Abstract
Cytokinins (CKs) are plant hormones that regulate a large number of processes associated with plant growth and development such as induction of stomata opening, delayed senescence, suppression of auxin-induced apical dominance, signaling of nitrogen availability, differentiation of plastids and control of sink strength. In maize, CKs are thought to play an important role in establishing seed size and increasing seed set under normal and unfavorable environmental conditions therefore influencing yield. In recent years, the discovery of isopentenyl transferase (IPT) genes in plants has shed light on the CK biosynthesis pathway in plants. In an effort to increase our understanding of the role played by CKs in maize development and sink-strength, we identified several putative IPT genes using a bioinformatics approach. We focused our attention on one gene in particular, ZmIPT2, because of its strong expression in developing kernels. The expression of the gene and its product overlays the change in CK levels in developing kernels suggesting a major role in CK biosynthesis for kernel development. We demonstrate that at 8–10 days after pollination (DAP) the endosperm and especially the basal transfer cell layer (BETL) is a major site of ZmIPT2 expression, and that this expression persists in the BETL and the developing embryo into later kernel development stages. We show that ectopic expression of ZmIPT2 in calli and in planta created phenotypes consistent with CK overproduction. We also show that ZmIPT2 preferentially uses ADP and ATP over AMP as the substrates for dimethylallyl diphosphate (DMAPP) IPT activity. The expression pattern of ZmIPT2 in the BETL, endosperm and embryo during kernel development will be discussed with an emphasis on the suggested role of CKs in determining sink-strength and grain production in crop plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytokinins are plant hormones that play a major role in cell differentiation and proliferation. They also influence different plant growth and developmental processes such as delayed leaf senescence, leaf expansion, release of apical dominance, control of shoot/root ratio, chloroplast formation and signaling of nutrient status (Mok 1994; Sakakibara 2006). From an agronomic perspective, CKs have been associated with crop productivity in soybean, rice and maize. In soybean, CKs have been shown to play an important role during flower and pod development. Exogenous benzyladenine (BA) applications to the raceme decrease abortion of flowers and/or pods (Dyer et al. 1988; Peterson et al. 1990; Mosjidis et al. 1993; Reese et al. 1995). Likewise, evidence supports a function for CKs in the regulation of flowering and seed set in soybean (Huff and Dybing 1980; Ghiasi et al. 1987; Peterson et al. 1990; Wiebold 1990; Mosjidis et al. 1993; Reese et al. 1995; Nagel et al. 2001). In rice, reduced expression of a cytokinin oxidase gene (OsCkx2) in some varieties was found to be associated with a quantitative trait locus (QTL) for grain production (Ashikari et al. 2005). Similarly, in maize, exogenous applications of CKs have been found to increase seed set and yield stability for heat-stressed kernels (Cheikh and Jones 1994; Dietrich and Morris 1995). Finally, a role of CKs in the regulation of source-sink relations has been evidenced (Roitsch and Ehness 2000; Balibrea-Lara et al. 2004). The discovery and characterization of CK metabolic enzymes such as the degradative enzyme cytokinin oxidase/dehydrogenase (Ckx) in Arabidopsis and maize (Houba-Herin et al. 1999; Morris et al. 1999; Werner et al. 2001; Brugière et al. 2003; Massonneau et al. 2004) and the biosynthetic enzyme IPT in Arabidopsis and Petunia (Kakimoto 2001; Takei et al. 2001; Zubko et al. 2002) have lead the way to a better understanding of the role played by CK in plant growth, development and productivity. In Arabidopsis, CK synthesis is catalyzed by a family of DMAPP:ADP/ATP isopentenyl transferases with different or overlapping patterns of expression (Miyawaki et al. 2004). The expression of some AtIPT genes was shown to be down-regulated by CKs whereas two were up-regulated in the roots by auxin application and one was up-regulated by nitrate application (Miyawaki et al. 2004; Takei et al. 2004). The existence of an AtIPT gene family and its differential regulation by different compounds raise the potential of a distinct physiological role for each IPT gene product (Takei et al. 2001; Sakakibara and Takei 2002). More recently, it was found that some plant IPT genes play a role in the formation and maintenance of meristems through their activation by Knotted-like homeobox (KNOX) proteins (Jasinski et al. 2005; Yanai et al. 2005; Sakamoto et al. 2006). The study of ATP/ADP and tRNA IPT mutants indicated that ATP/ADP IPTs are involved in the synthesis of the bulk of isopentenyladenine and trans-zeatin (t-Z) type CK whereas tRNA IPTs are needed for cis-zeatin (cis-Z) type CK production (Miyawaki et al. 2006).
In maize kernels, CK levels peak at approximately 10 days after pollination (DAP) (Dietrich and Morris 1995; Brugière et al. 2003). DMAPP:AMP IPT activity was detected in immature kernels extracts, but a CK biosynthetic enzyme was never purified (Blackwell and Horgan 1994). In this report, we demonstrate the existence of a gene family of isopentenyl transferases in maize. For one of these, ZmIPT2, we studied its expression pattern in detail and found that the gene is strongly and specifically expressed in developing kernels where its expression overlays changes in zeatin riboside (ZR) levels and coincides with well documented peak of endosperm cell division. We show that in 8 DAP kernels, the protein is detectable in the endosperm where it is more abundant in the BETL. We demonstrate the function of ZmIPT2 as a CK biosynthetic enzyme both in vivo and in vitro. The role of ZmIPT genes in CK biosynthesis and more specifically ZmIPT2 in endosperm cell division, embryo development and kernel sink strength will be discussed with an emphasis on possible applications for yield improvement.
Material and methods
Plant materials
Maize (Zea mays) B73 plants were used in this study. Self-pollinated kernels were harvested from 0 to 42 DAP. For each time point, duplicate ears were taken from each of four field replicates. Samples were collected between 10 am and 12 pm. Ovules of developing kernels were separated from the glumes and from 6 to 42 DAP, the lower part of the seed (pedicel enriched fraction) was separated from the rest of the kernel (Brugière et al. 2003). Samples from other organs were harvested from field-grown and stored at −80°C. Whole kernel samples were harvested every 5 days from 0 to 25 DAP and dissected by isolating whole kernels (0 DAP), pedicel, nucellus and pericarp (5 DAP), pedicel/placental chalazal/basal endosperm region (PPCBE), nucellus, endosperm/embryo sac and pericarp (10 DAP), or PPCBE region, embryo, endosperm and pericarp (15, 20 and 25 DAP). In each case tissues corresponding to 3–4 different plants were pooled. Arabidopsis thaliana ecotype Columbia-0 was used for Arabidopsis transformation studies.
Sequences identification and phylogenetic tree
The AtIPT1, AtIPT3–AtIPT8 protein sequences were used to identify maize candidate IPT proteins using a bioinformatics strategy. The AtIPT proteins were BLASTed (Altschul et al. 1997) against the six possible frames generated by the maize and rice genomic sequences and the proprietary EST database of Pioneer Hi-Bred, a DuPont business. The rice sequences having an E-score of at least 200 were then used for an additional BLAST survey of the Genome Sequence Survey (GSS) maize database. The maize genomic sequences obtained which had an E-score of at least 150 were pooled with the sequences obtained in the first round of BLAST search. At the time the search was done, the maize GSS database had not been assembled and GSS fragments were assembled using Sequencher (Gene Codes Corporation, Ann Arbor, MI). Identified sequences are summarized in Table S1.
The phylogenetic tree was calculated using the Unweighted Pair Group Method with Arithmatic Mean (UPGMA) method with Phylip (Phylogenetic Inference Package) Version 3.57c (Felsenstein 1989) based on a ClustalW alignment using the Blosum matrix. The resulting radial tree was displayed using TreeView (Page 1996).
PCR amplification of ZmIPT2
ZmIPT2 coding sequence was PCR amplified from B73 and Mo17 genomic DNA. Primers ZmIPT2-5′ (5′-ATCATCAAGACAATGGAGCACGGTG-3′) and ZmIPT2-3′ (5′-CGTCCGCTAGCTACTTATGCATCAG-3′) were designed based on the GSS contig sequence. ZmIPT2 was re-amplified using Gateway compatible primers (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGGAGCACGGTGCCGTCGCCG-3′) and (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATGCATCAGCCACGGCGGTG-3′), cloned in pDONR221 and sequenced (Invitrogen, Carlsbad, CA). In each case, a touchdown PCR was performed (GeneAmp PCR System 9700, Perkin-Elmer, Wellesley, MA), using the following cycling parameters: 94°C for 2 min (one cycle), 94°C for 30 s, 65°C for 45 s and 72°C for 1 min 30 s (5 cycles, annealing temperature reduced by 1°C per cycle), 94°C for 30 s, 60°C for 45 s and 72°C for 1 min 30 s (30 cycles), 72°C for 7 min, and termination at 4°C.
Northern blot analysis of ZmIPT2 gene expression
Northern blots for the study of ZmIPT2 expression were performed as previously described (Brugière et al. 1999). Hybridization was carried out at 65°C using ExpressHyb hybridization buffer (BD Clontech, San Jose, CA) and an α-32P dCTP labeled probe (Rediprime II, GE Healthcare, Little Chalfont, UK) corresponding to the ZmIPT2 gene coding sequence as described above and other probes such as 18S RNA and cyclophilin. Successive washes were performed as follows: three times at 25°C for 10 min each with 2× SSC; 0.1% (w/v) SDS (1× SSC is 150 mM NaCl and 15 mM sodium citrate) and twice for 20 min at 65°C with 0.1× SSC; 0.1% (w/v) SDS. Relative transcript abundance to cyclophilin (Marivet et al. 1995) or 18S RNA transcript levels was quantified using a phosphor-imager (Typhoon, Molecular Dynamics, Sunnyvale, CA) with imaging software (ImageQuant, Molecular Dynamics).
Calli and plant transformation
Media used for calli transformation were previously described (Kakimoto 1998). Sterilized A. thaliana seeds were germinated on GM medium and grown under continuous light at 23°C. For the above experiment, hypocotyls from 15-day-old seedlings were cut with a scalpel and placed on CIM (Callus Inducing Medium) for 10–12 days at 23°C under continuous light. Induced calli were soaked in a suspension of Agrobacterium tumefaciens (0.2 OD600) in AIM (Agrobacterium Infection Medium) medium for 5 min. The excess liquid was removed on filter paper, and calli were placed on CIM medium and grown in continuous light at 23°C for 2 days. Calli were then washed thoroughly in washing medium (WASHM) and placed on GM medium plus 100 mg/l cefotaxime, 50 mg/l carbenicilin, 3 mg/l Bialaphos and 0.3 mg/l indolebutyric acid (IBA), or GM medium plus 100 mg/l cefotaxime, 50 mg/l carbenicilin, 3 mg/l Bialaphos, 0.3 mg/l IBA and 1 mg/l trans-zeatin (t-Z) and cultured for 3–4 weeks.
In vivo transformation was achieved using a simplified Arabidopsis transformation protocol (Clough and Bent 1998) with an Agrobacterium tumefaciens strain (LBA4404) containing the 35S-AdhI-ZmIPT2-PinII construct (PHP24559). Seeds were sown in flats containing soil and incubated for 2 days at 4°C to optimize germination. After 10 days, transformants were selected by spraying the seedlings daily with a 1/1,000 dilution of FinaleTM herbicide (Farnam Companies, Inc., Pheonix, AZ) for 5 days.
Protein purification, Western blot and activity assay
The ZmIPT2 gene was amplified using gene specific primers with appropriate NdeI and NotI restriction site extensions (5′-GGCATATGGAGCACGGTGCCGTCGC-3′ and 5′-CCGCGGCCGCTCATCATGCATCAGCCACGGCGGTGA-3′). The resulting PCR product was digested using NdeI and NotI and cloned in pET30b (Novagen) using the same restriction enzymes. The sequence of the resulting plasmid, called PHP26978, was verified and transformed in BL21-Star E. coli competent cells (Invitrogen, Carlsbad, CA). Induction of ZmIPT2 protein synthesis was achieved at 16°C overnight using 1 mM IPTG in 1 l of culture (OD600 = ∼0.4). Bacterial cells were disrupted by sonication and purification of the protein carried out in native condition using a Talon® column (BD Biosciences, San Jose, CA) according to the manufacturer’s guidelines.
Polyclonal antibodies were raised in rabbits by Open Biosystems (Huntsville, AL) against purified C-terminal His-tagged recombinant ZmIPT2 protein. Fifteen micrograms of protein extracted from leaf, stalk, root and whole kernels harvested at different DAP were run using SDS-PAGE and blotted on a PVDF membrane. ZmIPT2 proteins were detected using anti-ZmIPT2 polyclonal antibodies as primary antibodies and anti-rabbit IgG antibodies raised in goat conjugated to an alkaline phosphatase (AP) as secondary antibodies (Sigma, St-Louis, MO) (Laemmli 1970). AP activity was detected using BCIP (5-Bromo-4-Chloro-3′-Indolyphosphate p-toluidine salt) and NBT (Nitro-Blue Tetrazolium Chloride) as substrate (Roche, Mannheim, Germany).
Purified protein was used to determine DMAPP:AMP and DMAPP:ATP isopentenyl transferase activities. Each purified protein extract was incubated in a reaction mixture containing 12.5 mM Tris–HCl (pH 7.5), 37.5 mM KCl, 5 mM MgCl2, 1 mM DMAPP and 1 mM AMP, ADP or ATP for 2 h at 30°C. The reaction was stopped by boiling the samples for 5 min. Half of the reaction mixture was treated with calf intestine alkaline phosphatase (CAIP) by adding one volume of 2× CAIP reaction buffer (0.45 M Tris–HCl pH 9, 10 mM MgCl2, 1,000 unit of CAIP/ml) and incubating for 1 h at 37°C. The reaction products were separated using reversed phase HPLC (Agilent 1100 system with diode-array-detector) using a C18-ODS2 column (Phenomenex) and a separation protocol using 0.1 M acetic acid pH 3.3 (Buffer A) and acetonitrile (Buffer B) as follow: 100% buffer A for 15 min, linear gradient of 1 ml/min. from 100% buffer A and 0% buffer B to 20% buffer A and 80% buffer B over 35 min. UV absorbance was monitored at 280 nm. Product retention times were compared to standards obtained from Sigma (St-Louis, MO) or OlChemIm (Olomouc, Czech Republic).
Immunolocalization of ZmIPT2 in 8 DAP kernels
Eight DAP kernels were embedded in LR White resin using the following procedure. Kernels were sliced longitudinally in 3–4 mm sections. Sections were fixed in 0.05 M phosphate buffer pH 7.2 containing 4% paraformaldehyde, 0.2% glutaraldehyde. Vacuum was applied at 15PSI for 2 h and sections were incubated at 4°C over night. After three washes of 3 h each in 0.05 M phosphate buffer at 4°C, sections were dehydrated in 30, 50, 70 and 95% ethanol for 3 h each, followed by three incubations in 100% ethanol for 3 h each. Infiltration was performed at room temperature using a 3:1 mixture of 100% ethanol and LR White overnight, followed by 1 day in a 1:1 mixture and overnight incubation in a 1:3 mixture of the same ingredients. Finally, sections were transferred to four changes of pure LR White resin of 8 h each at room temperature and cast in Beem capsules at 60°C. The serum containing ZmIPT2 polyclonal antibodies was purified against native purified ZmIPT2 protein immobilized on nitrocellulose. Nitrocellulose was cut in small pieces and washed 3 times in TBS buffer with 0.5% w/v milk powder to remove unbound protein. A total of 300 μl of serum was added to 1.7 ml of TBS/milk buffer containing the nitrocellulose pieces and the mixture incubated at room temperature for 6 h. The nitrocellulose was then washed three times with TBS and antibodies were released using Immunopure Elution buffer (Pierce). After 1 min, the solution was transferred to a microtube containing 1/5 vol of 1 M Tris pH 8 to restore pH to ∼7.5. One-micron sections were placed on drops of water on coverslips and dried on a slide warmer overnight. The localization was performed with the primary antibody at 1:10 and 1:5 dilutions for 2 h at room temperature. The secondary antibody was Alexa Fluor 488 conjugated goat anti-rabbit IgG. Imaging was performed on a Zeiss LSM 510 confocal microscope with a 488 nm laser line.
For quantification, kernels from mid-ear were harvested at 10 DAP, cut in half and fixed (4% paraformaldehyde, 0.1% glutaraldehyde in PBS, pH 7.4) for 1.5 h at room temperature under vacuum and then overnight at 4°C. Samples were rinsed in PBS, dehydrated in an ethanol series, infiltrated and cured in LR White resin. Sections were immunogold labeled followed by silver enhancement and imaged by electron microscopy.
Distribution of materials
Novel materials described in this publication may be available for non-commercial research purposes upon acceptance and signing of a material transfer agreement. In some cases such materials may contain or be derived from materials obtained from a third party. In such cases, distribution of material will be subject to the requisite permission from any third-party owners, licensors or controllers of all or parts of the material. Obtaining any permission will be the sole responsibility of the requestor. Transgenic Arabidopsis material will be made available at the discretion of the owner based on seed availability in accordance with all applicable governmental regulations.
Results
Identification of putative isopentenyltransferase genes from maize
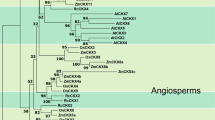
We used a computational method to identify putative CK biosynthetic genes from Zea mays based on similarity with Arabidopsis IPT proteins (Kakimoto 2001; Takei et al. 2001). Nine maize contigs encoding putative CK biosynthetic enzymes were identified (ZmIPT1–ZmIPT9) (see Supplemental Fig. 1S). ZmIPT1 was previously identified as an EST and the genomic sequence obtained from a proprietary Bacterial Artificial Chromosome (BAC) clone. The sequence was later found to have a coding region containing nine introns that was identical to ZmIPT3. The numbering of the genes was not changed for practical reasons. The ZmIPT9 contig did not display an ORF and therefore was dropped from further analysis. The remaining six contigs (ZmIPT2, 4, 5, 6, 7 and 8) showed an open-reading frame without introns. Whereas both Arabidopsis tRNA IPT genes possess several introns in their genomic coding region, only one cytokinin biosynthetic gene, AtIPT8, has an intron (Sun et al. 2003; Miyawaki et al. 2006). The gene structure of ZmIPT2, 4, 5, 6, 7 and 8 is therefore similar to the gene structure of most Arabidopsis CK biosynthetic genes. The size of the translated proteins corresponding to these putative maize genes ranged from 322 to 383 amino acids, which is similar to the reported size for other plant IPT proteins (Kakimoto 2001; Takei et al. 2001; Zubko et al. 2002). Accession numbers for the ZmIPT genes and corresponding MAGI and AZM contigs are given in Supplemental Table 1S. An alignment of the corresponding deduced protein sequences with AtIPT proteins is presented in Fig. 1. All the deduced proteins but ZmIPT8 contain a consensus pattern similar to the pattern used by Kakimoto (2001) to identify Arabidopsis IPT proteins (Fig. 1; asterisks). For ZmIPT8, aa number 14 of the consensus sequence was an A (another hydrophobic aa) instead of [VLI]. A putative ATP/GTP binding site (P-loop motif) was identified in all deduced protein sequences (Fig. 1; underlined aa). ZmIPT1 showed an additional ∼75 to 80 aa sequence in the middle of the alignment compared to other ZmIPT proteins. This feature had been reported for AtIPT2, a tRNA IPT (Kakimoto 2001; Takei et al. 2001). In addition, a typical consensus sequence found in tRNA IPT proteins (CxxCx(12,18)HxxxxxH) was identified in ZmIPT1. Eight putative IPT genes were recently identified in the rice genome (Sakamoto et al. 2006). The full-length protein sequences from Arabidopsis and rice IPT were aligned with ZmIPT proteins in order to generate a phylogenetic tree (Fig. 2) which would allow for the identification of the best Arabidopsis and rice ortholog(s) of each maize gene. Three major clades could be identified. One of them consisted of the AtIPT2, AtIPT9 tRNA IPT proteins and the putative tRNA IPT proteins ZmIPT1, OsIPT9 and OsIPT10. ZmIPT1 was 69.1% identical to OsIPT9 (Sakamoto et al. 2006) and the corresponding genes share a similar genomic structure with nine introns (Sakamoto et al. 2006) suggesting that it could play a role in tRNA isoprenylation and the production of cis-zeatin type CKs in maize (Miyawaki et al. 2006). OsIPT10 and AtIPT9 clustered separately from the other 3 tRNA IPT proteins indicating that the corresponding genes might have evolved independently from a common ancestor before speciation. Each of the remaining 2 clades consisted of maize, rice and Arabidopsis proteins with maize and rice proteins and Arabidopsis proteins clustering independently. It is unclear if this could represent functional differences between each group of proteins. The closest rice orthologs to maize ZmIPT2 were OsIPT1 and OsIPT2.
Alignment of maize putative IPT proteins (ZmIPT) with Arabidopsis IPT proteins (AtIPT). The positions of amino acid following the consensus pattern GxTxxGK[ST]xxxxx[ALI]x(7,8)[VI][VI]xxDxxQx(60,61)[VI][VLI]xGG[ST] similar to the one described by Kakimoto (2001) for other IPT proteins (where × denotes any amino acid residue, [ ] anyone of the amino acids shown in [ ], and x(m,n) m to n amino acid residues in number) are indicated by asterisks. Putative ATP/GTP-binding sites (P-loop) motif (prosite PS00017: consensus [AG]x(4)GK[ST]), are underlined. The tRNA binding site (CxxCx(12,18)HxxxxxH) of ZmIPT1 is denoted by black dots. Amino acids highlighted in yellow, blue and green respectively represent residues completely conserved, partially conserved and similar to consensus at a given position
Phylogenetic tree calculated using ZmIPT, OsIPT and AtIPT full-length proteins. Results indicate that tRNA IPT proteins of each species cluster together and in a separate clade than CK biosynthetic proteins. The tree also indicates strong similarities between ZmIPT1 and the tRNA IPT OsIPT9 and similarities between ZmIPT2 and OsIPT1 and OsIPT2 both expressed in rice flowers. Bar = 0.1 amino acid substitution per site
ZmIPT2 is strongly expressed in developing kernels
Transcript levels of ZmIPT genes were examined in our Massively Parallel Signature Sequencing (MPSS) Lynx libraries (Brenner and Corcoran 2000) representative of all corn tissues and were found to average 20 ppm except for ZmIPT2 in endosperm tissue (∼600 ppm) indicating an overall very low level of expression of ZmIPT genes. The elevated expression of ZmIPT2 in endosperm tissue of developing kernels suggested a possible involvement of the gene in CK biosynthesis, and the expression of this gene was therefore characterized in more detail.
The identification of proprietary B73 and Mo17 BAC clones for ZmIPT2 allowed us to position it on the physical map of maize. The gene was located on chromosome 2, bin 4. PCR of maize-oat addition lines together with Southern blot analysis confirmed the presence of only one gene on chromosome 2 (Supplemental Figs. 1S, 2S). ZmIPT2 was amplified from B73 genomic DNA and the amplified product cloned, sequenced and used as a probe for Northern blot analysis. Figure 3a shows the expression pattern of ZmIPT2 in root, leaf, stalk and developing kernels determined by Northern blot. Expression was quantified by normalizing ZmIPT2 expression to cyclophilin levels (Marivet et al. 1995). ZmIPT2 showed low expression levels in vegetative organs and 0 DAP kernels. Expression was found to be higher from 5 to 25 DAP with maximum expression attained at 10 DAP, which coincides with the peak of CK reported in developing kernels (Cheikh and Jones 1994; Dietrich and Morris 1995; Brugière et al. 2003). Because detection of IPT activity in developing kernels was found to be problematic (Blackwell and Horgan 1994), we decided to raise antibodies against the ZmIPT2 protein and examine protein levels in different organs. ZmIPT2 antibodies were raised in rabbits against a C-terminal His-tagged recombinant protein (see below). Results from the Western blot (Fig. 3b) show that ZmIPT2 protein levels parallel steady-state transcript levels and are strongest in developing kernels where it peaks at 10 DAP. Very low but detectable levels were found in leaf, stem and root. Altogether these data suggest that the ZmIPT2 protein level is likely controlled at the transcriptional level.
Expression of ZmIPT2 detected by Northern and Western blot analyses. (a) Northern blot and relative expression of ZmIPT2 in different vegetative organs and in whole kernels at different DAP. Transcript levels were measured in leaves (L), stalks (S), roots (R), and in whole kernels at 0, 5, 10, 15, 20 and 25 DAP, and quantified relative to abundance of cyclophilin transcripts. Thirty micrograms of total RNA were loaded in each lane (b) Western blot with protein extracts from vegetative organs and developing kernels. ZmIPT2 (arrow) was detected using anti-ZmIPT2 antibodies. Thirty micrograms of protein were loaded in each lane
ZmIPT2 expression is developmentally regulated in kernel tissues and coincides with cytokinin accumulation
To obtain a more precise view of the expression pattern of ZmIPT2 in kernels, steady-state levels of ZmIPT2 transcripts were measured in 0- to 5-DAP whole kernels and starting at 6 DAP the lower part of the kernel, consisting of the PPCBE region, was separated from the rest of the kernel. Analysis of the separate samples allowed for a comparison of ZmIPT2 expression with ZmCkx1 (Zea mays cytokinin oxidase 1) expression, which was found to be induced by CK in a previous study (Brugière et al. 2003). Figure 4 shows ZmIPT2 transcript levels normalized to cyclophylin transcript levels compared to ZR levels. Low levels of ZmIPT2 transcripts were detected in whole kernels between 0 and 5 DAP (Fig. 4a). In the upper part of the kernels, the expression gradually increases from 6 to 8 DAP, peaks at 8 DAP, and decreases gradually until 14 DAP before increasing again at later stages (15–34 DAP) (Fig. 4a). In the lower part of the kernel (Fig. 4b), transcript levels increase sharply from 6 to 10 DAP, peak at 10 DAP, and slowly decrease until 15 DAP. In the graph of Fig. 4b, the sample corresponding to the upper part of the seed at 9 DAP (S-9) was used as an internal control to allow the comparison with expression in the lower part of the seed at the same date. The relative expression of ZmIPT2 at 9 DAP is four times greater in the latter than in the rest of the seed. This difference is consistent with the fact that CK levels are nearly twice as abundant in the lower part of the kernel as in the rest of the seed (Brugière et al. 2003). When compared to ZR concentrations (red lines of Fig. 4) measured in the same samples used for Northern analysis, ZmIPT2 expression nicely parallels ZR accumulation in the lower part of the kernel (Fig. 4b), whereas in the rest of the kernel (Fig. 4a), it slightly precedes the peak in ZR concentration and decreases several days before ZR levels decrease. Altogether these results indicate that ZmIPT2 is expressed transiently during kernel development both in the lower part and the rest of the seed, and this expression pattern, which parallels ZR levels in the kernel, is consistent with the hypothesis that it is a CK biosynthetic gene.
Expression of ZmIPT2 and ZR levels in developing kernels. (a) Transcript levels were measured in 0–5 DAP whole kernels and in 6–34 DAP kernels without the pedicel/placental chalazal/basal endosperm (PPCBE) region. (b) Transcript levels measured in the PPCBE region between 6 and 42 DAP. The sample corresponding to the upper part of the seed at 9 DAP (S-9) was used to allow the comparison with expression in the lower part of the seed at the same date. Thirty micrograms of total RNA were loaded for each sample. Graphs display ZmIPT2 transcript levels relative to cyclophilin transcript levels. ZR levels (the most abundant CK in corn kernels) were previously measured in the same samples (Brugière et al. 2003) and are displayed as a red line
We decided to further characterize ZmIPT2 by looking at its expression pattern in dissected kernel samples from 0 to 25 DAP. B73 samples were collected and dissected into pedicel/placental chalazal/transfer cell region, nucellus, starchy endosperm/embryo sac, starchy endosperm, embryo and pericarp depending on the stage considered. The results shown in Fig. 5 confirm that ZmIPT2 transcript levels in the pedicel/placental chalazal/transfer cell region are more abundant than in the rest of the kernel. This is especially true at 15, 20 and 25 DAP where low expression is also detected in embryo samples. Strong ZmIPT2 expression was observed in the starchy endosperm/embryo sample at 10 DAP which coincides with the time of maximum cell division in the endosperm (Kowles and Phillips 1985). At 10 DAP, embryo cells are difficult to separate from the starchy endosperm and represent a very small percentage of the cells in this tissue (Kiesselbach 1949). Expression of ZmIPT2 in embryo at 10 DAP cannot be ruled out; however, immuno-localization results described later in this study, demonstrate the presence of the protein in the endosperm confirming that ZmIPT2 is expressed in endosperm tissue at this stage of development. Expression was absent from the nucellus and was low in the developing embryo at 15, 20 and 25 DAP compared to the pedicel/placental chalazal/transfer cell region and the starchy endosperm/embryo sample at 10 DAP. In the pericarp, expression was detectable at 5 and 10 DAP but undetectable at later stages. These results indicate that the pedicel/placental chalazal/transfer cell region is most likely a strong site of CK biosynthesis. The presence of ZmIPT2 transcripts in the endosperm/embryo sample at 10 DAP and in developing embryo at 15, 20 and 25 DAP was observed at times when cell division is the most active in these tissues. This again supports a role for ZmIPT2 as a CK biosynthetic protein, which would catalyze CK synthesis in fast dividing/developing tissues, but is also present in the pedicel/placental chalazal/transfer cell region where it could have a different physiological role.
Expression of ZmIPT2 in different tissues during kernel development. Transcript levels were measured in 0–25 DAP dissected kernels. “Pedicel region” refers to the pedicel/placental chalazal/transfer cells and “endosperm” refers to the starchy endosperm. Thirty micrograms of total RNA were loaded for each sample. The nylon membrane was stripped and re-hybridized with 18S RNA probe as a loading control
The ZmIPT2 protein is localized in the endosperm and is more abundant in endosperm transfer cells at 8–10 DAP
In order to identify the site of ZmIPT2 expression at the cellular level, we used sections of 8 DAP kernels to detect ZmIPT2 using immunolocalization with purified polyclonal antibodies raised against the protein. At this stage, the endosperm is quickly expanding (Olsen and Nichols 1999) and elevated CK levels have been detected (Cheikh and Jones 1994; Dietrich and Morris 1995; Brugière et al. 2003). Figure 6 shows that the protein can be found in the cytosol of endosperm cells with a stronger signal detected in the basal endosperm transfer cells (Fig. 6a, b). This signal could not be detected in the pedicel per se which is easily identifiable by the red autofluorescence of its cell walls. The control with preimmune serum showed weak cross reactivity localized on endosperm cell membranes but not in the endosperm cell cytosol (Fig. 6c) whereas control without primary antibody showed no signal (Fig. 6d). After immunogold labeling followed by silver enhancement, gold particles were found to be 5 times more abundant in transfer cells relative to starchy endosperm cells and 10 times greater in transfer cells relative to pedicel cells. The data indicate that the expression of ZmIPT2 observed in the pedicel/placental chalazal/transfer cell region at 10 DAP by Northern analysis is due to strong expression of the gene in the endosperm transfer cell layer.
Immunolocalization of ZmIPT2 protein in an 8 DAP kernel. (a, b) One micron sections of 8 DAP kernels embedded in LR white resin were used to detect ZmIPT2 (green fluorescence) with purified polyclonal antibodies raised against the protein as primary antibody. Cell walls (red autofluorescence) are clearly visible. (c) Result of immunolocalization with unpurified preimmune serum as primary antibody. (d) Result of immunolocalization with omitted primary antibody. (e) Gold particle counts in each cell type ± standard deviation (n = 74 for transfer cells, n = 22 for endosperm cells and n = 26 for pedicel cells). en = endosperm, tc = transfer cell, p = pedicel. Bar is 1 mm in (a) and (c) and 0.5 mm in (b) and (d)
ZmIPT2 over-expression in Arabidopsis calli and plants
To determine if the ZmIPT2 gene product functions as a CK biosynthetic enzyme, we used a callus transformation protocol which had been previously used to demonstrate the function of Arabidopsis IPT proteins (Kakimoto 1998, 2001). A construct was made consisting of the 35S promoter fused to the first intron of the maize alcohol dehydrogenase (ADH1) gene, fused to the ZmIPT2 coding sequence and a terminator. The resulting construct (35S-ADH1-ZmIPT2) was transformed into 10 day-old Arabidopsis calli which were transferred onto germination medium (GM) containing either auxin (IBA) or both auxin and CK (t-Z), as well as Bialaphos as the selection agent for transformed calli. A control construct, consisting of the 35S promoter fused to E. coli β-glucuronidase gene (GUS) followed by a terminator, was used to validate the transformation process (data not shown). Similarly a positive control construct was prepared consisting of the 35S promoter fused to the A. tumefaciens IPT gene followed by a terminator (35S-IPT). A clear phenotype could be observed 3 weeks after transformation and images of three representative regenerating calli per treatment were taken 4 weeks after transformation (Fig. 7). 35S-GUS, 35S-ADHI-ZmIPT2 and 35S-IPT calli grew identically on GM medium containing both auxin and CK (Fig. 7a). As expected, control calli transformed with the 35S-GUS construct were able to regenerate roots on GM medium containing only auxin (Fig. 7b). In contrast, calli transformed with the 35S-ADH1-ZmIPT2 construct, like calli transformed with the 35S-IPT construct, did not form any roots on this medium but were able to regenerate shoots (Fig. 7b). This implies that these calli are overproducing CKs due to the expression of the ZmIPT2 gene, which in turn decreases the auxin/CK ratio, thus preventing root formation (Kakimoto 2000). Identical results were obtained with two other putative CK biosynthetic genes, ZmIPT7 and ZmIPT8 (data not shown). To further prove that ZmIPT2 encodes a CK biosynthetic enzyme, we transformed the 35S-ADH1-ZmIPT2 construct into Arabidopsis. Fifteen days after selection of herbicide resistant transformants, several 35S-ADH1-ZmIPT2 plants had dark-green leaves and were strongly delayed in growth (Fig. 8a) compared to 35S-GUS transgenic controls (Fig. 8b). After 4 weeks, these plants were still delayed in growth and had shorter rosettes and serrated leaves (Fig. 8c, d) compared to controls (Fig. 8g). Some transformants showed an apparent normal growth pattern but displayed decreased apical dominance (Fig. 8f) compared to control (Fig. 8g). The most extreme phenotype was a transgenic event with a rosette of approximately 5 mm in diameter with very small curly leaves (Fig. 8e) covered by a large density of trichomes (Fig. 8h). Overall, over-expression of ZmIPT2 in Arabidopsis resulted in transgenic plants with phenotypes indicative of CK over-production.
Phenotypes of calli transformed with 35S-Adh1-ZmIPT2, 35S-GUS and 35S-IPT after transfer to medium containing (a) 300 ng/ml of IBA and 1,000 ng/ml of t-Z or (b) 300 ng/ml of IBA. Hypocotyls were grown on callus inducing medium and transformed with each construct. After transformation, calli were grown for 4 weeks on GM medium containing Bialaphos as the selection agent. Calli transformed with 35S-AdhI-ZmIPT2 construct, like calli transformed with the 35S-IPT construct did not regenerate roots on GM medium with auxin compared to transgenic controls 35S-GUS
Phenotypes of Arabidopsis plants transformed with 35S-AdhI-ZmIPT2. After selection of herbicide resistant transformants, several 35S-ADH1-ZmIPT2 plants had dark-green leaves and were strongly delayed in growth (a, red arrow) compared to 35S-GUS transgenic controls (b) (Bar = 5 mm). After 4 weeks, these plants were still delayed in growth and had shorter rosettes and serrated leaves (c, d) compared to control (g) (Bar = 2 cm). Transformants showing normal growth displayed decreased apical dominance (f) compared to control (g) (Bar = 2 cm). The most extreme phenotype was a transgenic event with a rosette of approximately 5 mm in diameter with very small curly leaves (e) (Bar = 2.5 mm) covered by a large density of trichomes (h) (Bar = 0.5 mm)
Determination of ZmIPT2 isopentenyl transferase activity in vitro
To further confirm the CK biosynthetic function of the ZmIPT2 gene product, we created a construct to express and purify ZmIPT2 as a C-terminal His-tagged recombinant protein. The protein was expressed in E. coli and purified using a Talon® column (Fig. 9a). The protein was found to be 95–100% pure based on quantification analysis of the Coomassie stained gel. A Western blot using antibodies raised against the 6× His tag was used to verify that the protein was appropriately tagged (data not shown). DMAPP:AMP and DMAPP:ATP transferase activity were assayed using purified ZmIPT2 protein. Figure 9b (top chromatogram) shows the profile obtained from the reaction mixture of ZmIPT2 purified enzyme incubated with DMAPP and 5′-AMP. The reaction product had the same retention time as isopentenyl adenosine mono-phosphate (iPMP)(Fig. 9b, middle chromatogram). Moreover, after alkaline phosphatase treatment, Ado (coming from the dephosphorylation of 5′-AMP) and isopentenyl adenosine (iPAR) were obtained confirming the identity of the reaction product as iPMP (Fig. 9b, lower chromatogram). The results show that the ZmIPT2 gene product is able to convert 5′-AMP to iPMP but with low efficiency as evidenced by the remaining 5′-AMP. The same reaction was carried out using 5′-ATP (Fig. 9c). After the reaction only one product was found by chromatography suggesting that all the 5′-ATP had been converted by the enzyme to isopentenyl adenosine tri-phosphate (iPTP) (Fig. 9c, upper chromatogram). The dephosphorylation product was found to have the same retention time as the iPAR standard (Fig. 9c, middle and lower chromatogram) consistent with the product of the reaction being iPTP. Similar results were obtained with 5′-ADP as a substrate and 1-Hydroxy-2-methyl-2-buten-4-yl 4-diphosphate (HDMAPP) was not a substrate for the enzyme (data not shown). Altogether these results prove that ZmIPT2 is a maize CK biosynthetic enzyme that functions as a DMAPP-isopentenyl transferase preferentially using 5′-ATP/ADP as substrates over 5′-AMP.
Purification of ZmIPT2 recombinant protein and determination of isopentenyl transferase activity. (a) Purification of His-tagged ZmIPT2 recombinant protein. Cells were lysed and separated into pellet (1) and supernatant (2) fraction by centrifugation. The supernatant was loaded on a Talon column and flow through collected (3). Recombinant protein was eluted using 10 mM imidazole (4), 20 mM imidazole (5), 50 mM imidazole (6) and 100 mM imidazole (7). Fractions 4–7 contained 82–100% pure recombinant ZmIPT2 protein based on Coomassie staining. (b) Determination of DMAPP:AMP isopentenyl transferase activity of recombinant ZmIPT2 protein. (c) Determination of DMAPP:ATP isopentenyl transferase activity of recombinant ZmIPT2 protein. iPMP = isopentenyl adenosine mono-phosphate, iPAR = isopentenyl adenosine, Ado = adenosine, iPTP = isopentenyl adenosine tri-phosphate
Discussion
IPT proteins are encoded by a multigenic family in Arabidopsis (Kakimoto 2001; Takei et al. 2001) and rice (Sakamoto et al. 2006). In addition, IPT genes have been identified in petunia (Zubko et al. 2002), hop (Sakano et al. 2004), soybean (Ye et al. 2006) and pea (Tanaka et al. 2006). Using a computational approach we identified a multigene IPT family from maize with corresponding deduced amino acid sequences containing a consensus sequence (Fig. 1) found in most tRNA IPT and IPT proteins (Takei et al. 2001).
Phylogenetic analysis identified OsIPT1 and OsIPT2 as the closest rice orthologs to ZmIPT2. These two genes have a specificity of expression towards rice flowers (Sakamoto et al. 2006) but their expression during rice grain development has not yet been documented. Three other ZmIPT genes, ZmIPT5, 7 and 8 were found to have low expression in developing kernels (unpublished data) suggesting that more than one gene may be responsible for CK synthesis in this organ. The expression pattern of ZmIPT2 in kernels is similar to the documented expression of AtIPT4 and AtIPT8 in the chalazal endosperm (CZE) of developing Arabidopsis seeds soon after fertilization (Miyawaki et al. 2004). However, whereas AtIPT4 and AtIPT8 were found to be addressed to plastids (Kasahara et al. 2004), immunolocalization of ZmIPT2 in 8 DAP kernels and results of an analysis using a subcellular localization prediction program (WoLF PSORT) indicate that the protein is most likely cytosolic.
ZmIPT2 is transiently expressed during kernel development and reaches maximum expression at 10 DAP (Fig. 4). This expression pattern coincides with the peak in rate of cell division (Lur and Setter 1993) and the peak of CK accumulation in the endosperm during kernel development (Fig. 4a; Jones et al. 1992; Lur and Setter 1993; Dietrich and Morris 1995). We believe that ZmIPT2 plays an important role in controlling endogenous CK levels necessary for active endosperm cell division. We hypothesize that the intensity and expression pattern of ZmIPT2, in conjunction with other hormones such as auxin and abscisic acid (ABA), could dictate the number of endosperm cells and amyloplasts established in the early phase of kernel development. These two parameters have previously been associated with kernel growth and size at maturity (Brenner and Cheikh 1995) and the manipulation of ZmIPT2 expression could increase our understanding of the role of CKs in the control of yield in cereal crops.
In the pedicel/placental chalazal/basal endosperm region, CK levels are 2–3 times higher than in the rest of the seed (Brugière et al. 2003). The expression of ZmIPT2 in this tissue not only parallels CK accumulation but comparative values of ZmIPT2 transcript levels between this region and the rest of the seed (Fig. 4b) are in agreement with CK level differences between the two tissues. Results of the immunolocalization of ZmIPT2 at 8 DAP indicate that the protein is below the limit of detection in the pedicel but is present at a high concentration in the endosperm transfer cell layer and at lower concentration in the starchy endosperm (Fig. 6). We demonstrated that ZmIPT2 is active as a DMAPP:ATP/ADP isopentenyl transferase which indicates a major role for ZmIPT2 in CK biosynthesis in developing kernels. ZmIPT2 most likely plays a role in endosperm cell division during the peak of mitotic activity in this tissue. But the presence of the protein in the endosperm transfer cell layer both during this period and at later stages of kernel development strongly suggests that it could also play a additional role in kernel sink-strength.
The expression of both maize cytokinin oxidase/dehydrogenase (ZmCkx) (Brugière et al. 2003; Massonneau et al. 2004; Galuszka et al. 2005) and ZmIPT2 in developing kernels suggest that regulation of CK levels is achieved by a balance between CK biosynthesis and degradation. Although other ZmCkx genes have been found to be expressed in developing kernels (Massonneau et al. 2004; Brugière et al. unpublished data), purification of a protein fraction with cytokinin oxidase/dehydroganase activity and micro-sequencing of the protein only identified one gene (ZmCkx1) (Houba-Herin et al. 1999; Morris et al. 1999) suggesting that ZmCkx1 is the most abundant cytokinin oxidase/dehydrogenase in developing seeds. Our previous analysis of ZmCkx1 expression showed that, at 8 DAP, the gene is expressed in the pedicel as well as the lower portion of the endosperm, and that its expression can be induced by CKs in 4 DAP kernels (Brugière et al. 2003). Our current data indicate that ZmCkx1 and ZmIPT2 are most likely the main players in the control of CK homeostasis in developing kernels. Regulation of the expression of the corresponding genes by environmental conditions could represent a means for the plant to regulate kernel growth under unfavorable growing conditions. For example, under either drought or heat stress when there is a reduction in photo-assimilates availability, ZmCkx1 induction in the kernel (Brugière et al. 2003) would result in reduced CK levels limiting sink strength and kernel development. It is possible that different pairs of ZmCkx and ZmIPT genes act in concert to control CK homeostasis in different tissues.
Because CKs have been associated with active cell division, growth and metabolism, a link between levels of this plant hormone and sink-strength and source-sink transition through regulation of photo-assimilate partitioning has been postulated (Kuiper 1993; Brenner and Cheikh 1995; Roitsch and Ehness 2000). Cytokinins are often found in high concentration in the endosperm of developing seeds where active cell division takes place, which needs to be fueled by an active supply of carbohydrates (Brenner and Cheikh 1995). In maize, we have found that stronger levels of CK can be found in both the pedicel/placental chalazal/transfer cell layer than in the rest of the seed (Fig. 4; Brugière et al. 2003), and that CKs are more abundant in the former tissue than in the latter. In maize, the pedicel and BETL are strongly associated with photo-assimilate unloading and sink-strength (Hanft and Jones 1986; Miller and Chourey 1992). Source–sink transitions are known to be promoted by CK, and recently extra-cellular invertase has been found to be an essential component of CK-mediated delay of senescence (Balibrea-Lara et al. 2004). In maize, the mn1 mutant affects insoluble invertase INCW2 which was found to suppress the cleavage of sucrose into fructose and glucose in both the pedicel and the BETL, resulting in aberrant pedicel and endosperm development (Miller and Chourey 1992). Cytokinins have been shown to stimulate unloading of photo-assimilates from excised bean seed coats (Clifford et al. 1986). Moreover, in Chenopodium rubrum and tomato, extracellular invertase transcript levels are highly up-regulated by physiological concentrations of CKs (Roitsch and Ehness 2000), and the similarity in expression timing and localization of Incw2 (Miller and Chourey 1992) and ZmIPT2 in the BETL in maize kernels is striking. Altogether these data suggest that CK synthesis by ZmIPT2 in the endosperm transfer cell layer, the starchy endosperm and in the developing embryo could not only play a role in cell-division but could also function in maintaining sink-strength during kernel development. We are currently studying a Mutator-tagged mutant of ZmIPT2 which should help shed light on the importance of CK biosynthesis in each function as well as its possible role on overall assimilate partitioning and grain filling capacity.
Abbreviations
- BETL:
-
Basal endosperm transfer layer
- cis-Z:
-
cis-zeatin
- IPT:
-
Isopentenyl transferase
- iPAR:
-
Isopentenyl adenosine
- iPMP:
-
Isopentenyl adenosine mono-phosphate
- iPTP:
-
Isopentenyl adenosine tri-phosphate
- PPCBE:
-
Pedicel/placental chalazal/basal endosperm
- t-Z:
-
trans-zeatin
- ZmIPT2:
-
Zea mays isopentenyl transferase 2
- ZR:
-
Zeatin riboside
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309:741–745
Balibrea Lara ME, Gonzalez Garcia M-C, Fatima T, Ehness R, Lee TK, Proels R, Tanner W, Roitsch T (2004) Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 16:1276–1287
Blackwell JR, Horgan R (1994) Cytokinin biosynthesis by extracts of Zea mays. Phytochemistry 35:339–342
Brenner M, Cheikh N (1995) The role of hormones in photosynthate partitioning and seed filling. In: Davies PJ (ed) Plant hormones. Kluwer, Dordrecht, The Netherlands, pp 649–670
Brenner S, Corcoran K (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18:630–634
Brugière N, Dubois F, Limami AM, Lelandais M, Roux Y, Sangwan RS, Hirel B (1999) Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 11:1995–2012
Brugière N, Jiao S, Hantke S, Zinselmeier C, Roessler JA, Niu X, Jones RJ, Habben JE (2003) Cytokinin oxidase gene expression in maize is localized to the vasculature, and is induced by cytokinins, abscisic acid, and abiotic stress. Plant Physiol 132:1228–1240
Cheikh N, Jones RJ (1994) Disruption of maize kernel growth and development by heat stress. Plant Physiol 106:45–51
Clifford PE, Offler CE, Patrick JW (1986) Growth regulators have rapid effects on photosynthate unloading from seed coats of Phaseolus vulgaris L. Plant Physiol 80:635–637
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dietrich J, Morris R (1995) Changes in cytokinins and cytokinin oxidase activity in developing maize kernels and the effects of exogenous cytokinin on kernal development. Plant Physiol Biochem 33:327–336
Dyer DJ, Cotterman JD, Cotterman CD, Kerr PS, Carlson DR (1988) Cytokinins as metabolic stimulants which induce pod set. In: Pharis RP, Rood SB (eds) Plant growth substances. Springer-Verlag, New York, NY, pp 457–467
Felsenstein J (1989) PHYLIP—Phylogeny inference package (Version 3.2). Cladistics 5:164–166
Galuszka P, Frebortova J, Luhova L, Bilyeu KD, English JT, Frebort I (2005) Tissue localization of cytokinin dehydrogenase in maize: possible involvement of quinone species generated from plant phenolics by other enzymatic systems in the catalytic reaction. Plant Cell Physiol 46:716–728
Ghiasi H, Dybing CD, Paech C (1987) Free amino acid content and metabolic activities of setting and aborting soybean ovaries. Plant Physiol 81:1069–1074
Hanft JM, Jones RJ (1986) Kernel abortion in Maize: I. Carbohydrate concentration patterns and acid invertase activity of maize kernels induced to abort in vitro. Plant Physiol 81:503–510
Houba-Herin N, Pethe C, d’Alayer J, Laloue M (1999) Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J 17:615–626
Huff A, Dybing CD (1980) Factors affecting shedding of flowers in soybean (Glycine max (L.) Merrill). J Exp Bot 31:751–762
Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15:1560
Jones RJ, McNeil K, Brenner ML, Foxon G (1992) Cytokinin levels and oxidase activity during maize kernel development. In: Kaminek M, Mok D, Zazimalova E (eds) Physiology and biochemistry of cytokinins in plants. SPB Academic Publishing bv, The Hague, The Netherlands, pp 235–239
Kakimoto T (1998) Genes involved in cytokinin signal transduction. J Plant Res 111:261–265
Kakimoto T (2000) CkI1, a histidine kinase homolog inplicated in cytokinin signal transduction. Science 274:1–8
Kakimoto T (2001) Identification of plant biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltranferases. Plant Cell Physiol 42:677–685
Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, Yamaguchi S, Sakakibara H (2004) Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J Biol Chem 279:14049–14054
Kiesselbach TA (1949) The structure and reproduction of corn. Univ Nebr Agric Exp Stn Res Bull 161:1–96
Kowles RV, Phillips RL (1985) DNA amplification patterns in maize endosperm nuclei during kernel development. Proc Natl Acad Sci USA 82:7010–7014
Kuiper D (1993) Sink strength: established and regulated by plant growth regulators. Plant Cell Environ 16:1025–1026
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lur H-S, Setter T (1993) Role of auxin in maize endosperm development. Plant Physiol 103:273–280
Marivet J, Frendo P, Burkard G (1995) DNA sequence analysis of a cyclophilin gene from maize: developmental expression and regulation by salicylic acid. Mol Gen Genet 247:222–228
Massonneau A, Houba-Hérin N, Pethe C, Madzak C, Falque M, Mercy M, Kopecny D, Majira A, Rogowsky P, Laloue M (2004) Maize cytokinin oxidase genes: differential expression and cloning of two new cDNAs. J Exp Bot 55:2549–2557
Miller ME, Chourey PS (1992) The maize invertase-deficient miniature-1 seed mutation is associated with aberrant pedicel and endosperm development. Plant Cell 4:297–305
Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37:128–138
Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103:16598–16603
Mok MC (1994) Cytokinins and plant development—an overview. In: Mok DWS, Mok MC (eds) Cytokinins—Chemistry, activity and function. CRC Press, Boca Raton, FL, pp 155–166
Morris R, Bilyeu K, Laskey JG, Cheikh N (1999) Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem Biophys Res Commun 255:328–333
Mosjidis COH, Peterson CM, Truelove B, Dute RR (1993) Stimulation of pod and ovule growth of soybean, Glycine max (L.) Merr., by 6-benzylaminopurine. Ann Bot 71:193–199
Nagel L, Brewster R, Riedell WE, Reese N (2001) Cytokinin regulation of flower and pod set in Soybeans (Glycine max (L.) Merr.). Ann Bot 88:27–31
Olsen O-A, Nichols S (1999) Developmental biology of the cereal endosperm. Trends Plant Sci 4(7):253–257
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Peterson CM, Williams JC, Kuang A (1990) Increased pod set of determinate cultivars of soybean, Glycine max, with 6-benzylaminopurine. Bot Gaz 151:322–330
Reese RN, Dybing CD, White CA, Page SM, Larson JE (1995) Expression of vegetative storage protein (VSP-{beta}) in soybean raceme tissues8. J Exp Bot 46:957–964
Roitsch T, Ehness R (2000) Regulation of source/sink relations by cytokinins. Plant Growth Regul 32:359–367
Sakakibara H (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57:431–449
Sakakibara H, Takei K (2002) Identification of cytokinin biosynthesis genes in Arabidopsis: a breakthrough for understanding the metabolic pathway and the regulation in higher plants. J Plant Growth Regul 21:17–23
Sakamoto T, Sakakibara H, Kojima M, Yamamoto Y, Nagasaki H, Inukai Y, Sato Y, Matsuoka M (2006) Ectopic expression of KNOX homeodomain protein induces expression of cytokinin biosynthesis gene in rice. Plant Physiol 142:54–62
Sakano Y, Okada Y, Matsunaga A, Suwama T, Kaneko T, Ito K, Noguchi H, Abe I (2004) Molecular cloning, expression, and characterization of adenylate isopentenyl transferase from hop (Humulus lupulus L.). Phytochemistry 65:2439–2446
Sun J, Niu Q-W, Tarkowski P, Zheng B, Tarkowska D, Sandberg G, Chua N-H, Zuo J (2003) The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol 131:167–176
Takei K, Sakakibara H, Sugiyama T (2001) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276:26405–26410
Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45:1053–1062
Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45:1028–1036
Werner T, Motyka V, Strnad M, Schmulling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 171:1–6
Wiebold WJ (1990) Rescue of soybean flowers destined to abscise. Agron J 82:85–88
Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N (2005) Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15:1566
Ye C, Wu S, Kong F, Zhou C, Yang Q, Sun Y, Wang B (2006) Identification and characterization of an isopentenyltransferase (IPT) gene in soybean (Glycine max L.). Plant Sci 170:542–550
Zubko E, Adams CJ, Machaekova I, Malbeck J, Scollan C, Meyer P (2002) Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J 29:797–808
Acknowledgments
The authors are indebted to Tom Davis for his help with ZmIPT2 recombinant protein purification and to Nic Bate and Shoba Sivasankar for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1S
ZmIPT Genbank accession numbers and corresponding MAGI and AZM contigs (XLS 22 kb)
Supplemental Fig. 1S
Southern blot using B73 and Mo17 genomic DNA and ZmIPT2 32P-dCTP as a probe. Only one band could be detected after hybridization when HindIII (H), EcoRI (E) or EcoRV (V) restriction enzymes were used suggesting that the gene is present at a single copy per haploid genome (PPT 925 kb)
Supplemental Fig. 2S
Chromosomic location of ZmIPT2 determined by PCR using oat-maize addition lines. ZmIPT2 was only found in the DNA of the oat-maize addition line containing maize chromosome 2 (OMAL 2). Position was further determined as chromosome 2 bin 4 using BAC clones and a proprietary genetic physical map (PPT 63 kb)
Rights and permissions
About this article
Cite this article
Brugière, N., Humbert, S., Rizzo, N. et al. A member of the maize isopentenyl transferase gene family, Zea mays isopentenyl transferase 2 (ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development. Plant Mol Biol 67, 215–229 (2008). https://doi.org/10.1007/s11103-008-9312-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9312-x