Abstract
Plants exposed to adverse environmental conditions are invariably compromised in their growth and development. The bZIP class of transcription factors (TF) form a large group among stress signalling components that regulate plant responses towards stress. We identified bZIP TF encoding genes that are expressed differentially in indica rice under stress and here we functionally characterize one such gene, OsbZIP16. Although, OsbZIP16 forms a clade with its orthologous monocot protein sequences, we find in our study that it can impart tolerance to abiotic stress in Arabidopsis. OsbZIP16 is expressed strongly upon dehydration, salt and ABA treatment in Oryza sativa cv. IR64 seedlings. It localizes in the cell nucleus and the gene product is capable of transcriptional activation, thus providing evidence for its capability as a functional TF. Upon overexpression in Arabidopsis, OsbZIP16ox plants show wild type morphology, however, these plants showed tolerance when subjected to drought stress at vegetative stage and set healthy seeds on recovery. The OsbZIP16ox seedlings showed reduced sensitivity to mannitol, ABA and sodium chloride during germination and also reduced ROS accumulation upon H2O2 exposure. Thus, OsbZIP16 regulates abiotic stress responses and is also a good candidate gene that can be utilised to impart tolerance in plants under water deficit conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants exposed to stress caused by drought, salinity, extreme heat or cold, have compromised growth and development. The stress hormone, ABA, key internal signal under desiccating cellular environment also regulates overall growth in plants (Finkelstein 2013). The production of secondary messengers on perception of stress triggers downstream signalling pathways bringing changes in gene expression among plethora of metabolic changes that eventually help the plant to acclimate with stress conditions. Gene expression changes brought in by transcription factors (TFs) remains an attractive choice that can be manipulated to design robust crops tolerant to fluctuating environmental conditions. Comprehensive research on genes belonging to DREB, ABF/AREB, bZIP, MYB or WRKY family of TFs has helped in deciphering molecular mechanisms underlying drought tolerance in plants (Osakabe et al. 2014). Binding of TF to its specific target sequence (including cis-acting elements) that lie usually upstream of transcription start site regulates expression of the target gene. Either an activation or repression in gene expression can be brought about by TFs acting alone or in combination. Members of bZIP TF family usually form dimers for their function while binding to target promoter sequence. Genome wide analyses have identified multigene families that invariably encode for TFs in both plants and animals (Riechmann et al. 2000).
The bZIP TFs represents a large and diverse multigene family in eukaryotes and are known to affect almost all phases of the plant life cycle. They have been shown to regulate number of plant processes such as seed development, light signalling, floral induction and flower development, biotic and abiotic stresses, ABA signalling and hormonal response (Chern et al. 1996; Bensmihen et al. 2005; Jakoby et al. 2002; Nijhawan et al. 2008). The bZIP TFs derive their name from a bZIP domain that is highly conserved and is made up of a basic region with a leucine zipper domain (Hurst 1995). The current number of bZIP gene models in rice genome has been predicted to be 140 and 94 for japonica and indica, respectively (Jin et al. 2017). Earlier, genome wide analysis in indica rice identified 89 bZIP protein encoding genes and their expression profile within various developmental stages and environmental stresses (Nijhawan et al. 2008). Functional analysis of bZIP genes in rice have shown their ability to regulate abiotic stress signalling and responses in Arabidopsis as well as in rice. In rice, OsbZIP23 is rapidly and strongly induced by drought, ABA, PEG and NaCl treatment and also confers tolerance to abiotic stress by regulating stress related gene expression in an ABA-dependent manner (Xiang et al. 2008). Similarly, OsbZIP46 is up-regulated to high levels under drought, heat and ABA treatment (Tang et al. 2012). The over-expression of OsbZIP46 increased sensitivity to ABA. However, its over-expression did not promote drought tolerance. Another ABA induced bZIP TF, OsBZ8 expressed strongly in salt-tolerant rice varieties compared to the sensitive ones (Nakagawa et al. 1996; Mukherjee et al. 2006). Among the core regulators of ABA sensitivity, OsABI5, a bZIP family member encodes for ABRE/G-box binding protein (Zou et al. 2007, 2008). OsABI5 protein localizes to nucleus, has trans-activity but negatively regulates stress tolerance on over-expression in rice. Similarly, OsbZIP16 in rice promotes ABA sensitivity but drought resistance in rice (Chen et al. 2012). OsbZIP52 is induced by low temperature (4 °C) and its over-expression increases susceptibility to cold and drought stress in rice (Liu et al. 2012).
Based on our earlier work on the expression profile of OsbZIP genes during abiotic stresses (Nijhawan et al. 2008), OsbZIP16 gene was selected for characterization and functional validation in the present study. Earlier, some initial work has been done by another group on the function of OsbZIP16 in rice (japonica) under drought stress (Chen et al. 2012) and in our laboratory too some of these observations could be replicated (Pandey 2015). OsbZIP16 was found to be stress inducible in rice cultivar IR64 as it showed elevated transcript levels in dehydration and salt stress treated rice seedlings. In the present study, OsbZIP16 was ectopically over-expressed in model plant Arabidopsis thaliana Col-0 (OsbZIP16ox). Morphologically, the over-expressing plants exhibited a phenotype similar to WT. However, seeds of these OsbZIP16ox when plated on mannitol/salt/ABA containing media performed better in seed germination assays than WT seeds. Our work while confirming the previous observations on role of OsbZIP16 under abiotic stress, extends its association with underlying redox mechanisms. The function of bZIP proteins in mediating redox changes under oxidative stress has not been characterised well. Our work suggests their involvement in regulation of genes involved in oxidative stress mitigation. The oxidative stress, seed germination and drought assays performed on OsbZIP16 (indica) over-expressing Arabidopsis plants confirmed the ability of OsbZIP16 in conferring dehydration and oxidative stress tolerance.
Materials and methods
Sequence analysis
The nucleotide and protein sequences were downloaded from NCBI (http://www.ncbi.nlm.nih.gov); RGAP (http://rice.plantbiology.msu.edu) and TAIR (http://www.arabidopsis.org) databases. The presence of putative nuclear localization signal in the protein sequence was predicted using NucPred tool (http://www.sbc.su.se/~maccallr/nucpred/cgi-bin/single.cgi).
Phylogenetic analysis
The orthologous genes of OsbZIP16 were initially identified using protein sequences (full-length) in Phytozome database (BLAST-proteome, http://www.phytozome.net). To authenticate, the searched sequence of the selected topmost (one or two) hits were rechecked in NCBI and rice array database. Finally, these confirmed protein sequences from different plant species were aligned in Clustal X software. Manual curation in the alignment was performed and final phylogenetic tree was constructed by neighbour joining method (bootstrap value 1000).
Abiotic stress and phytohormone treatments
For treatment with plant hormones, light grown rice seedlings (7 days old Oryza sativa cv. IR64) were grown on cotton soaked with yoshida medium and gently pulled out without injuring the roots (Borah et al. 2017). The treatment for stress or with different hormones was given as described previously (Jain et al. 2006a). After the treatment, the seedlings were snap-frozen using liquid nitrogen and stored at − 80 °C for isolation of RNA.
Real-time qPCR expression analysis
Total RNA from all the samples was isolated as described previously (Jain et al. 2006a) and real time qPCR was performed (Borah et al. 2017). The primer sequences are given in supplementary table 1. Ubiquitin5 gene (AK06198) was used as control for RT-qPCR analysis.
Molecular characterization of OsbZIP16
OsbZIP16 full CDS was amplified from rice cDNA prepared from 7-days old light grown seedlings that were dehydration stressed (Oryza sativa ssp indica cv. IR64). For transformation of Arabidopsis thaliana (Col-0), plants were grown in plastic pots filled with Soil rite under culture room conditions (temperature 22 ± 1 °C and daily cycle of 16 h light/8 h dark). The primary bolts were trimmed to induce the lateral bolts and when there were sufficient floral buds on the plant, transformation of plants was carried out by floral dip (Clough and Bent 1998). Further selection and analysis was performed as described (Jain et al. 2006b). Particle bombardment for localisation of fusion protein in onion epidermal cells and trans-activation in yeast was performed as described previously (Burman et al. 2017).
Stress assays
Seed germination assay was performed as described earlier (Jain et al. 2008). The number of germinated seeds was expressed as the percentage of total number of seeds. For dehydration stress assay, T4 homozygous generation OsbZIP16ox Arabidopsis seeds along with control WT were grown for 18 days in plastic pots filled with soilrite in culture room conditions. The plants were subjected to dehydration stress by water withdrawal for 14 days and recovered by watering again.
Estimation of ROS
The amount of ROS was detected by staining with DAB and NBT (Jabs et al. 1996; Schraudner et al. 1998). The 7-days-old seedlings grown on 1/2 MS medium were stressed using H2O2 (10 mM) treatment for 30 mins and then stained for 15 mins by NBT (2 mM NBT in 20 mM phosphate buffer) or overnight with DAB stain (Phosphate buffer (100 mM), Tween-20 (0.05 %), 200 mM Na2PO4 (200 mM), pH 3.0). The plants were washed once again with Miili-Q water and chlorophyll was removed by using 3:1:1 solution of ethanol, acetic acid and glycerol. Visualisation and image capture was done using a light microscope camera (Leica DFC295).
Results and discussion
Gene structure and phylogenetic analysis
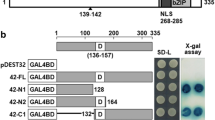
In our previous study, we identified 37 OsbZIP genes expressing differentially under drought, salt and cold stress (Nijhawan et al. 2008). The study identified 37 OsbZIP encoding genes, 26 up-regulated and 11 down-regulated in at least one of the stress condition. A drought and salt stress inducible gene, OsbZIP16 was selected from this expression analysis for further functional characterization. Drought and salinity severely effect plant physiology and metabolism and share a high degree of similarity due to the osmotic component (Sharma et al. 2015). OsbZIP16 gene (LOC_Os02g09830) encodes a bZIP transcription factor localised on the 2nd chromosome of rice. Alignment of genomic and full-length cDNA sequence revealed that the gene is intron less and a 513 bp CDS encodes for 170 amino acid residues. The bZIP domain is present between 70th and 134th residues, essential for transcriptional activity of bZIP proteins. Phylogenetic analysis shows OsbZIP16 forms a separate monocot clade while its other orthologous proteins clustered in a distinct clade (Supplementary Figure 1).
OsbZIP16 is strongly expressed by dehydration and salinity in rice
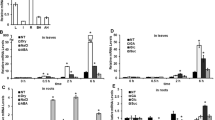
The expression profile generated through microarray analysis showed that OsbZIP16 is strongly induced under different abiotic stress conditions (Fig. 1a). The detailed expression analysis of OsbZIP16 done by RT-qPCR under stress conditions and different hormones treatment validates the microarray profile (Fig. 1b). Among the plant hormones tested, we find that only ABA has an inducing effect on the OsbZIP16 expression. OsbZIP family members OsbZIP12 and OsbZIP72 are also induced by and regulated in an ABA-dependent manner (Lu et al. 2009; Amir Hossain et al. 2010). Thus, OsbZIP16 is likely to be involved in the abiotic stress and ABA-related pathways especially under drought and salt stress conditions.
OsbZIP16 is nuclear localised and is capable of transcriptional activation
Being a transcription factor and to regulate target gene expression, OsbZIP16 should ideally be present inside the nucleus of cell. Bioinformatics analysis using protein sequence in NucPred software revealed presence of nuclear localization signal from 74 to 78 amino acid residues. The high score of 0.84 also predicted a nuclear localization. In-vivo localization of OsbZIP16 in onion epidermal peel was checked by particle bombardment. In the control samples, GFP fluorescence was distributed throughout the cell while OsbZIP16-GFP fusion construct was observed to be localized inside the nucleus (Supplementary Figure 2). OsbZIP proteins like OsbZIP23 and OsbZIP1 are also nuclear localised (Meng et al. 2005; Xiang et al. 2008).
Since OsbZIP16 is nuclear localized and is expected to work as a transcription factor, its transcription activation potential was checked in yeast. As the yeast cells transformed with the OsbZIP16::pGBKT7 constructs were able to grow on SD/-HW media similar to positive (+ve) control, it could be inferred that the OsbZIP16 could transcriptionally activate the histidine reporter gene in yeast, thus confirming its transcription activation potential (Fig. 2). This was in consonance with earlier reports of OsbZIP proteins being nuclear localised and having a transcriptional activation potential (Zou et al. 2008; Chen et al. 2012; Liu et al. 2012).
OsbZIP16 promotes abiotic stress tolerance in Arabidopsis
To gain an insight into the role of OsbZIP16 transcription factor in stress responses, Arabidopsis plants with over-expression of OsbZIP16 were raised to T4 generation before being assayed (Fig. 3). Confirmation of the transgene in Arabidopsis was done by PCR amplification that showed presence of an appropriate size band in the over-expressor Arabidopsis lines (OsbZIP16ox) (Fig. 3a). The transcript level checked by RT-qPCR analysis showed an expression of OsbZIP16 in the OsbZIP16ox Arabidopsis plants (Fig. 3b). The phenotypic analysis of OsbZIP16ox plants showed similarity in overall growth and plant morphology under normal conditions (Supplementary Figure 3). As the expression of OsbZIP16 was induced by dehydration, salt and ABA, the germination check was performed on OsbZIP16ox seeds under stress conditions (on ½ MS Agar + Mannitol/NaCl/ABA) and assessed for sensitivity in radicle growth. OsbZIP16ox seeds show higher germination percentage than wild type under ABA, mannitol and NaCl stimulated stress conditions (Fig. 4). The response of OsbZIP16ox plants towards drought stress conditions at vegetative stage was assayed by water withdrawal for 14 days. The assay revealed that OsbZIP16ox plants are drought tolerant as these plants could survive the stress and complete their life-cycle to set seeds after recovery phase. These assays using seeds and seedlings show that OsbZIP16 can alleviate stress arising due to dehydration and salinity in Arabidopsis. Since the OsbZIP16ox seedlings showed reduced sensitivity towards ABA, it could be speculated that the tolerance mechanism by OsbZIP16 also involves ABA-independent pathways. Many members of the bZIP TFs have been well characterized in both ABA dependent as well as ABA independent (RISBZ5/OsbZIP52) pathways of stress signalling in plants (Amir Hossain et al. 2010; Liu et al. 2012; Tang et al. 2012; Liu et al. 2014).
The OsbZIP16 over-expression plants in Arabidopsis were raised. a Confirmation of transgene integration in plants by CDS PCR amplification. b The real-time qPCR analysis of the three selected homozygous lines for OsbZIP16 expression. c Arabidopsis OsbZIP16 over-expression lines showing dehydration tolerance in comparison to WT plants after 14 days of drought stress
OsbZIP16ox seedlings accumulate lower ROS under oxidative stress
ROS is important for cellular signalling and sensing of cellular redox state (Mittler et al. 2011; Noctor et al. 2014). However, excess ROS is cause of oxidative stress that damages cellular membranes and compartments through lipid peroxidation. Thus, for survival of plants under stress, maintenance of ROS at optimum levels is an important factor. Since OsbZIP16 imparts tolerance under stress conditions, regulation of ROS level is crucial. We compared the amount of ROS generated by H2O2 treatment for 30 min in the seedlings grown under normal conditions. The seedlings were then stained with DAB (for H2O2) and NBT (for O2−). The staining intensity was used as an indicator for the amount of the ROS in the cotyledons of seedlings. The staining was comparatively less intense in the OsbZIP16ox seedlings (Fig. 5). Thus, OsbZIP16 could also possibly regulate antioxidant pathway genes that increase ROS mitigation under stressful conditions. We observed NBT stain only in form of patches and speckles in the seedlings showing only a localized accumulation of O2− under stress conditions. Such localized ROS production in the form of lesions or patches have been reported earlier in the tobacco leaves under stress conditions (Wohlgemuth et al. 2002).
Our previous study gave an exhaustive account of 89 bZIP protein coding genes identified in rice. Based on the expression data, number of bZIP genes have been selected and characterised for their different roles in plants including abiotic stress responses by various groups (Todaka et al. 2015; Burman et al. 2017). In the present study, a phylogenetically conserved bZIP transcription factor encoding gene has been characterised for its role under abiotic stress response. OsbZIP16 is a drought and salt stress inducible gene whose ectopic expression can positively promote tolerance towards abiotic stresses in plants. OsbZIP16 fusion protein from both indica as well as japonica rice as gene source localises in the nucleus and can also perform trans-activation in yeast confirming its capability as a functional transcription factor (Chen et al. 2012; Present study). A number of bZIP domain containing TFs have been characterised in the past and found to be involved in multiple abiotic stress responses. However, only very few studies have shown its association with redox changes in plants (Huang et al. 2010; Guo et al. 2011). Physiological mechanism involving downstream pathways such as redox changes and ROS scavenging still remains an unexplored area that we have attempted to address in the present study. Reduced ROS upon ectopic over-expression of OsbZIP16 in Arabidopsis suggests about its potential to regulate genes involved in ROS-scavenging and antioxidant related pathways. Although the functions of bZIP/ABFs have been characterised involving ABA related pathways, it remains to be seen if the ROS scavenging pathways are functionally under control of ABA that are in turn regulated by ABRE-bZIP system. It remains our endeavour to characterize the gene in rice through its over-expression and RNAi lines. One of the major challenges would be to find out the downstream target genes of such bZIP proteins. Much more definitive clue may emerge by performing chip assays using antibodies specific to these bZIP proteins. From perspective of crop improvement, bZIP genes associated with abiotic stress responses remain strong candidate genes that can be utilised for development of abiotic stress tolerant crops.
Abbreviations
- OsbZIP:
-
Oryza sativa basic leucine zipper
- ABA:
-
Abscisic acid
- ROS:
-
Reactive oxygen species
- TFs:
-
Transcription factors
References
Amir Hossain M, Lee Y, Cho J-I et al (2010) The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72:557–566. https://doi.org/10.1007/s11103-009-9592-9
Bensmihen S, Giraudat J, Parcy F (2005) Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J Exp Bot 56:597–603. https://doi.org/10.1093/jxb/eri050
Borah P, Sharma E, Kaur A et al (2017) Analysis of drought-responsive signalling network in two contrasting rice cultivars using transcriptome-based approach. Sci Rep 7:42131. https://doi.org/10.1038/srep42131
Burman N, Bhatnagar A, Khurana JP (2017) OsbZIP48, a HY5 transcription factor ortholog, exerts pleiotropic effects in light-regulated development. Plant Physiol. https://doi.org/10.1104/pp.17.00478
Chen H, Chen W, Zhou J et al (2012) Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci 193–194:8–17. https://doi.org/10.1016/j.plantsci.2012.05.003
Chern MS, Eiben HG, Bustos MM (1996) The developmentally regulated bZIP factor ROM1 modulates transcription from lectin and storage protein genes in bean embryos. Plant J 10:135–148
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Finkelstein R (2013) Abscisic acid synthesis and response. Arabidopsis Book 11:e0166. https://doi.org/10.1199/tab.0166
Guo M, Chen Y, Du Y et al (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus magnaporthe oryzae. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1001302
Huang XS, Liu JH, Chen XJ (2010) Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol. https://doi.org/10.1186/1471-2229-10-230
Hurst HC (1995) Transcription factors 1: bZIP proteins. Protein Profile 2:101–168
Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273:1853–1856. https://doi.org/10.1126/science.273.5283.1853
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006a) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651. https://doi.org/10.1016/j.bbrc.2006.04.140
Jain M, Tyagi AK, Khurana JP (2006b) Overexpression of putative topoisomerase 6 genes from rice confers stress tolerance in transgenic Arabidopsis plants. FEBS J 273:5245–5260. https://doi.org/10.1111/j.1742-4658.2006.05518.x
Jain M, Tyagi AK, Khurana JP (2008) Constitutive expression of a meiotic recombination protein gene homolog, OsTOP6A1, from rice confers abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Rep 27:767–778. https://doi.org/10.1007/s00299-007-0491-8
Jakoby M, Weisshaar B, Dröge-Laser W et al (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–11
Jin J, Tian F, Yang DC et al (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45:D1040–D1045. https://doi.org/10.1093/nar/gkw982
Liu C, Wu Y, Wang X (2012) BZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235:1157–1169. https://doi.org/10.1007/s00425-011-1564-z
Liu C, Mao B, Ou S et al (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84:19–36. https://doi.org/10.1007/s11103-013-0115-3
Lu G, Gao C, Zheng X, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229:605–615. https://doi.org/10.1007/s00425-008-0857-3
Meng X-B, Zhao W-S, Lin R-M et al (2005) Identification of a novel rice bZIP-type transcription factor gene, OsbZIP1, involved in response to infection ofMagnaporthe grisea. Plant Mol Biol Report 23:301–302. https://doi.org/10.1007/BF02772762
Mittler R, Vanderauwera S, Suzuki N et al (2011) ROS signaling: the new wave? Trends Plant Sci 16:300–309. https://doi.org/10.1016/j.tplants.2011.03.007
Mukherjee K, Choudhury A, Gupta B et al (2006) An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol 6:18. https://doi.org/10.1186/1471-2229-6-18
Nakagawa H, Ohmiya K, Hattori T (1996) A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J 9:217–227. https://doi.org/10.1046/J.1365-313x.1996.09020217.X
Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146:333–350. https://doi.org/10.1104/pp.107.112821
Noctor G, Mhamdi A, Foyer CH (2014) The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol 164:1636–1648. https://doi.org/10.1104/pp.113.233478
Osakabe Y, Osakabe K, Shinozaki K, Tran L-SP (2014) Response of plants to water stress. Front Plant Sci 5:86. https://doi.org/10.3389/fpls.2014.00086
Pandey AS (2015) Functional characterization of some rice bZIP transcription factor genes involved in stress tolerance and seed development (PhD thesis). University of Delhi
Riechmann JL, Heard J, Martin G et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110. https://doi.org/10.1126/science.290.5499.2105
Schraudner M, Moeder W, Wiese C et al (1998) Ozone-induced oxidative burst in the ozone biomonitor plant, tobacco Bel W3. Plant J 16:235–245. https://doi.org/10.1046/j.1365-313X.1998.00294.x
Sharma E, Sharma R, Borah P et al (2015) Emerging roles of auxin in abiotic stress responses. In: Pandey GK (ed) Elucidation abiotic stress signal plants funct genomics perspect. Springer, New York. https://doi.org/10.1007/978-1-4939-2211-6_11
Tang N, Zhang H, Li X et al (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158:1755–1768. https://doi.org/10.1104/pp.111.190389
Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front Plant Sci 6:84. https://doi.org/10.3389/fpls.2015.00084
Wohlgemuth H, Mittelstrass K, Kschieschan S et al (2002) Activation of an oxidative burst is a general feature of sensitive plants exposed to the air pollutant ozone. Plant Cell Environ 25:717–726. https://doi.org/10.1046/j.1365-3040.2002.00859.x
Xiang Y, Tang N, Du H et al (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148:1938–1952. https://doi.org/10.1104/pp.108.128199
Zou M, Guan Y, Ren H et al (2007) Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem Biophys Res Commun 360:307–313. https://doi.org/10.1016/j.bbrc.2007.05.226
Zou M, Guan Y, Ren H et al (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683. https://doi.org/10.1007/s11103-008-9298-4
Acknowledgements
This work was financially supported by a grant (BT/PR12394/AGIII/103/891/2014) from the Department of Biotechnology, Government of India. ASP, ES and NB are grateful to CSIR and NJ to UGC for the award of research fellowships. The infrastructure support provided by the Department of Science and Technology, Government of India, and the University Grants Commission, New Delhi, is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no competing financial conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pandey, A.S., Sharma, E., Jain, N. et al. A rice bZIP transcription factor, OsbZIP16, regulates abiotic stress tolerance when over-expressed in Arabidopsis. J. Plant Biochem. Biotechnol. 27, 393–400 (2018). https://doi.org/10.1007/s13562-018-0448-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-018-0448-8