Abstract
Many microorganisms interact with plants but information is insufficient concerning requirements for plant colonization and if interactions become beneficial or detrimental. Pretreatment of oilseed rape (Brassica napus) with Bacillus results in disease suppression upon challenge with pathogens. We have studied transcriptome effects on oilseed rape primed with the Bacillus amyloliquefaciens 5113 biocontrol strain and compared that with effects of the fungal pathogen Botrytis cinerea. Using the cDNA-AFLP technique 21,700 transcript fragments were obtained of which 120 were differentially expressed and verified by northern blot analysis for selected transcripts. Priming with Bacillus caused greater effect on leaf than root transcripts where sequencing and BLAST analysis suggested many of the transcripts to be involved in metabolism and bioenergy. Bacillus and Botrytis treatment also changed metabolic gene expression in addition to signaling and transcription control genes as well as a potential disease resistance (TIR-NBS-LRR) gene. The pathogen provoked non-primed plant profile was less dominated by metabolism than Bacillus and Bacillus–Botrytis treated plants. Several transcripts were homologues to unknown genes in the different treatments. Altogether Bacillus treatment of roots cause a systemic gene expression in leaves suggested to result in a metabolic reprogramming as a major event during priming.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil is a complex habitat containing a multitude of microorganisms that can interact with plants. Symbiotic interactions include microorganisms spanning from pathogens to mutualists that colonize the plants as endophytes or epiphytes. Rhizobacteria colonizing the root system of plants is an example of intricate and important interactions in the rhizosphere (Denison and Kiers 2004). Colonization of plants by microorganisms is complex and seems to be species specific. The microcosm present in the rhizosphere not only differs because of soil conditions but also depends on earlier and present plants (Garbeva et al. 2004). Certain bacteria of the Bacillus, Pseudomonas and Serratia families (Lucy et al. 2004) can improve plant growth. The Plant Growth Promoting Rhizobacteria (PGPR) effect can be due to e.g. production of plant hormones or by increasing the amount of minerals and nitrogen available for the plant (Bloemberg and Lugtenberg 2001).
Microorganisms can also confer disease suppression to plants by different mechanisms. Bacteria living in vascular tissue of plants, in the rhizosphere or phyllosphere are in an appropriate position to protect the plant from deleterious organisms. Competition for growth space and nutrients by the beneficial bacteria can indirectly protect the plant from harmful microorganisms (Idriss et al. 2002). Bacteria can produce antibiotics towards different fungi and bacteria (Raaijmakers et al. 2002). Alteration of the plant cell wall by certain bacteria causes an increased protection to pathogens (Benhamou et al. 1996; Walker et al. 2004). Bacteria can make the plant more tolerant to pathogens by stimulating defense as systemic acquired resistance (SAR) (Bostock 2005). Streptomyces, Pseudomonas and Bacillus have been shown to induce another plant defense system called induced systemic resistance (ISR). ISR is effective against many pathogens and has been shown to be operational many weeks after induction. In contrast to SAR, this defense system depends on jasmonic acid (JA) and ethylene (van Loon et al. 1998). SAR is dependent on salicylic acid (SA) and involves the production of pathogenesis related (PR) proteins not found in ISR. These systems both converge at the downstream control gene NPR1 (Bostock 2005) although alternative pathways exist (Ryu et al. 2004b). Rhizobacteria seem to prepare the response to pathogens by priming the plant to respond more rapidly to a pathogen rather than activating a constitutive defense. Genes important for resistance are induced faster and/or stronger than in unprimed control plants when attacked by a pathogen. A recent study of plant resource allocation has shown that priming has a lower fitness cost than a constitutive active defense even when under pathogen pressure (van Hulten et al. 2006).
The genus Bacillus is characterized by rod shaped, facultative aerobe, endospore forming bacteria that live in soil and often colonize the plant rhizosphere. Several Bacillus spp. produce antibiotics and some Bacillus strains, mainly B. subtilis, B. cereus and B. amyloliquefaciens, are known to mediate protection against pathogens on plants (Kloepper et al. 2004) and several commercial preparations are available (Schisler et al. 2004). B. amyloliquefaciens UCMB-5113 is a red pigmented strain originally isolated from soil, which is able to colonize oilseed rape (Brassica napus) (Reva et al. 2004) and provide protection against the fungal phytopathogens Alternaria brassicae, Botrytis cinerea, Leptosphaeria maculans and Verticillium longisporum (Danielsson et al. 2007).
The aim of this study was to evaluate the effects of treatment of oilseed rape with Bacillus UCMB-5113 on the Brassica transcriptome to improve our understanding of how a biocontrol bacterium mediates disease suppression in B. napus to fungal pathogens such as Botrytis. This information will be used as a basis to address how plants allow co-existence with certain microorganisms without deploying a defense program as for pathogens and also the relationship between belowground and aboveground responses in defense priming. For transcript fingerprinting we used the cDNA-AFLP technique, which is sensitive, robust and has high resolution. We have earlier established this technique for B. napus to analyze effects of wounding (Sarosh and Meijer 2007).
Materials and methods
Plant material
Oilseed rape (B. napus, cv. Westar) seeds were surface sterilized for 20 min in 20% sodium hypochlorite followed by a brief rinse with 50% methanol before planting in sterile soil into 11 × 11 × 5 cm pots. The pots were grown in controlled environment using a 16/8 h photoperiod with light of 200 μmol m−2 s−2 at 22/18°C. The pots received a specific amount of water to exclude differences between treatments.
Bacteria
Bacillus UCMB-5113 was grown in LB media at 28°C with agitation for 3 days to allow for production of spores. The bacterial cultures were heat treated at 75°C for 10 min to select for Bacillus spores and to kill possible contaminants. After centrifugation, spores were washed in sterile water and the concentration was determined by viable count analysis on LB plates and the stock solution was kept refrigerated until use.
Pathogen
B. cinerea (strain 30158) was grown on PDA plates (16/8 h photoperiod at 21/16°C), for a month or until spores had been produced. The spores were harvested and filtered through miracloth. The concentration of spores was measured with a Bürkner chamber and adjusted to 107 spores ml−1. The spore solution was stored at 4°C until use.
Biocontrol bacterial treatment
Plants were left to dry for 2 days before application of Bacillus spore solution (107 spores ml−1) by adding 100 ml to each pot 7 days before infection so that only soil but no aboveground parts received any bacterial spores.
Pathogen inoculation and sampling
Plants that had a similar size (five true leaves) were chosen from each treatment. Ten μl of B. cinerea solution was drop inoculated on the first leaf of each plant. The inoculated leaf was monitored for local effects while the non-inoculated second leaf was used to analyse systemic effects. Plants were kept in mini greenhouses 12 h before and 12 h after inoculation to maintain high humidity and facilitate infection. Plants were scored at 72 h as (1) uninfected; (2) <1\2 of the leaves infected; (3) >1\2 of the leaves infected; or (4) dead plants. Roots or leaves (local and systemic harvested separately where relevant) from three plants were pooled together and collected at 0, 12, 24 h and 3 d after infection. Water treated non-primed plants with or without pathogen inoculation were collected at the same intervals.
RNA isolation and cDNA-AFLP analysis
Total RNA was isolated from frozen leaf tissue (Chomczynski and Sacchi 1987) and amount and purity assessed by absorbance at 260 nm or by the A260/A280 ratio using a Nanodrop spectrophotometer. cDNA-AFLP analysis was carried out on two biological replicates. cDNA was synthesized using the mRNA Capture Kit (Roche Applied Science, Germany). The subsequent cDNA-AFLP analysis was performed as described by Breyne et al. (2002). mRNA was converted to cDNA from 5 μg of total RNA using a biotinylated oligo-dT primer in streptavidin-coated PCR tubes. The purified cDNA templates were first digested with BstYI restriction enzyme (New England Biolabs, Beverly, MA, USA). Subsequently, the 3′-ends of the cDNAs were captured with streptavidin-coated PCR tubes. Digestion with the second enzyme MseI (New England Biolabs), released the transcript tags. Pre-amplification was performed by a MseI primer without a selective nucleotide combined with a BstYI primer containing either T or C at the 3′ end. The pre-amplification reaction was carried out using 20 cycles (94°C for 30 s; 56°C for 1 min; 72°C for 1 min). The amplified reaction was diluted 600-fold and 5 μl was used for final selective amplification using a touchdown amplification programme (Vos et al. (1995). BstT and MseI primers and BstC and MseI primers with one selective nucleotide, respectively, were used for the cDNA-AFLP analysis and all 16 primer combinations were performed. Selective [33P]-ATP labeled amplification products were separated on a 6% polyacrylamide gel run at constant power (100 W) until 4,300 Vh was reached. Gels were dried before scanning with a Phosphor Imager (Bio-Rad) and exposure to Kodak Biomax film for 3 d.
Isolation, quantification and sequencing of transcript derived fragments
Gel profiles were quantified using Quantity One software (Bio-Rad). Lane-based background subtraction was carried out and the bands were then normalized to compensate for differences in any loading effects in the different lanes. The intensity of each band was quantified using volume units (intensity units × mm2).
Bands were excised from the gel and boiled in water for 5 min. The DNA was precipitated and reamplified using the BstYI(T)-0, BstYI(C)-0 and MseI-0 primers. For PCR, the same reaction conditions as in the preamplification were used.
The reamplified products were cloned into pbluescript SK+ vector and sequenced using the M13 F/R primers. Sequencing of the transcript derived fragments (TDFs) was carried out at Macrogen Inc., Korea. Database searches were performed using the BLASTN and BLASTX programs (Altschul et al. 1997) at NCBI, EMBL and TAIR. Only the best hit is presented in Tables 1, 2, 3, 4. Sequence data from this article can be found in the GenBank/EMBL data bases under accession numbers GH70926263778115–GH70930963778162.
Use of the Genevestigator software revealed the response profiles of genes to different stimuli or genes that respond to selected factors (Zimmermann et al. 2005).
Northern blot analysis
Northern analysis was carried out on the same RNA preparation used in the cDNA-AFLP analysis and repeated with two biological replicates. Ten μg of total RNA was fractionated on a 1.2% denaturing formaldehyde agarose gel and transferred onto Hybond N+ membranes (Amersham, UK) as described (Sambrook et al. 1989). cDNA clones were isolated after agarose gel electrophoresis of restriction digested plasmids. The PR-1a and PDF1.2 genes were used as probes (Uknes et al. 1992; Penninckx et al. 1996). The probes were labeled with [α-32P]-dATP using Rediprime II Random Prime Labelling System (Amersham Biosciences, Sweden). Prehybridization and hybridization were performed in 50% formamide at 42°C (Sambrook et al. 1989). After hybridization, the membranes were washed at 42°C in 0.5 × SSC, 0.1% SDS. A Phosphor Imager (Bio-Rad) was used for imaging and quantification.
Results
Partial transcriptome analysis of Brassica napus primed with Bacillus sp.
Application of B. amyloliquefaciens strain UCMB-5113 to oilseed rape plants indeed conferred protection to Botrytis (Fig. 1). Phenotypic evidence for disease suppression provided the basis for molecular studies of the underlying processes. Plants were collected and scored after 3 days and the disease suppression was easily discerned. Use of UCMB 5113, as well as several other closely related strains (Danielsson et al. 2007), result in approximately 40% decrease of disease symptoms 1 week after inoculation with Botrytis. This reduction was observed even during optimal conditions for the fungus, i.e., enclosure in mini-greenhouses providing high humidity as well as a very high inoculation dose. A detailed transcript profiling of B. napus roots in response to priming by B. amyloliquefaciens strain UCMB-5113 was carried out using the cDNA-AFLP technique. Roots and leaves from three plants were collected at 0, 12, 24 h and 3 d after priming with UCMB-5113. Water treated control plants were collected at the same intervals. RNA was extracted from three biological replicates and cDNA-AFLP profiles were obtained using 16 primer combinations. To identify genes involved during priming, TDFs resulting for the four time intervals using 16 primer combinations were analyzed. Bands were scored based on presence/absence and intensity of bands and quantified. A representative cDNA-AFLP autoradiogram developed due to priming in roots is shown in Fig. 2. Approximately 3,000 fragments were quantified with band sizes ranging from approximately 50–550 bp. Based on the quantification data, 27 TDFs in roots were selected that showed a two to three-fold difference compared to the control. Of these, 22 were up-regulated after priming and the remaining five fragments were found to be down-regulated. In total, 21 TDFs from roots were successfully cloned and sequenced.
Effects of Bacillus inoculation on Botrytis disease on B. napus leaves. B. napus leaves were detached from five-leaf stage plants primed with B. amyloliquefaciens (A), water treated (B), primed plants 3 days after inoculation with B. cinerea (C), and water treated control plants challenge inoculated with B. cinerea (D)
cDNA-AFLP analysis of B. napus roots and leaves after Bacillus treatment and challenge inoculation with Botrytis. cDNA-AFLP fragment profiles displayed after Bacillus treatment of roots of 3-week-old plants and subsequent B. cinerea inoculation. The samples correspond to: Lanes 1–4, root samples prior to Botrytis inoculation. 1, 3, Bacillus treated; 2, 4, control (water treated); Lanes 5–10, leaf samples 0 h after treatment; 5, control; 6, Bacillus treated; 7, B. cinerea inoculated local leaves; 8, B. cinerea inoculated systemic leaves; 9, Bacillus + B. cinerea local leaves; 10, Bacillus + B. cinerea systemic leaves; Lanes 11–16, leaf samples 24 h after treatment; 11, control; 12, Bacillus treated; 13, B. cinerea inoculated local leaves; 14, B. cinerea inoculated systemic leaves; 15, Bacillus + B. cinerea local leaves; 16, Bacillus + B. cinerea systemic leaves; Lanes 17–22, leaf samples 72 h after treatment; 17, control; 18, Bacillus treated; 19, B. cinerea inoculated local leaves; 20, B. cinerea inoculated systemic leaves; 21, Bacillus + B. cinerea local leaves; 22, Bacillus + B. cinerea systemic leaves
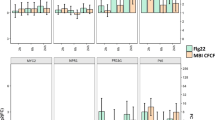
BLAST analysis of the sequenced TDFs resulted in identification of 16 homologues to known genes in the database and five TDFs corresponding to expressed proteins without any assigned function in Arabidopsis thaliana (Table 1). Of the 21 TDFs, seven were annotated to be involved in metabolism, two in signal transduction, two likely to be involved in energy generation and four transcription factors (Fig. 3A). A significant up-regulation of TDFs corresponding to metabolism and energy production was observed. Genevestigator program was used to correlate the role of Arabidopsis orthologs response during different stress challenges (Table 1). Genevestigator analysis of the BnBacR28B (expressed protein recently denoted as JAZ2), BnBacR20B (glycine-rich protein) and BnBacR13A (expressed protein) orthologs in Arabidopsis showed high induction by methyl jasmonate (MJ). Some TDFs were ethylene responsive like BnBacR08 (thiomethyltransferase), BnBacR6B (protein kinase) and BnBacR13A (expressed protein). Two TDFs, BnBacR15A (expressed protein) and BnBacR22A (glutamine synthetase) were induced by brassinosteroids.
Systemic gene expression in leaves after priming
Root treatment of B. napus with Bacillus strain UCMB-5113 resulted in a systemic gene expression in leaves. A representative cDNA-AFLP profile due to Bacillus treatment is shown in Fig. 2. Approximately 6,200 fragments were obtained using 16 primer combinations. In leaves, 41 TDFs corresponded to up-regulated genes and nine corresponded to down-regulated genes. In total, 35 TDFs were cloned and sequenced.
BLAST analysis using the TAIR tBLASTx tool on the differentially expressed TDFs indicated 31 TDFs with high identity to other known genes while unknown genes four TDFs were assigned to genes of expressed proteins without any assigned function in Arabidopsis (Table 2). The TDFs were annotated as: 12 predicted to be involved in metabolism, five in signal transduction, four in intracellular traffic, five in energy generation and four as transcription factors (Fig. 3B). The majority of the TDFs corresponded to genes involved in metabolism indicating a role in enhanced plant nutrition, growth and disease resistance. Differential expression of photosynthetic/chlorophyll genes was observed indicating that resources were being diverted to other metabolic pathways leading to plant nutrition and growth.
Based on the response viewer profiles of the Genevestigator, BnBacL16A (photosystem II), BnBacL04 (malate dehydrogenase), BnBacL23A (photosystem II), BnBacL27A (bZIP transcription factor), BnBacL13A (photosystem I), and BnBacL13B (leucine zipper transcription factor) orthologs were induced by brassinosteroids. Some TDFs were found to be elicited after MJ treatment like BnBacL5A (expressed protein) and BnBacL05 (multidrug resistant glycoprotein).
Differential gene expression in Bacillus treated and non-Bacillus primed plants challenged with the pathogen
B. amyloliquefaciens primed and non-primed plants were challenge inoculated with B. cinerea on the first leaves. Inoculated first leaves (local) and uninoculated second leaves (systemic) were harvested at 0, 12, 24 and 72 h post inoculation (hpi). RNA was extracted from the three replicates, each comprising three local and systemic leaves from three plants. Using 16 primer combinations the cDNA-AFLP analysis generated 12,500 fragments with an average of 50 fragments per lane (Fig. 2). The responses in local leaves were similar to those in systemic leaves. Quantification showed 24 differentially expressed TDFs in Bacillus treated-challenge inoculated plants. Of these, 19 genes were up-regulated and five genes down-regulated. Of the 10 TDFs identified in water treated-challenge inoculated plants, six were up-regulated and four down-regulated.
Sequence analysis of these TDFs revealed significant similarity to many known genes from GenBank (Table 3). A few unknown genes (expressed proteins) with unassigned function were also identified. Of the 19 TDFs with similarity to other known genes, eight were predicted to be involved in metabolism, four each in signal transduction and energy, one each in intracellular traffic and defense, and two likely to serve as transcription factors (Fig. 3C). Five TDFs (BnBacBc09, BnBacBc13A, BnBacBc25B, BnBacBc29B, BnBacBc6D) were annotated as ‘expressed proteins’ without any assigned function in Arabidopsis. BnBacBc15 is an TIR-NBS-LRR disease resistance protein ortholog.
The Genevestigator response profile, showed several of the TDFs to be elicited by brassinosteroids. These TDFs included BnBacBc04 (malate dehydrogenase), BnBacBc15 (disease resistance protein), BnBacBc17B (photosystem II), BnBacBc21B (chlorophyll A-B binding protein), BnBacBc19A (glycine rich protein) and BnBacBc6A (putative regulator of SNF1 related protein kinase). A few other TDFs were induced by MJ treatment—BnBacBc12 (WRKY transcription factor), BnBacBc25B (expressed protein), BnBacBc29B (expressed protein) and BnBacBc15A (bHLH protein). BnBacBc06 (Ulp1 protease) and BnBacBc14 (clathrin adaptor) were affected by SA.
Table 4 lists the 10 fragments differentially expressed in the non-primed plants inoculated with B. cinerea. The TDFs were assigned into functional categories; metabolism, signal transduction, energy, electron transport and intracellular traffic (Fig. 3D). Half of the TDFs were found to be sharing the metabolic and signal transduction functions. This is unlike the earlier trend, where a majority of the TDFs seemed to play a role in metabolism as observed in Bacillus treated or Bacillus treated-pathogen inoculated plants. Two TDFs (BnBc23A, BnBc21c) of unknown function were also found.
Northern blot analysis of expressed TDFs
To validate the differential expression of TDFs observed from the cDNA-AFLP analysis, selected clones were analyzed by northern blot analysis (Fig. 4). For Bacillus treated roots (Fig. 4A), BnBacR18A (expressed protein) showed strong induction in Bacillus treated roots in comparison with the water treated control at all time intervals tested. Transcript levels of BnBacR28B (expressed protein) were found to be up-regulated after 24 and 72 h of bacterial colonization in the roots. A MYB transcription factor (BnBacR3C) was observed to be strongly expressed at 12 h after priming compared to the control. Expression of a TDF (BnBacR4C) coding for an unknown protein was found to be highly up-regulated at later time point of 24 h after priming and maintained its course up to 72 h. Three TDFs were selected to confirm their systemic expression in leaves after Bacillus treatment (Fig. 4B). BnBacL19A (glycine rich protein), increased at 12 hpi and reached its highest expression at 72 hpi. BnBacL05 (multidrug resistant P-glycoprotein) showed a two-fold induction at 24 hpi compared to control. BnBacL6C (protein kinase) showed a four-fold increase at 12 hpi, indicating a role as an early player in priming. BnBacL9D (peroxidase) transcripts was found to be two-fold up-regulated in leaves at 12 h upon treatment and maintain upregulated to 72 hpi. TDF (BnBac5D) an unknown protein was observed to be down regulated at 24 h after priming compared to the control.
Northern blot analysis of TDFs expressed in B. napus tissues. Total RNA (10 μg) was separated on 1.2% formaldehyde agarose gels and stained with ethidium bromide to verify equal loading and transferred onto Hybond N+ membranes. A1 and A2 Root RNA after Bacillus priming was probed with the following TDFs; BnBacR18A (expressed protein) and BnBacR28B (expressed protein). Left panel shows control leaf samples, right panel shows UCMB-5113 treated root samples. B The blots containing B. napus leaf RNA after Bacillus priming in roots were probed with TDFs corresponding to; BnBacL19A (glycine-rich protein); BnBacL05 (Multidrug resistance P-glycoprotein) and BnBacL6B (expressed protein). The samples were water treated control (left) or UCMB-5113 primed (right) leaf samples. C The blots containing B. napus leaf RNA after Bacillus priming and challenge with B. cinerea were probed with TDFs corresponding to; BnBacBc09 (expressed protein); BnBacBc8B (endo-1,4-beta-glucanase) and BnBacBc25B (expressed protein). The samples were control leaves (left), challenged local leaves (middle) and systemic leaves (right). D The blots containing B. napus leaf RNA after challenge inoculation with B. cinerea were probed with TDFs corresponding to; BnBc23A (expressed protein); BnBc21C (expressed protein) and BnBc07 (Clathrin heavy chain). The samples were control leaves (left), challenged local leaves (middle), and challenged systemic leaves (right).
Northern blot analysis of TDFs from plants grown with or without Bacillus and challenged with Botrytis (Fig. 4C) showed increased levels in primed and challenged local and systemic leaves at 72 hpi for BnBacBc09 (expressed protein) and BnBacBc8B (glucanase). An augmented systemic expression was found with the BnBacBc09 indicating the systemic role of this expressed protein during priming and subsequent challenge inoculation. BnBacBc25B (expressed protein), was strongly up-regulated at 12–72 hpi in both local and systemic leaves but to a higher degree in local leaves.
Northern expression data (Fig. 4D) of the TDFs isolated after challenge inoculation with B. cinerea indicated that BnBc23A (expressed protein) was up-regulated in both local and systemic leaves but higher expressed in systemic leaves at 12–24 hpi. Expression analysis of the TDF BnBc21C (expressed protein) showed an augmented expression pattern in primed and challenged local leaves at 12–24 h after infection but a lower expression after 72 h compared with the control. A similar pattern of gene expression was observed in the systemic leaves but with a reduced amount compared to the local leaves. TDF BnBc07 (Clathrin), was more strongly expressed in local leaves after infection compared to both control and systemic leaves.
Bacillus priming seems to involve ISR
Gene expression of the PR-1a gene was investigated by northern analysis in leaves of Bacillus primed and Botrytis challenge inoculated plants (Fig. 5). The mRNA level of PR-1a was found to be very low or negligible in the UCMB-5113 primed plants but a low induction was observed in the UCMB-5113 primed and challenged plants. A high accumulation of PR-1a ortholog transcripts was observed in the B. cinerea challenged leaves, indicating that PR-1a is not playing a role during B. amyloliquefaciens mediated priming in oilseed rape. PDF1.2 mRNA was found to be elicited 2-fold in the 24 h old Bacillus primed leaves compared with the Bacillus primed and pathogen inoculated plants but with no detectable expression in the pathogen inoculated plants (Fig. 5). The lack of SA effect but presence of JA effect as a result of Bacillus treatment on B. napus suggests that ISR to Botrytis is occurring as a result of Bacillus priming.
Northern blot analysis using Arabidopsis probes on B. napus leaf RNA. Leaf samples were taken from B. napus plants pretreated with Bacillus and 2 days after challenge with Botrytis. The samples correspond to: C, control non-treated plants; B, Bacillus primed plants; BP, Bacillus primed and Botrytis challenged leaves; P, Botrytis challenged leaves. The blots were probed with Arabidopsis PR1a and PDF1.2 cDNA while actin served as a RNA control
Discussion
Priming of defense has been considered to be an efficient strategy to induce resistance in plants against a variety of pathogens (Beckers and Conrath 2007). Immunization of Arabidopsis by certain bacteria has been demonstrated to enhance defense capacity against a broad range of pathogens (Pieterse et al. 2002; Ahn et al. 2007). Bacteria mediated ISR is not associated with induced expression of PR genes in contrast to pathogen induced SAR (Pieterse et al. 1996; Verhagen et al. 2004). Literature till date on the molecular and physiological mechanisms of ISR has been concentrated on the Pseudomonads. The main objective of this study was to identify plant transcripts affected by beneficial Bacillus bacteria and pathogens for future studies to elucidate their role in plant–microbe interactions. Hence, the present study provides impetus on the mechanistic role of Bacillus primed defense in Brassicas. In order to identify genes associated with colonization, potential priming of ISR and concomitant disease suppression, we applied the cDNA-AFLP technique to analyze the transcript profile of roots and leaves of B. napus plants undergoing Bacillus UCMB 5113 mediated defense priming as visualized by the healthy phenotype observed after Botrytis challenge. cDNA-AFLP analysis of the different samples generated approximately 21,700 fragments, of which about 120 corresponded to differentially expressed genes. Several of these fragments in different samples probably represent the same gene so the number of unique genes is likely to be lower. In total, 21 TDFs from roots and 35 TDFs from leaves of Bacillus treated plants were cloned. Intriguingly, fewer transcripts were observed in roots compared to leaves of Bacillus treated plants showing a strong systemic effect in priming. Localized signals may be fewer in whole root, not detected because the changes in gene expression occurred prior to the time of sampling or that many genes just are slightly upregulated. Hence these transcripts were not amplified with the primers used or may rely more on post-translational modifications. We observed increased root biomass in the primed plants compared with the control indicating changed root morphology and physiology during Bacillus colonization. Root changes increase the capacity of the plants to absorb nutrients and enable compatible interactions in the rhizosphere (Lopez-Bucio et al. 2003). Rhizobacteria stimulate exudation of organic carbon by roots (Meharg and Killham 1995) suggesting enhanced photosynthetic rates to accommodate the sink in carbon source. Indeed changes in the expression of photosynthesis associated genes like photosystem I reaction center subunit II (BnBacL13A), PSBO2 photosystem II (BnBacL23A) and PSB01 photosystem II (BnBacL16A) were observed in the Bacillus primed plants.
ISR was originally described as the resistance conferred to Arabidopsis by the non-pathogenic root bacterium Pseudomonas fluorescens (Pieterse et al. 1996). In our experiments, the effectiveness of priming ISR like defense of oilseed rape by the Bacillus UCMB 5113 strain was tested towards B. cinerea. Under similar conditions Bacillus priming is also effective against A. brassicae, L. maculans and V. longisporum (Danielsson et al. 2007). Our studies indicated that PR-1a, dependent on SA, was not induced by Bacillus priming, while the elicitation of JA-dependent PDF1.2 indicated that B. amyloliquefaciens indeed mediated ISR in oilseed rape. Both JA and ethylene are needed for Bacillus UCMB5113 primed ISR in Arabidopsis to Pseudomonas syringae as deduced from mutant studies (Danielsson and Meijer, unpublished). Medium potent antibiotic substances in Bacillus extracts retard Botrytis growth in vitro (Danielsson et al. 2007). While such a direct effect can contribute to disease suppression, it seems more likely that the major effect is mediated through the plant as ISR considering the lower Bacillus density and spatial separation from Botrytis in a more natural plant soil system. ISR has been observed in other species e.g. tomato (Yan et al. 2002) and tobacco (Zhang et al. 2002) suggesting this to be a common phenomenon in plants. Certain bacteria also can provide protection to different plants and pathogens. For example Pseudomonas putida LSW17S protects tomato to Fusarium oxysporum but also prime and induce cellular and molecular defense mechanisms in Arabidopsis against P. syringae DC3000 (Ahn et al. 2007).
To investigate if ISR is associated with transcriptional changes only apparent after pathogen attack, we analyzed the expression profile in local and systemic leaves of primed plants upon challenge inoculation with B. cinerea vs. the primed non-challenged leaves. Of the 12,500 TDFs expressed, 30 TDFs were found to show an augmented change in ISR expressing leaves and 24 TDFs were sequenced. Blast analysis showed that genes involved in protection against pathogens and oxidative stress were activated systemically in leaves of colonized plants. Among the primed and pathogen responsive genes, the majority of the genes were predicted to be influenced by JA or ethylene, indicating that both signals play an important role.
Based on the Genevestigator profiles, BnBacR28B, homologous to an expressed protein (At1g74950) in Arabidopsis, showed an upward trend by MJ and wounding. This expressed protein has recently been designated as JAZ2 belonging to a family of Jasmonate Zim-Domain (JAZ) genes based on the JIM domains (Chinni et al. 2007; Thines et al. 2007). JAZ proteins have been shown to play an essential role in plant defense against insect herbivores (Chung et al. 2008). Hence, our studies indicate that Bacillus priming of the plants by root treatment might trigger these JAZ genes and in turn prepare the plant by eliciting the primary defense genes suggesting increased protein turnover to be an important early event in priming. Genvestigator analysis showed several of the genes that had differential expression in Bacillus treated plants to be induced also by brassinosteroids and other hormones triggering plant growth. Accordingly Bacillus colonization may affect metabolism of hormones that promote growth e.g. of root tissue. Other factors reported to mediate bacterial primed biocontrol are volatile organic compounds (Ryu et al. 2004a; Han et al. 2006). However, experiments conducted to study effects of volatiles from the Bacillus 5113 strain on Arabidopsis showed no protection to P. syringae suggesting volatiles not to be an important factor at least in that interaction (Danielsson and Meijer, unpublished). The role of plant candidate genes to study for their role in priming of ISR includes induced TDFs coding for methyl transferase (BnBacBc03A), F-box family protein (BnBacBc10), WRKY transcription factor (BnBacBc12), disease resistance protein (BnBacBc15), glycine-rich protein (BnBacBc19A), endo-1,4-β-glucanase (BnBacBc8B), MYC2 transcription factor (BnBacL11D and BnBacBc15A) and putative regulators of SNF1-protein kinase (BnBacBc6A). Recently it has been reported that qPCR analysis of the MYC2 transcript levels were upregulated in WCS417r-ISR expressing Arabidopsis plants. Functional analysis of the MYC2 impaired mutants jin1-1 and jin1-2 failed to develop ISR against P. syringae DC3000 or Hyaloperonospora parasitica (Pozo et al. 2008). Plant SNF1-related kinases are known to regulate the activity of rate limiting metabolic enzymes as well as the transcription of glucose and stress-regulated genes (Bhalerao et al. 1999). Studies of this regulator in primed plants could unravel the role in regulation of metabolism for growth or defense after bacterial priming. Very little is known about signals leading to ISR but mitogen-activated protein kinase3 has been proposed as a candidate for priming mediated signaling in Arabidopsis (Beckers and Conrath 2007). This study found several kinases to be affected by Bacillus treatment, which enable analysis of their specific role in priming. In our study we also observed an early up-regulated expression of a MYB transcription (BnBacR28B) in the roots upon colonization with the Bacillus. A recent study has shown that MYB72 is responsible for early induction of ISR in Arabidopsis roots upon treatment with P. putida WCS417r (van der Ent et al. 2008).
Another question is how Bacillus colonization is enabled without provoking a defense response by the innate immunity surveillance system operating in plants (Newman et al. 2007). If beneficial bacteria are recognised by pathogen- or microbe-associated microbial pattern receptors in the plant the bacteria must be able to suppress downstream signaling that otherwise would elicit negative defense factors. Recognition of lipopolysaccharides (LPS) and flagella proteins can trigger plant defense or elicit ISR when challenge inoculated with pathogens (Newman et al. 2007). LPS can suppress the hypersensitive response and programmed cell death associated with the defense responses induced by avirulent bacteria (Newman et al. 2007). In our study we identified a few genes as constituents in LPS mediated signaling during bacterial priming on Brassica. In addition, some of the genes of unknown function being up-regulated during priming may be involved in colonisation and suppression of defense. Further characterisation of these unknown genes would provide information regarding factors involved in priming. Bacterial components that could be involved in elicitation of ISR are e.g. lipopeptides (Ongena et al. 2007). The role and identity of bacterial factors in the specific interaction studied here remains to be elucidated.
This study showed that Bacillus colonization of oilseed rape roots cause a genetic reprogramming of plant cells both in local (root) and distal (leaf) tissues. Many of the genes affected seem to be involved in metabolism, energy generation and regulation. Other investigations also report that primed Arabidopsis plants underwent a transcriptional reprogramming that changed e.g. metabolic processes. Cartieaux et al. (2003) reported that Arabidopsis defense and carbon metabolism was affected with reduced carbon fixation after priming by Pseudomonas thivervalensis. They also observed small effects on root transcripts although overall morphological effects on roots were obvious. Another study (Verhagen et al. 2004) that analysed Arabidopsis primed by P. fluroescens WCS417r found a larger change in root transcripts but little effect on leaf transcripts. Many of these genes are controlled by JA or ethylene. Wang et al. (2005) primed Arabidopsis with P. fluorescens FPT9601-T5 and found shoot tissues to respond by differences mostly in genes connected with metabolism followed by transcription and communication. A recent investigation using Arabidopsis primed with Bradyrhizobium found effects on many genes in leaves regulated by JA or ethylene (Cartieaux et al. 2008). Most of the genes were down-regulated and belonged to several functional categories including metabolism and regulation to be common. It is clear that priming is not a universal response but seems to depend on the interaction studied. Obviously the resource allocation choice made by primed plants is a delicate balance to assure fitness (Bostock 2005; Heil 2001). In the Bacillus treated oilseed rape plants, genes related to metabolism were found to represent at least 40% of the genes isolated. The dramatic reduction in the proportion of the genes linked to metabolism and a subsequent increase of the disease resistance genes clearly illustrates a mechanism in which the plant recognizes the onset of pathogen attack and deviate the resources towards defense. These changes most likely alter metabolism and affect source–sink relationships and resource allocation in the plant and somehow prime defense as illustrated by disease suppression towards Botrytis. Metabolic re-programming during defense in Arabidopsis occurs following compatible and incompatible interactions (Scheidler et al. 2002). The precise mechanism of colonization and priming by beneficial bacteria is unclear but elucidating the role of the novel TDFs identified in this study may provide explanations for the molecular repertoire behind successful long term colonization and protection. This study also showed a clear difference in plant transcript profiles when exposed to beneficial vs. pathogenic microorganisms as a basis for studies of plant–microbe interactions and plant ability to differentiate among microorganisms and allow or counteract colonization. Future analysis of plant and bacterial factors can thus provide information about the role in colonization, priming and elicitation of ISR as well as the resource strategy of a primed plant.
Abbreviations
- hpi:
-
Hours post inoculation
- ISR:
-
Induced systemic resistance
- JA:
-
Jasmonic acid
- LPS:
-
Lipopolysaccharides
- PGPR:
-
Plant Growth Promoting Rhizobacteria
- SA:
-
Salicylic acid
- SAR:
-
Systemic acquired resistance
- TDF:
-
Transcript derived fragment
References
Ahn P, Lee SW, Suh SC (2007) Rhizobacteria-induced priming in Arabidopsis is dependent on ethylene, jasmonic acid and NPR1. Mol Plant Microbe Interact 20:759–768. doi:10.1094/MPMI-20-7-0759
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Beckers GJM, Conrath U (2007) Priming for stress resistance: from the lab to the field. Curr Opin Plant Biol 10:424–431. doi:10.1016/j.pbi.2007.06.002
Benhamou N, Kloepper JW, Quadt-Hallman A, Tuzun S (1996) Induction of defense-related ultrastructural modifications in pea root tissues inoculated with endophytic bacteria. Plant Physiol 112:919–929
Bhalerao RP, Salchert K, Bako L, Ökresz L, Szabados L, Murakana T, Machida Y, Schell J, Koncz C (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96:5322–5327. doi:10.1073/pnas.96.9.5322
Bloemberg GV, Lugtenberg BJJ (2001) Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol 4:343–350. doi:10.1016/S1369-5266(00)00183-7
Bostock RM (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43:545–580. doi:10.1146/annurev.phyto.41.052002.095505
Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, Rombauts S, Raes J, Cannoot B, Engler G, Inze D, Zabeau M (2002) Transcriptome analysis during cell division in plants. Proc Natl Acad Sci USA 99:14825–14830. doi:10.1073/pnas.222561199
Cartieaux F, Thibaud M-C, Zimmerli L, Lessard P, Sarrobert C, David P, Gerbaud A, Robaglia C, Somerville S, Nussaume L (2003) Transcriptome analysis of Arabidopsis colonized by a plant growth promoting rhizobacterium reveals a general effect on disease resistance. Plant J 36:177–188. doi:10.1046/j.1365-313X.2003.01867.x
Cartieaux F, Contesto C, Gallou A, Desbrosses G, Kopka J, Taconnat L, Renou JP, Touraine B (2008) Simultaneous interaction of Arabidopsis thaliana with Bradyrhizobium Sp. strain ORS278 and Pseudomonas syringae pv. tomato DC3000 leads to complex transcriptome changes. Mol Plant Microbe Interact 21:244–259. doi:10.1094/MPMI-21-2-0244
Chinni A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448:666–673. doi:10.1038/nature06006
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. doi:10.1016/0003-2697(87)90021-2
Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis Jasmonate ZIM-Domain genes in response to wounding and herbivory. Proc Natl Acad Sci USA 146:952–964
Danielsson J, Reva O, Meijer J (2007) Protection of oilseed rape (Brassica napus) toward fungal pathogens by strains of plant-associated Bacillus amyloliquefaciens. Microb Ecol 54:134–140. doi:10.1007/s00248-006-9181-2
Denison RF, Kiers ET (2004) Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol Lett 237:187–193. doi:10.1111/j.1574-6968.2004.tb09695.x
Garbeva P, van Veen JA, van Elsas JD (2004) Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42:243–270. doi:10.1146/annurev.phyto.42.012604.135455
Han SH, Lee SJ, Moon JH, Park KH, Yang KY, Cho BH, Kim KY, Kim YW, Lee MC, Anderson AJ, Kim YC (2006) GacS-Dependent production of 2R, 3R-butanediol by Pseudomonas chlororaphis O6 is a major determinant for eliciting systemic resistance against Erwinia carotovora but not against Pseudomonas syringae pv. tabaci in tobacco. Mol Plant Microbe Interact 19:924–930. doi:10.1094/MPMI-19-0924
Heil M (2001) The ecological concept of costs of induced systemic resistance (ISR). Eur J Plant Pathol 107:137–146. doi:10.1023/A:1008793009517
Idriss EE, Makarewicz O, Farouk A, Rosner K, Greiner R, Bochow H, Richter T, Borriss R (2002) Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 148:2097–2109
Kloepper JW, Ryu C-M, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266. doi:10.1094/PHYTO.2004.94.11.1259
Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6:280–287. doi:10.1016/S1369-5266(03)00035-9
Lucy M, Reed E, Glick BR (2004) Applications of free living plant growth-promoting rhizobacteria. Antonie Van Leeuwenhoek 86:1–25. doi:10.1023/B:ANTO.0000024903.10757.6e
Meharg AA, Killham K (1995) Loss of exudates from the roots of perennial ryegrass inoculated with a range of micro-organisms. Plant Soil 170:345–349. doi:10.1007/BF00010488
Newman M-A, Dow MJ, Molinaro A, Parrilli M (2007) Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J Endotoxin Res 13:69–84. doi:10.1177/0968051907079399
Ongena M, Jourdan E, Adam A, Paquot M, Brans A, Joris B, Arpigny JL, Thonart P (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol 9:1084–1090. doi:10.1111/j.1462-2920.2006.01202.x
Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Bucahala A, Metraux JP, Manners JM, Broekaert WF (1996) Pathogen induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid independent pathway. Plant Cell 8:2309–2323
Pieterse CMJ, Van Wees SCM, Hoffland E, Van Pelt JA, Van Loon LC (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8:1225–1237
Pieterse CMJ, Van Wees SCM, Ton J, Van Pelt JA, Van Loon LC (2002) Signaling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol 4:535–544. doi:10.1055/s-2002-35441
Pozo MJ, van der Ent S, van Loon LC, Pieterse CMJ (2008) Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180:511–523. doi:10.1111/j.1469-8137.2008.02578.x
Raaijmakers J, Vlami M, de Souza J (2002) Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81:537–547. doi:10.1023/A:1020501420831
Reva ON, Dixelius C, Meijer J, Priest FG (2004) Taxonomic characterization and plant colonizing abilities of some bacteria related to Bacillus amyloliquefaciens and Bacillus subtilis. FEMS Microbiol Ecol 48:249–259. doi:10.1016/j.femsec.2004.02.003
Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW (2004a) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026. doi:10.1104/pp.103.026583
Ryu C-M, Murphy JF, Mysore KS, Kloepper JW (2004b) Plant growth-promoting rhizobacteria systemically protect Arabidopsis thaliana against Cucumber mosaic virus by a salicylic acid and NPR1-independent and jasmonic acid-dependent signaling pathway. Plant J 39:381–392. doi:10.1111/j.1365-313X.2004.02142.x
Sambrook JE, Fristsch F, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sarosh BR, Meijer J (2007) Transcriptional profiling by cDNA-AFLP reveals novel insights during methyl jasmonate, wounding and insect attack in Brassica napus. Plant Mol Biol 64:425–438. doi:10.1007/s11103-007-9164-9
Scheidler M, Schlaich NL, Felleberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hohesel JD (2002) Monitoring the switch from housekeeping to pathogen defence metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem 277:10555–10561. doi:10.1074/jbc.M104863200
Schisler DA, Slininger PJ, Behle RW, Jackson MA (2004) Formulation of Bacillus spp. for biological control of plant diseases. Phytopathology 94:1267–1271. doi:10.1094/PHYTO.2004.94.11.1267
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signaling. Nature 448:661–666. doi:10.1038/nature05960
Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4:645–656
van der Ent S, Verhagen WMS, van Doorn R, Bakker D, Verlaan GM, Pel MJ, Joosten RG, Proveniers MC, van Loon LC, Ton J, Pieterse CM (2008) MYB72 is required in early signaling steps of Rhizobacteria-Induced systemic resistance in Arabidopsis. Plant Physiol 146:1293–1304. doi:10.1104/pp.107.113829
van Hulten M, Pelser M, van Loon LC, Pieterse CM, Ton J (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA 103:5602–5607. doi:10.1073/pnas.0510213103
van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483. doi:10.1146/annurev.phyto.36.1.453
Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, Van Loon LC, Pieterse CMK (2004) The transcriptome of rhizobacteria induced systemic resistance in Arabidopsis. Mol Plant Microbe Interact 17:895–908. doi:10.1094/MPMI.2004.17.8.895
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuper M, Zabeau M (1995) AFLP—a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414. doi:10.1093/nar/23.21.4407
Walker TS, Bais HP, Deziel E, Schweizer HP, Rahme LG, Fall R, Vivanco JM (2004) Pseudomonas aeruginosa-plant root interactions. pathogenicity, biofilm formation, and root exudation. Plant Physiol 134:320–331. doi:10.1104/pp.103.027888
Wang Y, Ohara Y, Nakayashiki H, Tosa Y, Mayama S (2005) Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol Plant Microbe Interact 18:385–396. doi:10.1094/MPMI-18-0385
Yan Z, Reddy MS, Ryu C-M, McInroy JA, Wilson M, Kloepper JW (2002) Induced systemic resistance against tomato late blight elicited by plant growth-promoting rhizobacteria. Phytopathology 92:1329–1333. doi:10.1094/PHYTO.2002.92.12.1329
Zhang S, Reddy MS, Kloepper JW (2002) Development of assays for assessing induced systemic resistance by plant growth-promoting rhizobacteria against blue mold of tobacco. Biol Control 23:79–86. doi:10.1006/bcon.2001.0992
Zimmermann P, Hennig L, Gruissem W (2005) Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci 10:407–409. doi:10.1016/j.tplants.2005.07.003
Acknowledgements
This work was supported by grants from the IMOP programme SLU, Carl Tryggers fund, FORMAS, Nilsson-Ehle fund, Helge Ax:son Johnson fund, and Persson fund. We are grateful to Johanna Lagensjö and Namita Wadke for assistance in TDF cloning.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sarosh, B.R., Danielsson, J. & Meijer, J. Transcript profiling of oilseed rape (Brassica napus) primed for biocontrol differentiate genes involved in microbial interactions with beneficial Bacillus amyloliquefaciens from pathogenic Botrytis cinerea . Plant Mol Biol 70, 31–45 (2009). https://doi.org/10.1007/s11103-009-9455-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9455-4