Abstract
RING finger proteins comprise a large family and play key roles in regulating growth/developmental processes, hormone signaling and responses to biotic and abiotic stresses in plants. A rice gene, OsBIRF1, encoding a putative RING-H2 finger protein, was cloned and identified. OsBIRF1 encodes a 396 amino acid protein belonging to the ATL family characterized by a conserved RING-H2 finger domain (C-X2-C-X15-C-X1-H-X2-H-X2-C-X10-C-X2-C), a transmembrane domain at the N-terminal, a basic amino acid rich region and a characteristic GLD region. Expression of OsBIRF1 was up-regulated in rice seedlings after treatment with benzothaidiazole, salicylic acid, l-aminocyclopropane-1-carboxylic acid and jasmonic acid, and was induced differentially in incompatible but not compatible interactions between rice and Magnaporthe grisea, the causal agent of blast disease. Transgenic tobacco plants that constitutively express OsBIRF1 exhibit enhanced disease resistance against tobacco mosaic virus and Pseudomonas syringae pv. tabaci and elevated expression levels of defense-related genes, e.g. PR-1, PR-2, PR-3 and PR-5. The OsBIRF1-overexpressing transgenic tobacco plants show increased oxidative stress tolerance to exogenous treatment with methyl viologen and H2O2, and up-regulate expression of oxidative stress-related genes. Reduced ABA sensitivity in root elongation and increased drought tolerance in seed germination were also observed in OsBIRF1 transgenic tobacco plants. Furthermore, the transgenic tobacco plants show longer roots and higher plant heights as compared with the wild-type plants, suggesting that overexpression of OsBIRF1 promote plant growth. These results demonstrate that OsBIRF1 has pleiotropic effects on growth and defense response against multiple abiotic and biotic stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants defend themselves against potential invading pathogens by activating a battery of defense mechanisms, in which thousands of defense-related genes are coordinately expressed. Extensive biochemical, genetic and genomic studies in Arabidopsis have revealed that different signaling pathways, e.g. salicylic acid (SA)-dependent and jasmonic acid (JA)/ethylene (ET)-dependent signaling pathways, are responsible to activate disease resistance responses by coordinately regulating expression of defense-related genes. Generally, the level and speed of de novo biosynthesis and accumulation of novel proteins (e.g. pathogenesis-related proteins, transcriptional factors, and other regulatory proteins) after pathogen infection are correlated to the degree and strength of the defense responses. In fact, most of the studies that have been made so far take advantage of an approach based on identification of novel proteins resulted from induction of gene expression during defense responses in order to elucidate the molecular basis of plant disease resistance. On the other hand, recent studies have also shed a new window that degradation of proteins is one of most important biochemical and physiological events that play critical roles in regulating almost all aspects of cellular processes.
The ubiquitin/26S proteasome system is a primary pathway for degradation of cellular proteins among eukaryotes (Glickman and Ciechanover 2002). Biochemical process of this system basically starts with ubiquitination of substrate proteins, which are then targeted for degradation by 26S proteasome (Hershko and Ciechanover 1998; Smalle and Vierstra 2004). Ubiquitination of proteins requires a cascade of enzymatic reactions involving three enzymes including ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin protein ligase E3 (Glickman and Ciechanover 2002). Among them, E3s interact with specific substrates and thereby determine specificity to the degradation process. Generally, E3s are grouped into four classes according to the characteristic domains: RING zinc finger, PHD finger, U-Box and HECT (Fang and Weissman 2004; Moon et al. 2004; Smalle and Vierstra 2004). The RING finger domain is defined by the consensus sequence CX2CX (9-39)CX(1-3)HX(2-3)C/HX2CX(4-48) CX2C, where X is any amino acid and the number of X residues varies in different fingers, and is further subcategorized into RING-HC and RING-H2 based on the presence of cysteine and histidine at the fifth coordination site, respectively (Saurin et al. 1996; Freemont 2000; Joazeiro and Weissman 2000).
RING finger proteins comprise a large family, which ubiquitously exist in all of eukaryotes including yeast, animals and plants. Database searches have identified 469 RING-finger proteins from the Arabidopsis genome and 218 cDNA clones in the rice full-length cDNA database (Stone et al. 2005; Katoh et al. 2005). Large number of the RING-finger proteins and their nature in determining specificity of the degradation process imply the diversity of their biological functions. The involvement and functions of RING finger proteins in plant growth/development and hormone responses have been well established recently. Genetic and biochemical studies have identified a number of RING finger proteins that play key roles in regulating growth and developmental processes. These include Arabidopsis COP1 and CIP8 (COP1-interacting protein 8) in photomorphogenesis, BIG BROTHER in organ size, RIE1 in seed development, SHA1 in shoot apical meristem maintenance, PEX10 in peroxisome biogenesis, and Arabidopsis XBAT32 and rice EL5 in root development (Hardtke et al. 2000, 2002; Osterlund et al. 2000; Ma et al. 2002; Lechner et al. 2002; Holm et al. 2002; Saijo et al. 2003; Xu and Li 2003; Seo et al. 2003, 2004; Duek et al. 2004; Nodzon et al. 2004; Jang et al. 2005; Yang et al. 2005; Disch et al. 2006; Koiwai et al. 2007; Sonoda et al. 2007; Schumann et al. 2007). There are many examples demonstrating that some RING finger proteins are directly involved in regulation of hormone signaling pathways by targeting of specific proteins for degradation. Arabidopsis SINAT5 attenuates the auxin signaling by targeting NAC1 for degradation (Xie et al. 2002). AIP2 negatively regulates ABA signaling by modulating its substrate ABI3 stability (Zhang et al. 2005), while SDIR1 positively regulates ABA signaling (Zhang et al. 2007). The Arabidopsis BRH1 gene was rapidly down-regulated by brassinolide in wild-type plants but not in a BR-insensitive mutant plants, indicating a BR-dependent manner for its expression (Molnar et al. 2002). Furthermore, RING finger proteins also participate in regulation of symbiotic nodulation in Lotus (Nishimura et al. 2002; Shimomura et al. 2006).
In addition, recent studies have also showed that a number of RING finger proteins play important roles in regulating defense responses against abiotic and biotic stresses. The Arabidopsis HOS1, a negative regulator of cold signal transduction, physically interacts with ICE1, a transcription factor activating the expression of CBF genes, and mediates the ubiquitination of ICE1 (Lee et al. 2001; Dong et al. 2006). Cold stress induces the degradation of ICE1 in plants, and this degradation requires HOS1, indicating that HOS1 is a negative regulator of cold responses by mediating the degradation of the ICE1 protein (Dong et al. 2006). SDIR1 and XERICO have been shown to be positive regulators of drought tolerance through altering the homeostasis of ABA signaling pathway (Ko et al. 2006; Zhang et al. 2007). RING finger proteins are also involved in plant defense response. Arabidopsis ATL2 is induced rapidly and transiently by chitin and cellulase treatments (Salinas-Mondragon et al. 1999) and the eca mutants with constitutive expression of ATL2 gene exhibited up-regulated expression of defense-related genes and SA- and JA-responsive genes (Serrano and Guzman 2004). A T-DNA insertion mutant of Arabidopsis ATL9 results in increased susceptibility to powdery mildew disease (Ramonell et al. 2005). Arabidopsis RIN2 and RIN3 and tobacco ACRE132 are involved in disease resistance gene-specific hypersensitive response (Kawasaki et al. 2005; Durrant et al. 2000). Furthermore, ectopic expression of genes encoding RING finger proteins in Arabidopsis alters disease resistance phenotype and expression of defense-related genes (Cheung et al. 2007; Hong et al. 2007).
Despite of large number of RING finger proteins in rice, only few of them has been identified so far for their possible functions (Katoh et al. 2005). The EL5 gene, encoding a RING finger E3 ligase, is transiently induced by N-acetylchitoheptaose elicitor in rice cells, and functional analysis suggests that EL5 is involved in root development through the maintenance of cell viability (Takai et al. 2001, 2002; Katoh et al. 2003; Koiwai et al. 2007). OsCOIN is strongly induced by some abiotic stress factors including low temperature, salt and drought and overexpression in transgenic rice significantly enhanced their tolerance to cold, salt and drought stresses (Liu et al. 2007). OsRING-1 and OsRHC1 are induced by pathogen infection and defense-related signal molecules and overexpression of OsRHC1 in Arabidopsis results in enhanced disease resistance (Meng et al. 2006; Cheung et al. 2007). Here we report the cloning and characterization of a novel rice RING-H2 finger protein gene, OsBIRF1. Expression of OsBIRF1 was induced by defense-related signal molecules and during an incompatible interaction between rice and Magnaporthe grisea. Ectopic expression of OsBIRF1 in transgenic tobacco plants results in enhanced disease resistance against viral and bacterial pathogens and constitutive expression of defense-related genes. Moreover, OsBIRF1 transgenic tobacco plants also showed enhanced tolerance to drought and oxidative stress. Our results indicate that OsBIRF1 has pleiotropic effects on growth and defense response against multiple abiotic and biotic stresses.
Materials and methods
Plant growth and treatments
Rice cv. Yuanfengzao (Oryza sativa L. subsp. indica) and a pair of isogenic lines (H8R and H8S) were used in this study. Seedlings were grown in a growth room under a 14 h/10 h day/night regime at 22–27°C for 3 weeks. Seedlings of cv. Yuanfengzao were treated by foliar spraying with solutions of 0.3 mM benzothidiazole (BTH, Novartis Crop Protection Inc., USA), 1.5 mM salicylic acid (SA, pH 6.5) (Sigma-Aldrich, St. Lousi, USA), 100 μM jasmonic acid (JA) (Sigma-Aldrich, St. Lousi, USA) or 100 μM 1-amino cyclopropane-1-carboxylic acid (ACC) (Sigma-Aldrich, St. Lousi, USA), respectively. JA and ACC were dissolved in 0.1% ethanol. The controls were treated in the same way as spraying with 0.1% ethanol or distilled sterilized water. Inoculation of seedlings of H8R and H8S with Magnaporthe grisea (strain 85-14B1, race ZB1) was carried out as described previously (Luo et al. 2005a). Leaf samples were collected at different time points as indicated and stored at −80°C until use.
Tobacco plants (Nicotiana tabacum) were cultivated in a growth room under a 14 h/10 h day/night regime at 20–25°C.
Cloning of the OsBIRF1 cDNA
A differentially expressed cDNA clone, BIHN-w1, was obtained previously from a suppression subtractive hybridization library (Song and Goodman 2002). BIHN-w1 appears to be a part of a gene encoding a putative RING finger protein, but lacks most part of sequence at 5′-end. To obtain the full-length cDNA, BIHN-w1-specific primers BIHN-w1-1R (5′-ATCTCCAGTAGCATCTCTCT-3′) and BIHN-w1-2R (5′-CAG AGC TTA TGC CAG TAG CT-3′) and a vector primer T3-2 (5′-CCT GCA GGTCGACACTAGTG-3′) were used to amplify the 5′-end sequence using phage DNA prepared from a rice cDNA library as template. Amplified PCR products were purified using the DNA Gel Purification Kit (Sangon, Shanghai, China) and cloned into pUCm-T-vector (Sangon, Shanghai, China) by T/A cloning. A pair of gene-specific primers, BIHN-w1-orf-1F (5′-TCTCCTCCCAATCCGAGATTGC-3′) and BIHN-w1-orf-1R (5′-TAATCCTGAACAATCACAAGCA-3′), located in 5′- and 3′-UTRs, were designed and used to amplify the full-length cDNA. Plasmid containing the full-length cDNA was designated as pUCm-BIHN-w1-1.
DNA sequencing and sequence analysis
DNA sequencing was performed by Invitrogen sequencing company (Shanghai, China). Similarity searches were carried out using BLAST at the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignments and phylogenetic tree construction were performed using ClustalW program in DNAstar software (LaserGene, Madison, WI, USA).
Construction of binary vector and transformation of tobacco
The coding sequence was amplified using primers, BIHN-w1-orf-2F (5′-GGA TCC ATG GTT GCC GCA GTA GCA GCT-3′) (underlined is BamHI site) and BIHN-w1-orf-2R (5′-GTC GAC TCA CAT ATT TGA CTT ATC TCC AGT-3′) (underlined is SalI site), with plasmid pUCm-BIHN-w1-1 as template. PCR product was cloned and confirmed by sequencing, yielding plasmid pUCm-BIHN-w1-2. The ORF sequence was released from pUCm-BIHN-w1-2 by digestion with BamHI/SalI and ligated into BamHI/SalI sites of the plant binary vector CHF3, obtaining plasmid CHF3-BIHN-w1, which was then introduced into Agrobacterium tumefaciens strain EHA105 by electroporation using a GENE PULSER II Electroporation System (Bio-Rad Laboratories, Hercules, CA, USA). Agrobacteria were grown at 28°C with shaking (200 rpm) in YEP broth containing 100 μg/ml streptomycin and 100 μg/ml spectinomycin and collected by centrifugation, followed by resuspending in MS medium to a final OD600 of approximately 0.8. Transformation of tobacco was performed using Agrobacterium-mediated leaf disc transformation as described previously (Luo et al. 2005b). Re-generated shoots were rooted on MS medium containing 200 μg/ml kanamycin and 250 μg/ml carbenicillin, and transferred to soil and grown in greenhouse. Seeds of the transgenic lines were germinated on 1/2 MS medium containing 200 μg/ml kanamycin and the survivals were transferred into natural soil and grown to set seeds. The transgenic lines were allowed to grow to three generations and homozygous and single-copy lines were screened and confirmed by segregation of antibiotic resistance marker.
Disease resistance assays
Eight-week-old wild-type and transgenic tobacco plants were used to assay disease resistance against tobacco mosaic virus (TMV). To prepare the inoculum, TMV-infected tobacco leaves were ground to a fine powder in a potassium phosphate buffer (50 mM, pH 7.0). Tobacco plants were inoculated with TMV using a standard mechanical rubbing method. Briefly, fully expanded leaves were dusted with dry carborundum and inoculated by gently rubbing the upper leaf surface with 100 μl of the viral suspension inoculum, followed by rinsing with distill sterilized water immediately. After inoculation, plants were maintained at 23–26°C under continuous illumination provided by fluorescent lamps. Lesion numbers on the inoculated leaves were counted 5 day after inoculation.
Pseudomonas syringae pv. tabaci was cultured in King’s B broth at 28°C for 2 days, and a bacterial suspension was prepared in 10 mM MgCl2 (5 × 104 colony-forming units ml−1). Inoculation was performed by infiltration of the bacterial suspension using a 1-ml syringe without a needle into leaves of 8-week-old wild-type and transgenic tobacco plants. To determine bacterial titers, 10 infected leaves were collected after inoculation. Leaf discs of the same size made using a hole puncher were homogenized in 10 mM MgCl2, then serial-diluted by 1:10, and plated onto King’s B medium. Plates were incubated at 28°C for 2 days, and bacterial colonies appeared were counted.
Abiotic stress tolerance assay
Assays for oxidative stress tolerance were performed as follow. Fully expanded leaves from 8-week-old plants of wild-type and transgenic lines were briefly washed in sterile distill water, and leaf discs (13 mm in diameter) were punched out using a hole puncher. For methyl viologen (MV) treatment, leaf discs were floated on a solution containing different concentrations of MV and 0.1% Tween-20 in the dark for 1 h, followed by illumination at moderate light intensity (200 μmol m−2 s−1) for 18 h at 25°C. For H2O2 treatment, leaf discs were incubated in MES buffer containing different concentrations of H2O2 for 1 day under the light of 200 μmol m−2 s−1. Chlorophyll contents in leaf discs at 18 h after MV treatment or at 24 h after H2O2 treatment were measured as described previously (Veronese et al. 2003). Leaf disc samples were harvested 5 h after treatments with MV (2 μM), H2O2 (50 mM) or water as control and were subject to extraction of total RNA for analysis of gene expression by semi-quantitative RT-PCR.
ABA sensitivity and drought tolerance were assayed. Surface-sterilized seeds were sown on 1/2 MS medium containing different concentrations of ABA and seedlings were grown under16 h/8 h light/night regime at 22–25°C. Root lengths of the transgenic and wild-type seedlings were measured 2 weeks after germination and at least 40 individual seedlings were measured. To evaluate drought tolerance, transgenic and wild type seeds were placed on filter paper saturated with or without 200 mM mannitol or different concentrations of PEG6000 and incubated under16 h/8 h light/night regime at 22–25°C.
Gene expression analysis by RT-PCR
Total RNA was extracted from rice and tobacco leaf samples using TRIZOL reagent (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. Total RNA (500 ng) was reverse-transcribed using the SuperScript III Kit (Invitrogen, Shanghai, China). One microliter of the RT reaction and 10 pmol of each primer were used for semi-quantitative RT-PCR in a total volume of 25 μl. PCR conditions were set as 94°C 30 s, 48–65°C 30 s and 72°C 30 s for 25–30 cycles based on the abundance of transcript for each gene, followed by 7 min of final extension at 72°C. PCR products were electrophoresed on a 1.2% agarose gel. Information on gene-specific primers used was listed in Table 1.
Results
Cloning and characterization of rice OsBIRF1, encoding a RING finger protein
In our previous studies aimed at elucidating the molecular biology of rice defense response induced by BTH, a defense-related differentially expressed cDNA clones, BIHN-w1, was isolated and identified through approach of suppression subtractive hybridization (Song and Goodman 2002). Sequencing and BLAST similarity searching against the GenBank database revealed that BIHN-w1 is a part of a gene encoding a putative RING-H2 finger protein. The cDNA insert in BIHN-w1 seems to encode the C-terminal of this putative RING finger protein. The full-length cDNA of this putative RING finger gene was obtained by amplification of 5′-end sequences flanking the insert in clone BIHN-w1 and was named as OsBIRF1 (Oryza sativa L. BTH-induced RING finger protein 1). The full-length cDNA of OsBIRF1 is 1,873 bp with an open reading frame (ORF) of 1,191 bp. The OsBIRF1 gene corresponds to locus Os02g50930 as annotated by the TIGR Rice Genome Annotation program, which is located on chromosome 2 of the rice genome and consists of a single exon without intron.
The OsBIRF1 gene was predicted to encode a 396 amino acid protein with a calculated molecular weight of 43 kD and isoelectric point of 8.13. Database searches for conserved domains and alignment with ATL1D and ATL5I, two previously characterized Arabidopsis RING-H2 finger proteins, revealed that OsBIRF1 contains a highly conserved RING-H2 finger domain (C-X2-C-X15-C-X1-H-X2-H-X2-C-X10-C-X2-C) (Fig. 1a). In addition, a putative transmembrane domain from 58-A to 80-V, a basic amino acid region from 101-H to 124-S, and GLD domain from 125-G to 140-R, were identified (Fig. 1a). Protein similarity searches and phylogenetic tree analysis indicated that OsBIRF1 has high levels of similarity to Arabidopsis ATL1D and ATL5I, showing identity of 42–48% (Fig. 1b). The features of conserved domain organization in OsBIRF1 protein are similar to the characteristic regions present in the ATL family proteins (Salinas-Mondragon et al. 1999; Serrano et al. 2006), and therefore OsBIRF1 encodes a rice RING-H2 finger protein belonging to the ATL family.
OsBIRF1 is a RING finger protein. (a) Alignment of OsBIRF1 with Arabidopsis thaliana ATL1D (AAQ55274) and ATL5I (BAD44076). The putative transmembrane domain (TM), basic amino acid rich region, conserved ATL family GLD region and RING finger domain is indicated above the sequence and the conserved amino acid residues in the RING finger domain are indicated by filled triangles. (b) Phylogenetic tree analysis of OsBIRF1. The plant RING finger proteins used are Arabidopsis thaliana ATL1D (At1g23980, AAQ55274), ATL5I (At5g40250, BAD44076), AtATL3 (AT1G72310, AAM61130), AtRHA3b (At2g17450, AAM64643), AtATL4 (At3g60220, NP_191581) and AtATL5C (At5g05810, Q5EAE9); Oryza sativa Os12g40460 (ABA99194)
Expression patterns of OsBIRF1 in rice disease resistance responses
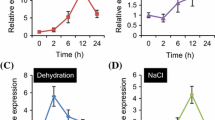
The differentially expressed cDNA clone BIHN-w1 was originally obtained from a suppression subtractive hybridization library constructed by subtraction of cDNAs from BTH-induced and pathogen-infected rice leaf samples with those from water control (Song and Goodman 2002), suggesting that the gene corresponding to BIHN-w1 might be induced during rice defense response. We therefore analyzed by semi-quantitative RT-PCR whether expression of OsBIRF1 in 3-week-old rice seedlings was induced by treatments with some of defense-related signal molecules including BTH, SA, ACC or JA. The results showed that expression of OsBIRF1 was induced by all these defense-related signal molecules with similar kinetics. Up-regulation of OsBIRF1 expression was detected at 12 h, peaked at 48 h, and maintained at a relatively high level during 12–72 h after BTH treatment (Fig. 2a). Likely, expression of OsBIRF1 was induced in 6–72 h by ACC and JA, and in 24–72 h by SA. No significant induced expression of OsBIRF1 was observed in water-treated seedlings during the experiment period, except for a low level of basal expression (Fig. 2a). These results indicate that OsBIRF1 gene is responsible to multiple defense-related signal molecules, suggesting a role for OsBIRF1 in induced defense response in rice.
Expression of OsBIRF1 in rice disease resistance response. (a) Expression of OsBIRF1 induced by BTH, ACC, SA and JA. Three-week-old rice seedlings were treated by foliar spraying with 0.3 mM BTH, 1.5 mM SA, 100 μM ACC, 100 μM JA or water. (b) Differential expression of OsBIRF1 in interactions between rice and Magnaporthe grisea. Three-week-old rice seedlings of H8R and H8S were inoculated with M. grisea and leaf samples were collected at each time point (h) as indicated. Total RNA was extracted and used for semi-quantitative RT-PCR analysis
To further elucidate the involvement of OsBIRF1 in disease resistance response, the expression pattern of OsBIRF1 in incompatible and compatible interactions between a pair of near-isogenic lines, H8R and H8S, and the blast fungus, M. grisea, was analyzed and compared. Once inoculated with spores of strain 85-14B1 of M. grisea, seedlings of H8R and H8S showed incompatible and compatible interactions, resulting in disease resistance and susceptible responses, respectively. In leaves of H8R seedlings, expression of OsBIRF1 was activated at 24 h, maintained a high level during 24–60 h after inoculation with M. grisea (Fig. 2b). However, expression of OsBIRF1 was maintained unchanged in leaves of H8S seedlings after pathogen inoculation (Fig. 2b). These results indicate that expression of OsBIRF1 is involved in the incompatible interaction between rice and M. grisea.
Generation of OsBIRF1-overexpressing transgenic plants
To study further the biological role of OsBIRF1 in defense responses, a functional analysis of OsBIRF1 in transgenic tobacco plants was performed. The coding sequence of OsBIRF1 was cloned into a plant binary vector CHF3 under the control of CaMV 35S promoter and was introduced into tobacco using the Agrobacterium-mediated leaf disc transformation method. A total of 21 independent transgenic plants were obtained by screening of kanamycin and by PCR detection of the transgene with genomic DNA as template and five independent transgenic lines with single copy of OsBIRF1 were obtained through screening based on segregation of antibiotic resistance marker in progenies. These single copy transgenic lines were allowed to grow for three generations and two homozygous lines were selected to for further studies. RT-PCR analysis indicated that OsBIRF1 was expressed in these transgenic tobacco lines (Fig. 3a).
Generation and growth of OsBIRF1 transgenic tobacco plants. (a) Expression of OsBIRF1 in transgenic tobacco plants. Total RNA was extracted from transgenic and wild-type plants grown under normal condition and used for semi-quantitative RT-PCR analysis. (b, c) Comparison of root elongation and growth of the transgenic and wild-type seedlings grown on 1/2 MS for 3 weeks (b) or in soil for 10 weeks (c). (d) Root length of the transgenic and wild-type seedlings grown on 1/2 MS for 3 weeks. (e) Height of the transgenic and wild-type plants grown in soil for 10 weeks. Values shown are the means ± SD from three independent experiments. Different letters above the columns indicate significant differences (P < 0.05). WT, wild-type plants; #5 and #8, independent transgenic lines
Overexpression of OsBIRF1 promotes growth of transgenic tobacco plants
During screening the transgenic lines, we noted that the T0 transgenic plants grew faster and had more leaves than the wild-type plants. This implied that overexpression of OsBIRF1 affected growth and development of the transgenic tobacco plants. We thus analyzed and compared growth and development phenotypes of the OsBIRF1 transgenic plants with wild-type plants. The transgenic seedlings grown on 1/2 MS medium showed some morphological changes as compared with the wild-type seedlings (Fig. 3b, c). The transgenic seedlings had longer roots by 62–75% increase in root length and more leaves by 1–2 leaves than those of the wild-type seedlings when grown on 1/2 MS medium for 3 weeks (Fig. 3b, d). Heights of 3-week-old seedlings grown on 1/2 MS and of 10-week-old plants grown in soil also were significantly higher than those of wild-type plants (Fig. 3c, e). These results reveal that OsBIRF1 might have a function in regulation of growth and development in transgenic tobacco plants as well as in rice.
Enhanced disease resistance in OsBIRF1-overexpressing transgenic plants
To clarify the possible role of OsBIRF1 in defense responses, disease assays were performed to evaluate the disease resistance levels of the OsBIRF1 transgenic tobacco plants against two different types of pathogens, TMV and P. syringae pv. tabaci. In TMV disease resistance assays, necrotic lesions were typically observed on the leaves of wild-type and the OsBIRF1 transgenic plants 3 days after inoculation, and timing of symptom appearance was similar between the transgenic and wild-type plants (Fig. 4a). However, the lesion numbers in the leaves of the OsBIRF1 transgenic plants was significantly reduced as compared with that in wild-type plants, resulting in reductions of 45 and 63% in transgenic lines #5 and #8, respectively (Fig. 4b). In addition, the disease resistance of the OsBIRF1 transgenic plants against P. syringae pv. tabaci was also tested. Under our experimental conditions, disease symptom was observed around 3 days after inoculation on the leaves of wild type plants with yellowish chlorotic and necrotic areas, and the large necrotic lesions were formed at end of the experiments (Fig. 4c). On the transgenic plant leaves, the symptom was only visible 4–5 days after inoculation with relatively small necrotic areas (Fig. 4c). Measurement of bacterial titers in inoculated leaves of the OsBIRF1 transgenic and wild-type plants further confirmed the observed phenotypes in OsBIRF1 transgenic plants. The bacterial titers in inoculated leaves of the transgenic plants were markedly reduced as compared with those in wild-type plants at 4 day and 7 day after inoculation (Fig. 4d). These results suggest that overexpression of OsBIRF1 in transgenic tobacco plants confers an enhanced disease resistance against TMV and P. syringae pv. tabaci.
Enhanced disease resistance and elevated expression levels of defense-related genes in OsBIRF1 transgenic tobacco plants. (a, b) Symptom and severity of disease caused by tobacco mosaic virus in transgenic and wild-type plants. Photos were taken 5 days after inoculation. Disease severity as indicated by lesion numbers on the OsBIRF1 transgenic and wild-type plants. (c, d) Symptom and severity of disease caused by Pseudomonas syringae pv. tabaci in transgenic and wild-type plants. Photos were taken 7 days after inoculation. Bacterial titers in inoculated leaves of the OsBIRF1 transgenic and wild-type plants were measured at different time points after inoculation. Values shown are the means ± SD of three independent experiments. Different letters above the columns indicate significant differences (P < 0.05). (e) Expression of PR genes in transgenic and wild-type plants. Leaf samples were collected from transgenic and wild-type plants grown in soil under normal condition and expression of PR genes was analyzed by semi-quantitative RT-PCR. WT, wild-type plants; #5 and #8, independent transgenic lines
Up-regulated expression of defense-related genes in OsBIRF1 transgenic plants
To ascertain whether the observed enhanced disease resistance in OsBIRF1 transgenic tobacco plants was resulted from activation of defense response by expression of OsBIRF1, we analyzed and compared the expression levels of some selected defense-related genes, e.g. PR-1, PR-2, PR-3 and PR-5, in OsBIRF1 transgenic and wild-type plants. As shown in Fig. 4e, under normal growth condition, no significant expression of PR genes was observed in the wild-type plants. However, up-regulated expression of PR-1, PR-2 and PR-3 were detected in the OsBIRF1 transgenic plants (Fig. 4e), and relatively higher levels of expression for PR-1 and PR-3 in transgenic line #8 was observed. PR-5 had slight increased expression in OsBIRF1 transgenic plants as compared with that in wild-type plants (Fig. 4e). These results indicate that overexpression of OsBIRF1 activates expression of PR genes in transgenic tobacco plants, resulting in constitutive activation of defense responses.
Enhanced oxidative stress tolerance in OsBIRF1 transgenic plants
Possible roles of OsBIRF1 in oxidative stress was further studied by testing the tolerance of leaves discs from 8-week-old transgenic and the wild-type plants to exogenous H2O2 or MV. As shown in Fig. 5a, c, no significant bleaching or chlorosis was observed in leaf discs from both transgenic and wild-type plants in normal medium without H2O2 or MV during the experimental period. After incubation in different concentrations of H2O2 or MV, symptoms of bleaching or chlorosis were observed in leaf discs both from transgenic and wild-type plants, but bleaching or chlorosis of leaf discs from wild-type plants was much more severe than those from transgenic plants (Fig. 5a, c). This was further confirmed by measuring chlorophyll contents in leaf discs from the transgenic and wild-type plants after H2O2 or MV treatments. Relative chlorophyll contents in leaf discs of the transgenic plants were markedly higher than those from the wild-type plants after treatments with different concentrations of H2O2 or MV (Fig. 5b, d).
Increased oxidative stress tolerance in OsBIRF1 transgenic plants. (a) Comparison of sensitivity to methyl viologen (MV) between leaf discs from transgenic and wild-type plants. Leaf discs were collected from 8-week-old transgenic and wild-type plants and floated on solutions containing different concentrations of MV in dark for 1 h, and then under illumination at moderate light intensity (200 μmol m−2 s−1) for 18 h at 25°C. Photos were taken 19 h after treatment. (b) Relative chlorophyll contents in leaf discs at 18 h after MV treatments. (c) Comparison of tolerance to H2O2 between leaf discs from transgenic and wild-type plants. Leaf discs were collected from 8-week-old transgenic and wild-type plants and floated on MES buffer containing different concentrations of H2O2 under illumination at moderate light intensity (200 μmol m−2 s−1) for 24 h at 25°C. Photos were taken 24 h after treatment. (d) Relative chlorophyll contents in leaf discs at 24 h after H2O2 treatments. Values shown are the means ± SD from three independent experiments. Different letters above columns indicate significant differences (P < 0.05). (e) Expression of oxidative stress-related genes. Leaf discs were collected from 8-week-old transgenic and wild-type tobacco plants were treated with MV (2 μM) or H2O2 (50 mM) for 5 h. Leaf discs were harvested for extraction of total RNA and analysis of gene expression by semi-quantitative RT-PCR. WT, wild-type plants; #5 and #8, independent transgenic lines
Expression of oxidative stress-related genes, including APX, CAT and GST, was analyzed to gain insights into the possible mechanisms of the enhanced oxidative stress tolerance in OsBIRF1 transgenic plants. Leaf discs from both transgenic and wild-type plants were treated by incubation in MV, H2O2 or water, and samples were collected to analyze expression of oxidative stress-related genes by RT-PCR. In water-treated controls, CAT and APX showed an increased expression in transgenic plants, and no significant expression of GST was detected in both wild-type and transgenic plants (Fig. 5e). After incubation with H2O2 or MV, expression of APX, GST and CAT was up-regulated in leaf discs of wild-type plants, indicating an oxidative stress was applied to leaf discs (Fig. 5e). However, the expression levels of APX, GST and CAT in leaf discs of the transgenic plants were higher than those in leaf discs of the wild-type plants (Fig. 5e). It was noted that expression of CAT showed no significant difference between the transgenic and wild-type plants after treatment with MV, but showed markedly increase in transgenic plants after treatment with H2O2 (Fig. 5e). It was thus concluded that overexpression of OsBIRF1 in transgenic tobacco plants improves oxidative stress tolerance through up-regulating expression of oxidative stress-related genes.
Reduced ABA sensitivity and increased drought tolerance of the OsBIRF1 transgenic plants
Previous studies have shown that RING finger proteins are involved in ABA signaling pathway and thus play an important role in drought tolerance (Ko et al. 2006; Zhang et al. 2005). To gain information on the function of OsBIRF1 in drought tolerance, we further analyzed the sensitivity of OsBIRF1 transgenic plants to exogenous ABA and drought tolerance. As mentioned above, roots of the OsBIRF1 transgenic seedlings on 1/2 MS without ABA were longer than the wild-type seedlings (Fig. 6a). On 1/2 MS with exogenous ABA, root elongation of the wild-type seedlings was significantly inhibited; however, root elongation of the OsBIRF1 transgenic seedlings was less inhibited (Fig. 6a). Compared with the wild-type seedlings, root growth of the transgenic seedlings was less inhibited by exogenous ABA, and this was much evident at high concentration of ABA supplemented in the medium (Fig. 6b). With the increase of ABA concentrations, inhibition of root length was much obvious in wild-type seedlings than in transgenic seedlings. Relative root length of wild-type seedlings grown on 1/2 MS containing 4 μM ABA was measured only ∼10% of those grown on 1/2 MS without ABA, while relative root length of the transgenic seedlings on ABA-containing MS was ∼40% of those grown on ABA-free medium (Fig. 6b). These results suggest that overexpression of OsBIRF1 in transgenic tobacco plants results in a reduced ABA sensitivity.
Reduced ABA sensitivity and increased drought tolerance in OsBIRF1 transgenic plants. (a, b) Inhibition of root elongation by ABA. (a) Growth of the transgenic and wild-type seedlings on 1/2 MS medium containing different concentrations of ABA. (b) Root lengths of the transgenic and wild-type seedlings 2 weeks after treatment with ABA. (c, d) Increased drought tolerance in OsBIRF1 transgenic plants. Seed germination on 1/2 MS medium containing different concentration of PEG6000 (c) or mannitol (d). Values shown are means ± SD from three independent experiments. Different letters above the columns indicate significant differences (P < 0.05). WT, wild-type; #5 and #8, independent transgenic lines
Drought tolerance of the OsBIRF1 transgenic plants was studied by analyzing and comparing seed germination on PEG6000- or mannitol-containing medium with the wild-type. Germination rates of seeds from the transgenic and wild-type plants were comparable under normal condition without PEG6000 or mannitol (Fig. 6c, d). In the presence of PEG6000, germination rates of the wild-type seeds were significantly reduced, germination rates of the transgenic seeds were markedly higher than the wild-type on medium containing 15% PEG6000, giving 125–170% increase over the wild-type seeds. Under 20% PEG6000 condition, the wild-type seeds did not germinate, while there were 20–34% of the transgenic seeds germinated (Fig. 6c). Similarly, under 200 mM mannitol, only ∼36% of the wild-type seeds germinated, while 74–78% of the transgenic seeds germinated, giving an increase of 105–117% over the wild-type seeds. These results indicate that overexpression of OsBIRF1 in transgenic tobacco plants leads to an enhanced drought tolerance.
Discussion
RING finger proteins, comprising of a large protein family, ubiquitously exist in all of eukaryotes and, as a kind of ubiquitin E3 ligases, play key roles in ubiquitination of specific proteins for degradation by the ubiquitin/26S proteasome system (Freemont 2000; Joazeiro and Weissman 2000). Among the RING finger proteins, a specific family, called ATL family, was first identified in Arabidopsis (Salinas-Mondragon et al. 1999) and later found to distribute widely in plant species, for example, the Arabidopsis and rice ATL family contain 80 and 121 members, respectively (Serrano et al. 2006). In addition to a typical RING finger domain, the ATL family proteins contain common characteristic structural features, e.g. a predicted transmembrane domain, a basic amino acid rich region, a conserved GLD region and a highly diverse region in the C-terminal (Salinas-Mondragon et al. 1999). The fact that OsBIRF1 not only contains the above-mentioned characteristic structural features but also is phylogenetically related to Arabidopsis ATLs clearly demonstrates that it is a member of the rice ATL family. OsBIRF1 is an intronless gene, which is also similar to the previous observation that 90% of the Arabidopsis ATL genes are intronless (Serrano et al. 2006). Although more than 100 members were identified for the rice ATL family, only one member, EL5, has been studied in detail for its biochemical and biological function (Takai et al. 2001, 2002; Katoh et al. 2003; Koiwai et al. 2007). Our functional analysis in transgenic tobacco suggests important roles for OsBIRF1 in growth and defense responses against biotic and abiotic stresses. These findings provide new insights into the biological functions of the rice ATL family.
It was previously found that expression of several ATL genes in Arabidopsis is induced by fungal elicitors (Serrano et al. 2006). Similarly, the EL5 gene is transiently induced by N-acetylchitoheptaose elicitor in rice cells (Takai et al. 2002). However, there is no further evidence supporting a role for EL5 in rice disease resistance response. OsBIRF1 is up-regulated by some well-known defense-related signal molecules including SA and JA, which are believed to mediate the SA-dependent pathway and the JA/ET pathway in defense responses, respectively (Glazebrook 2005). Importantly, OsBIRF1 is also differentially induced in incompatible but not in compatible interactions between rice and the blast fungus, which is in agreement with the notion that expression of ACRE132, a tobacco ATL gene, is induced in Avr9- and Cf9-mediated incompatible interaction (Durrant et al. 2000). The induced expression feature during defense responses provides preliminary evidence supporting a role for OsBIRF1 in regulating disease resistance response in rice. Direct evidence supporting this conclusion came from our functional analysis of OsBIRF1 in transgenic tobacco plants. In this study, the OsBIRF1-overexpressing transgenic tobacco plants showed enhanced disease resistance against at least two different types of pathogens, virus (TMV) and bacteria (P. syringae pv. tabaci), as revealed by reduced disease severity and bacterial population in the transgenic plants. Accompanied with the enhanced disease resistance phenotype in the OsBIRF1 transgenic tobacco plants is the up-regulated expression of some defense-related genes in the absence of pathogen infection or elicitor induction. It was recently found that the Arabidopsis eca mutants, which showed constitutive expression of the ATL2 gene, exhibits elevated expression levels of defense-related genes and SA- and JA-responsive genes and knockout mutation with T-DNA insertion in ATL9 results in increased susceptibility to powdery mildew disease (Serrano and Guzman 2004; Ramonell et al. 2005). However, it is not clear yet whether the eca mutant plants have enhanced disease resistance phenotype and the ATL9 T-DNA knockout plants impairs activation of defense-related genes in responding to pathogen infection (Serrano and Guzman 2004; Ramonell et al. 2005). The tomato ATL6-mediated ubiquitin/proteasome system contributes to fungal elicitor-activated defense response via the JA-dependent signaling pathway (Hondo et al. 2007). It was also reported that ectopic expression of the rice OsRHC1, encoding a RING finger protein other than the ATL family members, in Arabidopsis shows enhanced disease resistance and elevated expression of defense-related genes (Cheung et al. 2007). On the other hand, overexpression of the CaRFP1 gene in transgenic Arabidopsis plants leads to increased disease susceptibility and reduced PR gene expression (Hong et al. 2007). Therefore, the fact that enhanced disease resistance is associated with up-regulated PR gene expression in the transgenic tobacco plants suggests that the enhanced disease resistance in OsBIRF1 transgenic plants is most likely to be the results of activation of defense-related gene expression.
It has been demonstrated that some of the RING finger proteins play important roles in regulation of abiotic stress tolerance (Dong et al. 2006; Ko et al. 2006; Zhang et al. 2007; Kam et al. 2007; Sahin-Cevik and Moore 2007). In this study, the OsBIRF1 transgenic tobacco plants show enhanced tolerance to oxidative stress, as revealed by lowering in bleaching and chlorophyll loss of leaf discs after treatments with exogenous MV and H2O2. We also noted that in OsBIRF1 transgenic tobacco plants, expression of oxidative stress-related genes, e.g. APX, CAT and GST, was up-regulated. It is well established that enhanced oxidative stress tolerance is associated with high levels of expression of genes that are involved in oxidative stress responses in plants (Mittler et al. 2004, 2006). Interestingly, a further induction of APX, CAT and GST genes was observed in leaf discs of the transgenic plants after challenged with exogenous MV or H2O2, when compared to the expression levels in leaf discs without stress. This result suggests that (1) OsBIRF1 itself in the transgenic plants can elevate oxidative stress tolerance by activating expression of the oxidative stress-related genes in the absence of stress, and (2) OsBIRF1 can also potentitate or amplify the signal(s) required for activating expression of the oxidative stress-related genes when exposure to exogenous stress.
Some of RING finger proteins have been demonstrated to regulate ABA signaling and affect drought tolerance (Zhang et al. 2005, 2007). ABA treatment has been suggested to inhibit seed germination and seedling growth in plants (Finkelstein et al. 2002). The OsBIRF1 transgenic plants show reduced ABA sensitivity, as revealed by reduced inhibition of primary root elongation with exogenous ABA treatments. This is similar to that of the Arabidopsis AIP2, whose constitutive expression in Arabidopsis resulted in insensitivity to ABA for primary root growth (Zhang et al. 2005), but is contrast to that of the ATL43 gene, whose mutation led to ABA insensitivity (Serrano et al. 2006). Furthermore, results from seed germination on PEG6000- or mannitol-containing medium demonstrate that the OsBIRF1 transgenic plants also have increased tolerance against drought stress. ABA is an essential mediator in triggering plant responses to most of the common abiotic stresses, including drought stress (Finkelstein et al. 2002). Reduced ABA sensitivity and increased drought tolerance were observed in the OsBIRF1 transgenic tobacco plants, which is different from previous observation that constitutive expression of the Arabidopsis SDIR1 resulted in increased ABA sensitivity and enhanced tolerance to multiple abiotic stresses including drought stress (Zhang et al. 2007). This may suggest that RING finger proteins have functions in ABA signaling and drought response through different mechanisms.
A number of RING finger proteins have been shown to regulate different aspects of growth and developmental processes (Moon et al. 2004). Transgenic alfalfa and Arabidopsis overexpressing the alfalfa MsRH2-1, an ortholog of Arabidopsis ATL4, exhibit shorter plant heights and abnormal lateral root development (Karlowski and Hirsch 2003). We also found that the OsBIRF1 transgenic tobacco plants have longer primary roots and larger plant heights as compared with the wild-type plants. Interestingly, constitutive expression and suppressing by RNAi of EL5 in rice did not cause any obvious phenotypic changes (Koiwai et al. 2007). Auxin signaling has been shown to be involved in RING finger proteins-mediated regulation of plant growth and development processes (Xie et al. 2002; Nodzon et al. 2004). The involvement of auxin or other phytohormones, if possible, in promotion of the OsBIRF1 transgenic plants needs further investigation.
Taken together, our studies suggest that OsBIRF1 plays important roles in regulation of plant growth and defense response against biotic and abiotic stresses. Overexpression of OsBIRF1 results in constitutive activation of defense-related genes in transgenic tobacco plants and thus OsBIRF1 appears to be a positive regulator of defense response to multiple environmental stresses. Considering that OsBIRF1 encodes a putative RING-H2 finger ubiquitin E3 ligase, OsBIRF1 protein might target potential factors that are negative regulators of defense responses in plants. Further identification and functional analysis of the downstream targets will lead to a better understanding of the roles of OsBIRF1 in the cellular defense response to pathogen infection and environmental stresses. Generation of transgenic rice plants overexpressing or knocking down the OsBIRF1 gene is underway. Results from these analyses will provide new insights into the biological functions of this gene in rice growth/development and defense responses against biotic and abiotic stresses.
Abbreviations
- ABA:
-
Abscisic acid
- ACC:
-
1-Amino cyclopropane-1-carboxylic acid
- BTH:
-
Benzothiadiazole
- CaMV:
-
Cauliflower mosaic virus
- ET:
-
Ethylene
- H2O2 :
-
Hydrogen peroxide
- JA:
-
Jasmonic acid
- MV:
-
Methyl viologen
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- PEG:
-
Polyethylene glycol
- PR:
-
Pathogenesis-related
- RT-PCR:
-
Reverse transcription-PCR
- SA:
-
Salicylic acid
- TMV:
-
Tobacco mosaic virus
References
Cheung MY, Zeng NY, Tong SW, Li FWY, Zhao KJ, Zhang Q, Sun SM, Lam HM (2007) Expression of a RING-HC protein from rice improves resistance to Pseudomonas syringae pv. tomato DC3000 in transgenic Arabidopsis thaliana. J Exp Bot 58:4147–4159
Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol 16:272–279
Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103:8281–8286
Duek PD, Elmer MV, van Oosten VR, Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14:2296–2301
Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG (2000) cDNA-AFLP reveals a striking overlap in race specific resistance and wound response gene expression profiles. Plant Cell 12:963–977
Fang S, Weissman AM (2004) A field guide to ubiquitylation. Cell Mol Life Sci 61:1546–1561
Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(suppl):S15–S45
Freemont PS (2000) RING for destruction? Curr Biol 10:R84–R87
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82:373–428
Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19:4997–5006
Hardtke CS, Okamoto H, Stoop-Myer C, Deng XW (2002) Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J 30:385–394
Hershko A, Ciechanover A (1998) The ubiquitin system. Ann Rev Biochem 67:425–479
Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16:1247–1259
Hondo D, Hase S, Kanayama Y, Yoshikawa N, Takenaka S, Takahashi H (2007) The LeATL6-associated ubiquitin/proteasome system may contribute to fungal elicitor-activated defense response via the jasmonic acid-dependent signaling pathway in tomato. Mol Plant Microbe Interact 20:72–81
Hong JK, Choi HW, Hwang IS, Hwang BK (2007) Role of a novel pathogen-induced pepper C3-H-C4 type RING-finger protein gene, CaRFP1, in disease susceptibility and osmotic stress tolerance. Plant Mol Biol 63:571–588
Jang IC, Yang JY, Seo HS, Chua NH (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19:593–602
Joazeiro CA, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552
Kam J, Gresshoff P, Shorter R, Xue G-P (2007) Expression analysis of RING zinc finger genes from Triticum aestivum and identification of TaRZF70 that contains four RING-H2 domains and differentially responds to water deficit between leaf and root. Plant Sci 173:650–659
Karlowski WM, Hirsch AM (2003) The over-expression of an alfalfa RING-H2 gene induces pleiotropic effects on plant growth and development. Plant Mol Biol 52:121–133
Katoh S, Hong C, Tsunoda Y, Murata K, Takai R, Minami E, Yamazaki T, Katoh E (2003) High precision NMR structure and function of the RING-H2 finger domain of EL5, a rice protein whose expression is increased upon exposure to pathogen-derived oligosaccharides. J Biol Chem 278:15341–15348
Katoh S, Tsunoda Y, Murata K, Minami E, Katoh E (2005) Active site residues and amino acid specificity of the ubiquitin carrier protein-binding RING-H2 finger domain. J Biol Chem 280:41015–41024
Kawasaki T, Nam J, Boyes DC, Holt 3rd BF, Hubert DA, Wiig A, Dangl JL (2005) A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. Plant J 44:258–270
Ko JH, Yang SH, Han KH (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47:343–355
Koiwai H, Tagiri A, Katoh S, Katoh E, Ichikawa H, Minami E, Nishizawa Y (2007) RING-H2 type ubiquitin ligase EL5 is involved in root development through the maintenance of cell viability in rice. Plant J 51:92–104
Lechner E, Goloubinoff P, Genschik P, Shen WH (2002) A gene trap dissociation insertion line, associated with a RING-H2 finger gene, shows tissue specific and developmental regulated expression of the gene in Arabidopsis. Gene 290:63–71
Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15:912–924
Liu K, Wang L, Xu Y, Chen N, Ma Q, Li F, Chong K (2007) Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta 226:1007–1116
Luo H, Song F, Goodman RM, Zheng Z (2005a) Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol 7:459–468
Luo H, Song F, Zheng Z (2005b) Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. J Exp Bot 56:2673–2682
Ma L, Gao Y, Qu L, Chen Z, Li J, Zhao H, Deng XW (2002) Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell 14:2383–2398
Meng XB, Zhao WS, Lin RM, Wang M, Peng YL (2006) Molecular cloning and characterization of a rice blast-inducible RING-H2 type zinc finger gene. DNA Seq 17:41–48
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) The reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580:6537–6542
Molnar G, Bancoş S, Nagy F, Szekeres M (2002) Characterisation of BRH1, a brassinosteroid-responsive RING-H2 gene from Arabidopsis thaliana. Planta 215:127–133
Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16:3181–3195
Nishimura R, Ohmori M, Fujita H, Kawaguchi M (2002) A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc Natl Acad Sci USA 99:15206–15210
Nodzon LA, Xu WH, Wang YS, Pi LY, Chakrabarty PK, Song WY (2004) The ubiquitin ligase XBAT32 regulates lateral root development in Arabidopsis. Plant J 40:996–1006
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466
Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138:1027–1036
Sahin-Cevik M, Moore GA (2007) Isolation and characterization of a novel RING-H2 finger gene induced in response to cold and drought in the interfertile Citrus relative Poncirus trifoliate. Physiol Plant 126:153–161
Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17:2642–2647
Salinas-Mondragon RE, Garciduenas-Pina C, Guzman P (1999) Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol Biol 40:579–590
Saurin AJ, Borden KL, Boddy MN, Freemont PS (1996) Does this have a familiar RING? Trends Biochem Sci 21:208–214
Schumann U, Prestele J, O’Geen H, Brueggeman R, Wanner G, Gietl C (2007) Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc Natl Acad Sci USA 104:1069–1074
Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423:995–999
Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18:617–622
Serrano M, Guzman P (2004) Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 167:919–929
Serrano M, Parra S, Alcaraz LD, Guzman P (2006) The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J Mol Evol 62:434–445
Shimomura K, Nomura M, Tajima S, Kouchi H (2006) LjnsRING, a novel RING finger protein, is required for symbiotic interactions between Mesorhizobium loti and Lotus japonicus. Plant Cell Physiol 47:1572–1581
Smalle J, Vierstra RD (2004) The ubiquitin 26s proteasome proteolytic pathway. Ann Rev Plant Biol 55:555–590
Song F, Goodman RM (2002) OsBIMK1, a rice MAP kinase gene involved in disease resistance responses. Planta 215:997–1005
Sonoda Y, Yao SG, Sako K, Sato T, Kato W, Ohto MA, Ichikawa T, Matsui M, Yamaguchi J, Ikeda A (2007) SHA1, a novel RING finger protein, functions in shoot apical meristem maintenance in Arabidopsis. Plant J 50:586–596
Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137:13–30
Takai R, Hasegawa K, Kaku H, Shibuya N, Minami E (2001) Isolation and analysis of expression mechanisms of a rice gene, EL5, which shows structural similarity to ATL family from Arabidopsis, in response to N-acetylchitooligosaccharide elicitor. Plant Sci 160:577–583
Takai R, Matsuda N, Nakano A, Hasegawa K, Akimoto C, Shibuya N, Minami E (2002) EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor-responsive ubiquitin-conjugating enzyme, OsUBC5b. Plant J 30:447–455
Veronese P, Narasimhan ML, Stevenson RA, Zhu JK, Weller SC, Subbarao KV, Bressan RA (2003) Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J 35:574–587
Xie Q, Guo HS, Dallman G, Fang SY, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419:167–170
Xu R, Li QQ (2003) A RING-H2 zinc-finger protein gene RIE1 is essential for seed development in Arabidopsis. Plant Mol Biol 53:37–50
Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17:804–821
Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19:1532–1543
Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19:1912–1929
Acknowledgements
We are grateful to Dr Zuhua He, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Science, for the rice near-isogenic lines H8R and H8S, and Mr Rongyao Chai, Zhejiang Academy of Agricultural Science, for the Magnaporthe grisea isolate 85-14B1. This study was supported by National Natural Science Foundation of China (grants no. 30571209 and 30771399), the National High-Tech (“863”) Project (2006AA10Z430), the National Key Basic Research and Development Program (2006CB101903) and the Fund for the New Century Talent Program from MOE of China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, H., Zhang, H., Yang, Y. et al. Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol Biol 68, 17–30 (2008). https://doi.org/10.1007/s11103-008-9349-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9349-x