T, Mauch F, Luan S, Zou G, Whitham SA, Budworth PR, Tao Y, Xie Z, Chen X, Lam S, Kreps JA, Harper JF, Heinlein M, Kobayashi K, Hohn T, Dangl JL, Wang X, Zhu T (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14:559–574
Chen W, Zhu T (2004) Networks of transcription factors with roles in environmental stress response. Trends Plant Sci 9:591–596
Chern MS, Eiben HG, Bustos MM (1996) The developmentally regulated bZIP factor ROM1 modulates transcription from lectin and storage protein genes in bean embryos. Plant J 10:135–148
Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451
Cho JI, Ryoo N, Eom JS, Lee DW, Kim HB, Jeong SW, Lee YH, Kwon YK, Cho MH, Bhoo SH, Hahn TR, Park YI, Hwang I, Sheen J, Jeon JS (2009) Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol 149:745–759
Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275:1723–1730
Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E (2006) Integration of abscisic acid signalling into plant responses. Plant Biol 8:314–325
de Vetten NC, Ferl RJ (1995) Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J 7:589–601
Finkelstein RR, Lynch T (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Fode B, Siemsen T, Thurow C, Weigel R, Gatz C (2008) The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20:3122–3135
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17:3470–3488
Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96:15348–15353
Hurst HC (1994) Transcription factors 1. bZIP proteins. Protein Profile 1:123–168
Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413:383–389
Izawa T, Foster R, Nakajima M, Shimamoto K, Chua NH (1994) The rice bZIP transcriptional activator RITA-1 is highly expressed during seed development. Plant Cell 6:1277–1287
Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, Kim J, Shin M, Jung M, An G (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45:123–132
Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14:343–357
Karimi M, Inze D, Depicker A (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195
Kato-Noguchi H, Morokuma M (2007) Ethanolic fermentation and anoxia tolerance in four rice cultivar. J Plant Physiol 164(2):168–173
Kim SY (2007) Recent advances in ABA signaling. J Plant Biol 50:117–121
Kim SY, Chung HJ, Thomas TL (1997) Isolation of a novel class of bZIP transcription factor that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J 11:1237–1251
Kim S, Kang J, Cho D, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40:75–87
Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49:199–222
Liu L, White MJ, MacRae TH (1999) Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur J Biochem 262:247–257
Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32:317–328
Lu G, Gao C, Zhong X, Han B (2008) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229:605–615
McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2:163–171
Meshi T, Iwabuchi M (1995) Plant transcription factors. Plant Cell Physiol 36:1405–1420
Minami M, Huh GH, Yang P, Iwabuchi M (1993) Coordinate gene expression of five subclass histones and the putative transcription factors, HBP-1a and HBP-1b, of histone genes in wheat. Plant Mol Biol 23:429–434
Nakagawa H, Ohmiya K, Hattori T (1996) A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J 9:217–227
Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26:627–635
Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3 and ABA responsive genes including RD29B and RD29A in seeds, germinating embryos and seedlings of Arabidopsis. Plant Mol Biol 60:51–68
Nakashima K, Tran lam-son P, Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Nantel A, Quatrano RS (1996) Characterization of three rice basic/leucine Zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J Biol Chem 271:31296–31305
Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146:333–350
Niu X, Renshaw-Gegg L, Miller L, Guiltinan MJ (1999) Bipartite determinants of DNA-binding specificity of plant basic leucine zipper proteins. Plant Mol Biol 41:1–13
Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351
Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim JK (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol 150:1368–1379
Park HY, Kang IS, Han JS, Lee CH, An G, Moon YH (2009) OsDGE10 encoding a small RNA-binding protein is involved in abiotic stress signaling. Biochem Biophys Res Commun 380:597–602
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3:423–434
Rodriguez-Uribe L, O’Connell MA (2006) A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris). J Exp Bot 57:1391–1398
Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR (1992) Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J 11:1261–1273
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji K, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full length cDNA microarray. Funct Integr Genomics 2:282–291
Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K (2003) Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnol 14:194–199
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97:11632–11637
Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of basic leucine zipper transcription factor family for conferring Abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148:1938–1952
Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic and cold-stress-responsive promoters. Trends Plant Sci 10:88–94
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yanagisawa S (1998) Transcription factors in plants: physiological functions and regulation of expression. J Plant Res 111:363–371
Zheng X, Chen B, Lu G, Han B (2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379:985–989
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683
Acknowledgments
We thank Dr. Soo Young Kim of Chonnam National University, Kwangju, Korea for providing the yeast strains. This work was supported by grants from the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2005-070-C00128), and from the World Class University (R33-2008-000-10168-0) and Crop Functional Genomics Center (CG2111-2) programs funded by the Korean Ministry of Education, Science, and Technology.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2009_9592_MOESM1_ESM.pdf
Supplementary Fig. S1. Multiple sequence alignment and phylogenetic analysis of the OsABF1 protein in comparison with other bZIP proteins. Amino acid alignment of OsABF1 with the bZIP proteins, OsbZIP40 (AAT07607), GBF4 (BAF00453), and AtbZIP13 (NP_199221) is shown. Blast analysis was performed using the NCBI-P blast program. The Clustal W-PBI program was used for multiple sequence alignments. The double underline represents the basic region and leucine repeat whereas the single underlines indicate the conserved regions. (PDF 84 kb)
Rights and permissions
About this article
Cite this article
Amir Hossain, M., Lee, Y., Cho, JI. et al. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72, 557–566 (2010). https://doi.org/10.1007/s11103-009-9592-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9592-9