Abstract
To identify novel genes in petal and stamen development, a genetic screen was carried out for enhancers of the unusual B class mutant pistillata-5 (pi-5). In pi-5 flowers, second whorl organs develop as sepals rather than petals, but third whorl stamens are normal. One pi-5 enhancer, dornröschen-like-2 (drnl-2), results in third whorl positions developing as filamentous organs. In addition to enhancing the pi-5 phenotype, drnl-2 mutants also exhibit a phenotype in a wild-type PI background. Although stamen primordia are morphologically visible during early stages of flower development, they fail to enlarge in drnl-2 mutants. DRNL, which encodes a single AP2 domain protein, is expressed in a dynamic pattern in the embryo, seedling, and flower. Analysis of both the drnl-2 mutant phenotype and the DRNL expression pattern in flowers suggests that DRNL plays a critical role in stamen emergence in Arabidopsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The most complex structure in a flowering plant is the flower. The Arabidopsis thaliana flower initially forms on the flanks of the shoot apical meristem after the vegetative to reproductive transition (Smyth et al. 1990). During the initial stages of flower development, the flower primordium consists of a small cluster of morphologically undifferentiated cells. As the floral primordium enlarges, the floral organ anlagen first become morphologically distinct then organ identity is specified. The flower consists of four organ types that develop in distinct floral whorls: four sepals in the first whorl, four petals in the second whorl, six stamens in the third whorl, and two fused carpels in the fourth whorl. Within each of these floral organs, a variety of tissue and cell types differentiate.
Over the past 15 years, considerable progress has been made in understanding the function of a small set of floral homeotic genes that specify floral organ identity in Arabidopsis and other flowering plants (Krizek and Fletcher 2005). These genes were initially identified in loss-of-function mutants that exhibited homeotic transformations of floral organs. Genetic analysis of these mutants led to the well-known ABC model in the early 1990s (Bowman et al. 1991; Coen and Meyerowitz 1991). A, B, and C refer to distinct activities that function in adjacent whorls of the flower. For example, B class activity is postulated to specify the identity of second whorl petals and third whorl stamens. In B class mutants such as apetala3-3 (ap3-3) and pistillata-4 (pi-4), second whorl petals develop as sepals, and third whorl stamens as carpels. Conversely, ectopic expression of the two B class genes AP3 and PI results in conversions of sepals to petals in whorl 1, and carpels to stamens in whorl 4 (Krizek and Meyerowitz 1996). Together, AP3 and PI are postulated to be necessary and sufficient for B class activity. To date, there are no examples of petal or stamen development in Arabidopsis in the absence of functional AP3 and PI. Similarly, there are no examples of AP3 functioning in the absence of PI, or PI functioning in the absence of AP3. Both AP3 and PI encode MADS transcription factors. Considerable biochemical and genetic evidence supports a model whereby AP3 and PI form a DNA binding heterodimer that functions to specify petal and stamen identity (Schwarz-Sommer et al. 1992; Goto and Meyerowitz 1994; Jack et al. 1994; Hill et al. 1998).
Although much is known about how floral organ identity is specified, less is known about what controls the initial morphological appearance of the floral organ primordia. These two processes are separable because mutants that affect floral organ identity specification do not affect the ability of the floral organ primordia to form. For example, in pi mutants, the primordia for the second and third whorl organs develop normally during early stages of flower development demonstrating that floral organ identity genes are not necessary for the emergence floral organ primordia from the meristem (Hill and Lord 1989). Experiments utilizing temperature-sensitive B class alleles have demonstrated that floral organ identity genes are required throughout flower development to specify floral organ identity, but not the initial morphological appearance of the floral organs (Bowman et al. 1989; Zachgo et al. 1995).
Most of the key ABC genes that specify floral organ identity encode MADS transcription factors. The one exception is the A class gene APETALA2 (AP2), which is the founding member of a large plant-specific transcription factor family, the AP2/ERF family (Jofuku et al. 1994; Kim et al. 2006). The AP2/ERF genes are defined by conservation of the AP2 domain, a 70 amino acid sequence-specific DNA binding motif (Riechmann and Meyerowitz 1998). There are approximately 145 AP2/ERF genes in the Arabidopsis genome, and the function of the majority of these genes has not been elucidated (Magnani et al. 2004; Kim et al. 2006). The AP2/ERF family can be divided into two broad subclasses. The first subclass, which contains approximately 14 members, includes proteins that contain two AP2 domains. In this subclass are AP2 itself as well as other genes involved in various aspects of development, including AINTEGUMENTA (ANT), which functions in flower and ovule development (Elliott et al. 1996; Klucher et al. 1996; Krizek et al. 2000), and the PLETHORA1 and PLETHORA2 genes, which function redundantly to specify cell types in the root (Aida et al. 2004). The second and much larger subclass of AP2/ERF genes includes proteins with only a single AP2 domain. The single AP2 domain proteins that have been studied to date exhibit a wide range of functions. For example, the DREB/CBF proteins play a critical role in cold tolerance control (Cook et al. 2004; Sakuma et al. 2006), the ethylene response factors function in hormone signaling pathways and disease resistance (Gutterson and Reuber 2004), and LEAFY PETIOLE is a developmental control gene involved in seed germination (van der Graaff et al. 2000; Ward et al. 2006).
In this study, we characterize a loss-of-function allele of the DORNRÖSCHEN-LIKE (DRNL) gene, and identify a novel function of DRNL in stamen development in Arabidopsis. In drnl-2 mutants, third whorl organs are either missing or are converted to small filamentous structures. DRNL encodes a single AP2 domain protein that plays a key role in stamen primordia emergence during early stages of flower development. DRNL is the first single AP2 domain protein demonstrated to play a key role in flower development. Although DRNL has been described previously (Kirch et al. 2003; Ikeda et al. 2006; Marsch-Martinez et al. 2006; Ward et al. 2006), no loss-of-function floral phenotypes were described in earlier reports; thus this is the first description of a loss-of-function floral phenotype.
Materials and methods
Plant growth conditions
Plants were grown in a 2:1:1 mixture of promix:perlite:vermiculite at 23°C under continuous light conditions.
pi-5 modifier screen
pi-5 homozygous seeds were mutagenized with 0.2% ethylmethanesulfonate (EMS). Seeds were collected from 1,500 individual M1 plants. 8000 M2 plants were scored for enhancement or suppression of flower phenotype, and mutants were named B class modifier (bcm) mutants. The enhancers displayed either absence of stamens or conversion of stamens to carpelloid organs in whorl three. The suppressors developed petals or petal-like organs in place of sepals in whorl two. All bcm mutants were retested in the M3 to test the consistency of the phenotype. bcm1/drnl-2 was backcrossed to Ler twice prior to phenotypic scoring. From the Ler backcross, it was evident that drnl-2 mutants exhibit a phenotype in both a pi-5 and a PI+ background (305 wt: 98 pi-5: 105 drnl-2: 32 pi-5 drnl-2, a 9:3:3:1 ratio (Chi-square P < 0.05) indicating that drnl-2 is unlinked to PI and segregates as a single gene recessive nuclear mutation.
Map-based cloning of DRNL
For mapping, drnl-2 was crossed to wild-type Columbia, and DNA was prepared from individual mutant F2 plants (Dellaporta et al. 1983). A pool of 34 drnl-2 mutant DNA samples was subjected to bulk-segregant analysis (Lukowitz et al. 2000). DRNL was found to be tightly linked to ciw12 on chromosome 1. DNA was prepared from a total of 182 drnl-2 mutants. Each individual DNA sample was subjected to analysis with SSLP or CAPS markers and linkage was evaluated. Markers were generated using information available on TAIR (http://www.arabidopsis.org) or polymorphisms identified by CEREON (Jander et al. 2002). Annotations of genes from http://www.arabidopsis.org were used to define candidate genes.
SEM
Floral tissue was fixed for 24 h in 4% glutaraldehyde, coated with 1% osmium tetroxide in 20 mM phosphate buffer for 48 h, followed by critical-point drying, dissection, mounting, and gold-palladium sputter-coating. Images were taken at an accelerating voltage from 1 to 15 kV using a FEI XL-30 ESEM-FEG scanning electron microscope.
Plasmid constructs
Details on the constructs for overexpression (35S::DRNL), promoter–reporter fusion (pDRNL::GUS), and phenotypic rescue are available on request.
GUS staining
β-glucuronidase staining was performed for 1–48 h at room temperature or 37°C using 2 mM X-Gluc from Gold Biotechnology using established protocols (Campisi et al. 1999).
In situ hybridization
In situ hybridization analysis was performed using DIG-labeled RNA probe synthesized using RNA labeling kit from Roche (Catalog number NC9250811) using standard protocols (Long and Barton 1998). Wild-type Columbia DNA was amplified using TJ884 (5′-GGTCAACCATGGAAGAAGCAATCA-3′) and TJ885 (5′-GATAAGCACGTAAAAAGTAGAACA-3′) and cloned in pGEM7z(+) to create plasmid pD1973. pD1973 was linearized using EcoRI (full length probe) or AccI (AP2 domain deleted probe), and in vitro transcribed with SP6 RNA polymerase to generate DRNL antisense probes. A full-length DRNL sense probe was synthesized using T7 RNA polymerase from HindIII digested pD1973.
Results
A screen for modifiers of the floral phenotype of pi-5
To identify additional genes in the AP3/PI pathway, an enhancer/suppressor screen was performed in a pi-5 background. pi-5 was initially isolated as an enhancer of the weak ap3 allele ap3-11 (Yi and Jack 1998; Yang et al. 2003). pi-5 has a phenotype different from other B class mutants in Arabidopsis, in that organ identity defects are observed primarily in a single whorl of the flower. Most B class mutants (e.g., ap3-3 and pi-4) exhibit floral organ identity transformations of both second whorl petals to sepals and third whorl stamens to carpels. In pi-5 mutants, third whorl stamens are almost completely wild type but second whorl organs resemble sepals (Fig. 1A).
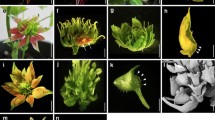
drnl-2 mutant flowers. (A) pi-5 flower. Sepals develop in whorl 2 and stamens develop in whorl 3. (B) drnl-2 pi-5 flower. Third whorl organs are most often missing. (C) drnl-2 mutant flower. Arrow points to filamentous third whorl organ. (D) drnl-2 mutant flower. Arrow points to mosaic petal–sepal first whorl organ. (E) SEM of a drnl-2 mutant flower. Three sepals and one petal have been manually dissected away to allow visualization of third whorl organs. Arrow points to a third whorl filamentous organ. (F) SEM of a drnl-2 flower with whorl 1 and 2 organs removed, leaving three filamentous third whorl organs surrounding the central gynoecium. Asterisks mark the nectaries. (G) SEM of a drnl-2 inflorescence apex. Numbers indicate floral stage. The stage 4 flower on the left contains five first whorl primordia (arrowheads). In the stage 4 flower on the right, the petal primordia are visible (arrows) but the stamen primordia are not visible. H) SEM of a stage 7 drnl-2 flower. Three petal primordia are indicated (arrows). In wild type at this stage the stamen primordia are considerably larger than the petal primordia. In drnl-2, the third whorl primordia (arrowheads) do not develop beyond small bulges, and remain as a ring of undeveloped organs surrounding the carpel. (I) Rescued drnl-2 mutant. A 3.2 kb genomic clone containing the DRNL gene rescues the stamen defects of drnl-2 mutants. (J) drnl-1 mutant flower that develops only four stamens. (K) SEM of a third whorl organ from drnl-1 showing a fusion of two stamen filaments with two anthers. (L) drnl-2 seedling that exhibits a fused cotyledon phenotype. Size bar: 200 μM in (E), (K), 100 μM in (F, G), 25 μM in (H)
We mutagenized homozygous pi-5 seeds with ethylmethanesulfonate (EMS) and screened 8000 M2 progeny from 1500 M1 plants for recessive enhancers and suppressors of the pi-5 floral phenotype. Enhancers were identified as plants that contained flowers where the third whorl stamens were either missing or developed as carpels or staminoid carpels. Enhancers exhibited increased rates of male sterility as reflected in decreased seed set. This aspect of the phenotype of the enhancers was in sharp contrast to pi-5 plants, which are self-fertile and exhibit normal seed set. From putative enhancers, selfed seeds were collected in the M2. M3 plants were examined to ascertain the consistency of the enhanced phenotype.
This manuscript focuses on the molecular and genetic characterization of one of the pi-5 enhancers that we initially named B c lass m odifier 1 (bcm1). After cloning the gene, we realized that the gene had previously been described as DORNRÖSCHEN-LIKE (DRNL) (Kirch et al. 2003). Based on this, we refer to the bcm1 allele that we isolated in the pi-5 enhancer screen as drnl-2.
drnl-2 mutants exhibit dramatic defects in stamen development
In a pi-5 background, the drnl-2 enhancer exhibits a conversion of stamens in whorl three to filamentous structures. Frequently in pi-5 drnl-2 double mutants third whorl organs are missing (Fig. 1B). Fertile stamens are never observed; thus pi-5 drnl-2 plants are male sterile.
drnl-2 exhibits a phenotype in a PI+ background as evidenced by the segregation of plants with a novel floral phenotype in the F2 of the backcross to Ler (Fig. 1C–H). The most dramatic phenotypic defects are in the third whorl of the flower where the stamens are frequently converted to filamentous structures (Fig. 1F, arrows Fig. 1C, E). Careful analysis of these flowers reveals that the phenotype in whorl 3 is variable. The average number of organs that develop in third whorl positions is 3.11 (wild-type average 5.88 organs) indicating that frequently, there is a failure of organ development in the third whorl of drnl-2 mutants. Most commonly, third whorl organs are filaments (average 2.89 filaments/flower, Table 1) that resemble stamen filaments, but are most often shorter. In most cases there is no anther or anther-like structure at the distal end of the filament, though in rare cases, an anther-like structure does develop, and very rarely, fertile stamens develop. The morphologically normal stamens produce viable pollen, and thus it is possible to isolate a small number of homozygous drnl-2 mutant seeds.
Although the most dramatic phenotypic defects are in the third whorl of the flower, the second whorl petals exhibit subtle defects in drnl-2 mutants. Second whorl organ number is not affected in drnl-2 mutants (Table 1), and the organs that develop in second whorl positions always strongly resemble petals. However, many of the petals that develop in drnl-2 mutants are not identical to wild-type petals: they are shorter, more crinkled, and the blade is less lobed than in wild-type petals. Frequently the petals within a single flower are variable in height; some petals are of normal height, others are only about two-thirds the height of the central gynoecium.
First whorl organs also exhibit defects in drnl-2 mutants. Occasionally, first whorl organs develop as petals, or as petal-sepal mosaic organs (arrow Fig. 1D, Table 1). Organ number is not dramatically altered in the first whorl of drnl-2 mutants. Although most fourth whorl carpels are normal, occasionally misshapen and bent carpels are observed.
Analysis of developing drnl-2 flowers in the scanning electron microscope (SEM) reveals that many stamen primordia fail to develop. Similar to wild type, in drnl-2 mutants, petal and stamen primordia become morphologically distinct from the floral meristem at stage 4. There are two unusual features of stage 4 drnl-2 flowers. First, some develop with five first whorl primordia (arrowheads, stage 4 flower, Fig. 1G). It is possible that flowers with five first whorl primordia will go on to develop the sepal-petal mosaic organs that develop in drnl-2 mutants (arrow Fig. 1D). Second, the petal primordia are larger than the stamen primordia (arrows Fig. 1G); this is the reverse of what is observed in wild type where stamens grow much more rapidly than petals. The retarded development of the stamens is more obvious by stage 7 (Fig. 1H). In wild-type flowers at stage 7, the medial stamens are about as tall as the central gynoecium. In drnl-2, third whorl organs fail to enlarge (arrowheads in Fig. 1H), and second whorl petals (arrow in Fig. 1H) are larger than the third whorl stamen primordia. In summary, this analysis shows that third whorl organs in drnl-2 mutants most often fail to enlarge from very early floral stages.
DRNL encodes an AP2 domain protein
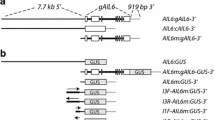
Using standard map-based cloning approaches (Jander et al. 2002), drnl-2 was mapped to a 221 kb region at the top of chromosome 1 (Fig. 2A, B). This 221 kb region contains 54 predicted genes. To identify candidates, we focused first on genes that encode transcription factors, and second on genes that were expressed in young flowers. Since the phenotype that we observe in drnl-2 mutants is most dramatic in the flower, we thought it would be very likely that the gene would be expressed in flowers, specifically in stamens where the phenotypic defects are strongest. One gene in this interval, At1g24590, fit these criteria in that it was shown to be expressed in developing petals and stamens by RNA in situ hybridization (Kirch et al. 2003). Expression in stamen primordia correlates well with the drnl-2 mutant phenotype where stamen development is disrupted beginning at early floral stages. This expression pattern strongly suggested that At1g24590 was the site of mutation in drnl-2.
DRNL locus. (A) Using standard map-based cloning approaches, DRNL was mapped to a 221 kilobase (kb) region at the top of chromosome 1. The numbers on the left refer to chromosomal position (in kb), the numbers on the right indicate the number of recombinant chromosomes over the total number of chromosomes analyzed. (B) DRNL is a small gene with no introns. The black bar indicates the genomic fragment used in rescue experiments. The hatched bars indicate the location of consensus auxin response elements (AREs). (C) The drnl-2 allele contains a missense mutation in the AP2 domain, a change from alanine to valine. The drnl-1 allele contains an En transposon insertion at amino acid 257
At1g24590 encodes a 306 amino acid protein that contains a single AP2 domain. The closest homolog in the Arabidopsis genome is the single AP2 domain protein variously called ENHANCER OF SHOOT REGENERATION 1 (ESR1) (Banno et al. 2001) or DORNRÖSCHEN (DRN) (Kirch et al. 2003), which is postulated to function in shoot regeneration and meristem maintenance. DRN and DRNL are 91% identical (51/56 positions) in the AP2 domain, but outside the AP2 domain the similarity is much less; overall DRN and DRNL are 31% identical. DRN and DRNL also share a common gene structure; specifically, neither DRN nor DRNL contain introns. Despite the similarity of these two genes, the spatial and temporal expression patterns of DRN and DRNL are not identical.
We sequenced At1g24590/DRNL in drnl-2 mutants and found a missense mutation in a highly conserved alanine residue in the AP2 domain (A93V) (Fig. 2C). This mutation is not present in the pi-5 strain that was the starting point for the mutagenesis. One line of evidence that this position is important comes from alignments of the AP2 domains from a large number of plant proteins (Fig. 2C). The alanine residue in the AP2 domain is conserved in almost all AP2 domains from a wide range of plant species, suggesting that this mutation is functionally important (Magnani et al. 2004). A second line of evidence that this position is important comes from studies on the DREB-III-4 protein of Brassica Napus. DREB-III-4, which contains a naturally occurring valine at this position, is unable to bind to AP2 target DNA sites in vitro (Liu et al. 2006). Convincing evidence that At1g24590 is DRNL comes from mutant rescue experiments. A 3.2 kb genomic fragment that contains the entire DRNL open reading frame plus 1.7 kb of 5′ sequence rescues the third whorl stamen defects in drnl-2 mutants (Fig. 1I, 2B). Based on this evidence, we are confident that drnl-2 is an allele of DRNL/At1g24590.
drnl-2 is allelic to drnl-1
A previously described allele of DRNL, drnl-1, contains an En transposon insertion in the open reading frame of At1g24590, at amino acid 257 (Fig. 2C) (Marsch-Martinez et al. 2006; Chandler et al. 2007), but a floral phenotype was not reported. Our analyses reveal that drnl-1 plants exhibit a weak floral phenotype (Fig.1J, K). As with drnl-2, the most obvious defects in drnl-1 are in the third whorl of the flower where stamen number is reduced (4.49 on average compared to 5.88 in wild type). Rarely, third whorl positions are converted to filamentous structures or stunted sterile stamens (Table 1). Despite the third whorl defects, multiple fertile stamens develop in drnl-1 plants, and flowers produce a normal seed set. In drnl-1, second whorl petals are often heterogeneous in size (some are taller than others), and the blade is often not fully expanded. Unlike drnl-2, we did not observe any first whorl petaloid organs in drnl-1 mutant flowers.
One developmental abnormality that was observed in drnl-1 homozygotes (10 occurrences in 71 flowers scored), but only rarely in drnl-2, was the development of organs where two stamens were fused entirely along the length of the stamen filament. At first glance, these organs look like they are comprised of two anthers on a single filament, but on closer inspection, the filament is broader than a normal stamen filament, and clearly consists of two fused filaments as well as two anthers (Fig. 1K).
The drnl-1/drnl-2 transheterozygote exhibits a phenotype intermediate between drnl-1 and drnl-2 demonstrating that drnl-1 and drnl-2 are allelic (Table 1). drnl-1/drnl-2 plants produce more stamens (1.62 on average) than drnl-2, but fewer than drnl-1. Similarly, phenotypic defects in whorls one and two are less severe than observed in drnl-2, but more severe than that observed in drnl-1. Since both alleles are recessive, and appear to be hypomorphic alleles, in allelic series, we conclude that drnl-2 is the stronger allele, and drnl-1 the weaker allele.
drnl-1 and drnl-2 exhibit seedling defects
In addition to the floral defects, both drnl-1 and drnl-2 homozygotes exhibit a fused cotyledon phenotype in young seedlings (Fig. 1L) (Chandler et al. 2007). In drnl-1 homozygotes this phenotype is observed in about 2% of the plants, while in drnl-2 homozygotes fused cotyledons are observed in 31% of seedlings. Despite these early defects, seedlings with fused cotyledons continue to develop, and go on to produce normal vegetative leaves and inflorescences.
DRNL is expressed in restricted regions throughout plant development
To gain insight into the spatial and temporal expression pattern of DRNL, we constructed a pDRNL::GUS transgenic line. We are confident that this line reflects the endogenous expression of DRNL because in the flower and seedling, the GUS activity pattern matches the expression pattern of the DRNL RNA as determined by RNA in situ hybridization (see below).
In the embryo, GUS activity is detected at the distal tips of the developing cotyledons in pDRNL::GUS (Fig. 3A). In very young seedlings, GUS activity is detected in the shoot apical meristem as well as in the cotyledons (Fig. 3B). In older seedlings, GUS activity is detected throughout young leaf primordia, but not in the shoot apical meristem (SAM) itself (Fig. 3C, D). As the leaf primordia enlarge, strong GUS activity is detected at the distal tip of the developing leaf primordia (Fig. 3C, D). In young leaves, GUS activity becomes detectable in the hydathodes (arrows Fig. 3C). Strong expression in hydathodes is observed as the signal at the distal tip of the leaf is decaying (Fig. 3C). As the leaves mature, the intensity of the GUS signal in the hydathodes decreases. In older leaves, GUS activity is not detectable.
DRNL is expressed broadly in development. (A) pDRNL::GUS embryo. GUS activity is detected at high levels at the tips of the cotyledons. (B) Very young pDRNL::GUS seedling, prior to leaf emergence, that exhibits GUS activity in the shoot apical meristem (arrow) as well as at the tips of the cotyledons. (C) pDRNL::GUS seedling. GUS activity is detected at high levels in leaf primordia. In young leaves, GUS activity is detected at the tips of the developing leaves. In slightly older leaves, GUS activity is detected in the hydathodes (arrows). (D) Longitudinal section through a pDRNL::GUS seedling. GUS activity is detected throughout very young leaf primordia. Arrowhead marks the shoot apical meristem. (E) pDRNL::GUS inflorescence. High levels of GUS activity is detectable in flowers through stage 9. (F–L) In situ hybridization. (F–K), antisense DRNL probe. (L), sense control probe. (F) In the inflorescence, DRNL RNA is detected in the shoot apical meristem and throughout stage 1–4 flowers. (G) DRNL RNA accumulates throughout stage 3 flower. At stage 6, DRNL RNA is detected in developing petals, stamens, and carpels. (H) At stage 9, DRNL RNA accumulates at high levels in developing anthers. DRNL RNA is also detected at the apical tip of developing petals (arrow). (I) Higher magnification view of expression in the anther. DRNL RNA is detected in developing microspores and tapetum. (J) Cross-section through a stage 9 flower, showing high-level expression in anthers. DRNL RNA is also detectable in developing ovules. (K) In the ovule, DRNL RNA is detectable in the endothelium. (L) DRNL sense control
GUS staining of inflorescences reveals a high level of GUS activity in developing petals and stamens (Fig. 3E). To determine the details of the DRNL expression pattern in young flowers, we performed RNA in situ hybridization. DRNL RNA accumulates throughout the inflorescence meristem, in cells of both the central and peripheral zones, and in all three-cell layers (Fig. 3F). During floral stages 1 and 2, prior to differentiation of the floral organ primordia from the floral meristem, DRNL RNA accumulates in the anlagen for all four floral organs, and the level of expression is higher than in the SAM (Fig. 3F). During stages 3 and 4, when the sepals become morphologically distinct from the floral meristem, DRNL RNA continues to be detected throughout the flower primordium (Fig. 3F, G). At stage 5, the petals and stamens become distinct from the floral meristem, and DRNL RNA is detectable throughout the developing petals, stamens, and carpels, but the expression level in sepals is lower compared to earlier stages (Fig. 3F, G). By stage 9, when the four floral organs are distinct, and each organ has differentiated into various tissues and cell types, DRNL RNA is detectable in a subset of cells of petals, stamens, and carpels (Fig. 3H). In the petals, highest level of DRNL is present at the distal tip of the developing petal (arrow Fig. 3H). In the stamen, a very high level of expression is detectable in developing microspores and tapetum, but is absent from the epidermis, endothecium, middle layer, stamen filament, and connective (Fig. 3H–J). High level of expression of DRNL continues throughout microspore development. In the carpel, DRNL RNA becomes restricted to ovule primordia beginning at stage 7 (Fig. 3J). As the ovules develop, RNA is detected throughout the ovule. At the time of fertilization, DRNL RNA is expressed at high levels in the endothelium, but expression is not detected in the integuments or in the funiculus (Fig. 3K).
Ectopic expression of DRNL results in dramatic defects in plant development
To ascertain the effects of ectopic expression, transgenic plants were constructed that misexpress DRNL under the control of the strong and broadly expressed CaMV 35S promoter. 35S::DRNL plants exhibit a range of phenotypic defects. In the strongest 35S::DRNL lines, the plants develop as extreme dwarfs with severe defects in internode elongation (Fig. 4A, B). Leaf and flower morphology is also abnormal in strong 35S::DRNL plants. Both rosette and cauline leaves are small, dark green, and abnormally curled (Fig. 4B). The flowers in 35S::DRNL have defects in all four floral whorls (Fig. 4C, D). First whorl positions are occupied by thickened green organs that resemble bracts. The fourth whorl carpel develops as a flattened cylinder. Both second whorl petals and third whorl stamens are stunted, and never achieve a height greater than 50% of the height of the carpel. The stamens in strong 35S::DRNL lines do not produce pollen and are male sterile. The strongest lines are also female sterile. Complete male and female sterility results in the inability to maintain the strongest lines after they are generated. In weaker 35S::DRNL lines, both the dwarf and sterility phenotypes are less severe. Weaker lines are self-fertile allowing lines to be maintained. Overall, the range of phenotypes observed is similar to what has been observed in other studies that analyzed DRNL overexpression (Kirch et al. 2003; Ikeda et al. 2006; Marsch-Martinez et al. 2006; Ward et al. 2006).
Ectopic expression of DRNL causes dramatic defects in plant development. (A) A 35S::DRNL plant (in the Col background) (left) and a wild-type Columbia plant (right) of approximately the same age. (B) The 35S::DRNL plant is a severe dwarf with internode elongation defects, curled leaves, and abnormal flowers. (C–D) 35S::DRNL flowers consist of thickened bract-like first whorl organs, stunted petals and stamens in whorls two and three, and a flattened misshapen carpel with reduced stigmatic papillae
The initial expression pattern of AP3 is normal in drnl-2 mutants
We were interested in determining the regulatory relationship between DRNL and the B class genes AP3 and PI. In situ hybridization experiments demonstrate the DRNL RNA accumulates in stage 1–2 flowers, prior to the initial appearance of AP3 and PI RNA at stage 3. To determine whether AP3 or PI play a role in regulating the DRNL expression pattern, we analyzed DRNL expression using pDRNL::GUS in ap3-3 and pi-4 mutants. ap3-3 and pi-4 are strong loss-of-function alleles. In these mutants, GUS activity is detectable throughout early stage flowers, but high-level expression in older flowers (older than stage 7–8) is not observed (Fig. 5A–C). The fact that GUS activity is detected during early floral stages suggests that AP3 and PI do not play a role in activating DRNL in floral meristems and young flowers. Similarly, GUS activity is observed in young flowers in pDRNL::GUS drnl-2 flowers (Fig. 5D) demonstrating that DRNL is not required for its own expression.
DRNL and AP3/PI are activated independently. (A) pDRNL::GUS inflorescence. (B) pDRNL::GUS ap3-3 inflorescence. (C) pDRNL::GUS pi-4 inflorescence. (D) pDRNL::GUS drnl-2 inflorescence. (E) pAP3::GUS inflorescence. (F) pAP3::GUS drnl-2 inflorescence. (G) pAP3::GUS drnl-2 flower. The third whorl filaments (arrowhead) in drnl-2 exhibit GUS activity. (H) First whorl petaloid-sepal organ in a pAP3::GUS drnl-2 flower. The petaloid sectors of the first whorl organ exhibit GUS activity
We also examined the expression of AP3 in drnl-2 utilizing a pAP3::GUS transgene. In wild-type flowers, AP3 expression is established in petal and stamen primordia at stage 3. As the flowers mature, AP3 expression is detected specifically in developing petals and stamens (Fig. 5E). In drnl-2 mutants, the early expression of AP3 is unaffected demonstrating that DRNL does not play a role in the establishment of AP3 expression (Fig. 5F). In older flowers, GUS activity is detected in the third whorl filaments that develop in drnl-2 mutants (Fig 5G) suggesting that these organs have an identity similar to stamen filaments. GUS activity is also observed in the petaloid sectors of the first whorl petaloid-sepal organs that occasionally develop in drnl-2 flowers (Fig. 5H).
DRNL expression is not altered in arf mutants
The similarity of the DRNL spatial and temporal expression pattern to the regions of the embryo and seedling that exhibit the highest concentration of active auxins suggested that DRNL might function in the auxin response. The auxin response factors (ARFs) are transcription factors that are activated in response to auxin. Considerable functional redundancy exists among the 23 members of the ARF gene family in Arabidopsis. To test whether the ARFs control DRNL expression, we analyzed pDRNL::GUS in several arf mutant backgrounds. We reasoned that the most likely ARFs to be controlling DRNL are those that are expressed in flowers, and exhibit a mutant phenotype in stamens, such as arf1-5 arf2-8 and arf6-2 arf8-3 mutants, which exhibit stamen filament elongation defects. However, the DRNL::GUS expression pattern was not altered in arf1-5 arf2-8 and arf6-2 arf8-3 mutants demonstrating that it is unlikely that ARF1, ARF2, ARF6, and ARF8 transcriptionally regulate DRNL in the flower (Supplementary Fig. 1).
Discussion
DRNL is critical for proper development of stamens. In drnl-2 mutants, third whorl positions either do not develop or are converted to small filamentous structures. Less dramatic phenotypic defects are also observed in seedlings and in whorls 1, 2, and 4 of drnl-2 mutant flowers. Analysis of flowers during very early stages of flower development demonstrates that third whorl organs fail to enlarge and differentiate demonstrating that DRNL is critical for stamen emergence.
The phenotype of drnl-2 mutants is reminiscent of the unisexual female flowers that develop in a wide range of species such as cucumber, Silene latifolia (white campion), and asparagus (Lebel-Hardenack and Grant 1997; Knopf and Trebitsh 2006; Zluvova et al. 2006). In these species, the stamen primordia form, but stamen development aborts resulting in a unisexual female flower. At present, the molecular mechanism for how stamen abortion occurs is not well understood in any species. Alteration of the levels of DRNL activity provides a possible mechanism for how stamen abortion could occur.
DRNL is expressed throughout the floral meristem and young flowers, at a stage when the floral organ primordia first become morphologically visible. At later stages, DRNL is expressed at high levels in cells in the anther and ovule. In the B class mutants ap3-3 and pi-4 mutants, the expression of DRNL is normal during early floral stages. Similarly, the expression of the B class gene AP3 is normal in drnl-2 mutant. These results suggest that there is no transcriptional relationship between DRNL and AP3; i.e., DRNL does not result in defects in stamen development by altering the initial expression pattern of the B class genes. Thus, DRNL must be functioning during the early stages of flower development in an AP3/PI-independent manner to affect stamen development.
DRNL encodes an AP2 domain transcription factor. DRNL is a member of the subgroup of AP2/ERF proteins that contains a single AP2 domain. Other AP2 domain transcription factors that play an important role in flower development, such as AP2 (Jofuku et al. 1994) and AINTEGUMENTA (Elliott et al. 1996; Klucher et al. 1996), are members of a subgroup that contains two AP2 domains. DRNL is the first single AP2 gene demonstrated to play a role in flower development.
Other studies on DRNL
DRNL has been described by several other groups. Kirch et al. (2003) initially named the gene DRNL because it is related to a closely related AP2 gene called DORNRÖSCHEN (DRN). DRN is the gene most similar to DRNL in the Arabidopsis genome. DRN was identified from an activation-tagged line that exhibited a meristem arrest phenotype resulting from ectopic expression of DRN. Kirch et al. (2003) reported that 35S::DRNL lines exhibited a dwarf and silique-shape phenotype (Kirch et al. 2003). DRNL was also studied by Ikeda et al. (2006) who named the gene ENHANCER OF SHOOT REGENERATION 2 (ESR2). Overexpression of DRNL/ESR2 results in enhanced shoot regeneration in the absence of cytokinin in tissue culture. Two groups have described activation-tagged alleles of DRNL. In one case, the activation-tagged allele sob2-D suppressed the long hypocotyl phenotype of a phyB mutant (Ward et al. 2006). Adult sob2-D phyB4 plants exhibit curled leaves without petioles and abnormal siliques. In a second case, the activation-tagged allele named bolita-D results in a small leaf phenotype (Marsch-Martinez et al. 2006). Another recent study of DRN and DRNL, utilizing loss-of-function alleles, demonstrates that these two genes function redundantly in embryo development (Chandler et al. 2007). In addition, both DRN and DRNL proteins interact in vitro and in vivo with class III HD-ZIP proteins such as PHAVOLUTA, PHABULOSA, and REVOLUTA (Chandler et al. 2007).
In this study, we characterize a loss-of-function allele of the DRNL gene. Previous studies on this gene primarily focused on the overexpression phenotypes (Ikeda et al. 2006; Marsch-Martinez et al. 2006; Ward et al. 2006), which may not reflect the actual function of this gene. Although previous studies had access to either the weak drnl-1 allele (Marsch-Martinez et al. 2006) or RNAi lines for DRNL (Ikeda et al. 2006), no loss-of-function floral phenotype was reported. This study clearly demonstrates that DRNL plays an important role in flower development. Most importantly, DRNL function is required for proper stamen development.
Is drnl-2 a null allele?
There are two extant loss-of-function DRNL alleles: drnl-2 and drnl-1. Our genetic analysis indicates that drnl-2 exhibits more severe seedling and flower phenotypes than drnl-1. The drnl-2/drnl-1 transheterozygote exhibits a flower phenotype intermediate between drnl-2 and drnl-1 confirming that both drnl-2 and drnl-1 are loss-of-function alleles, and that drnl-2 is the stronger allele. At first glance, this is a bit surprising because drnl-1 contains a transposon insertion in the open reading frame, at the C-terminal end of the protein (at amino acid 257), and likely results in the truncation of the C-terminal 50 amino acids of DRNL. The stronger allele, drnl-2, contains a missense mutation in the AP2 domain, replacement of one non-polar amino acid (alanine) with another (valine). Since alanine is conserved at this position in the AP2 domain in the majority of plant AP2 domains, it suggests that the larger valine R group may result in diminished protein activity. The importance of an alanine at this position is indicated by studies on the Brassica Napus AP2 protein DREB-III-4, which contains a naturally occuring alanine to valine mutation. DREB-III-4 is unable to bind to consensus AP2 DNA binding sites in vitro suggesting that an alanine at this position is important for DNA binding (Liu et al. 2006).
At present, it is not known if drnl-2 is a null allele. Three lines of indirect evidence suggest that drnl-2 may not be a null, and we speculate that drnl nulls may be lethal. First, no T-DNA insertional alleles have been reported in DRNL. Although this could be due to chance, it is also true that lethals are lost from T-DNA pools since, for example, in the Salk collection, T-DNA insertions are not selected for after they are generated (Alonso et al. 2003). Second, the DRNL RNAi lines that have been generated have very weak floral and seedling phenotypes, suggesting that lines with a stronger phenotype may not be recoverable (T. Jack and A. Nag, unpublished). Third, a floral phenotype similar to what we observe in drnl-2 has not been described to date. drnl-2 exhibits a dramatic floral phenotype in an otherwise wild-type background. Over the past 20 years, many screens have been done to identify floral mutants (Koornneef et al. 1983; Komaki et al. 1988; Liu and Meyerowitz 1995; Bowman and Smyth 1999; Pruitt et al. 2003), yet a phenotype similar to drnl-2 has not been described prior to this report. Perhaps this is due to the fact that drnl-2 contains a specific, but rare, missense mutation that results in partial loss-of-function, while complete loss-of-function, which can be generated many different ways, is lethal. In the case of a strong loss-of-function mutation in DRNL, the floral phenotype would not be able to be visualized if the mutation resulted in embryonic or seedling lethality. In drnl-2, we hypothesize that the alanine to valine missense mutation in the AP2 domain leads to a partially functional protein that possesses sufficient activity to direct proper embryonic and seedling development, but does not possess sufficient activity to direct proper stamen development. There are many examples of this in developmental biology, though not so many in plants. For example, in Drosophila, the key components that specify the identity of the photoreceptor cells in the Drosophila eye are members of the Ras pathway. Null mutations in these components result in embryonic lethal phenotypes, but when these genes are mutated specifically in the eye, dramatic developmental phenotypes result (Simon et al. 1991).
It is also possible that the reason drnl-2 null alleles have not been identified is because DRNL is a small target for mutagenesis (306 amino acids, no introns). This debate will be resolved by isolation and characterization of a true RNA or protein null for DRNL.
Potential mechanisms of DRNL function
Other studies on DRNL have suggested that DRNL may function in the cytokinin response pathway. Two groups have shown that ectopic expression of DRNL results in shoot regeneration in tissue culture in the absence of cytokinin (Ikeda et al. 2006; Marsch-Martinez et al. 2006). In one study, shoot regeneration occurred in the absence of cytokinin even in a cre1/ahk4 cytokinin receptor mutant suggesting that DRNL functions in the cytokinin response pathway downstream of the receptor (Ikeda et al. 2006).
DRNL overexpression results in small plants with fewer cells and smaller cells (Marsch-Martinez et al. 2006). This phenotype suggests that DRNL normally plays a role in suppressing cell division and cell expansion. Based on this, the prediction would be that drnl loss-of-function mutant plants would be larger than normal, or would possess more cells or larger cells. However, this is not what is observed in drnl mutants as drnl mutant plants have normal sized organs, and there is no evidence that cell size or cell number is affected in most organs. The most dramatic defect in drnl mutants is a failure in stamen development. Perhaps the failure to observe a more dramatic phenotype is due to the fact that DRNL functions redundantly with related AP2 genes such as DRN. Insights into potential redundancy will be gained by analysis of floral phenotypes drn drnl double mutants.
It is also possible that the DRNL overexpression phenotype is not reflective of the normal function of the gene. Deceiving pleiotropic overexpression phenotypes are often observed when transcription factors are overexpressed under the control of strong promoters such as the CaMV 35S promoter. For example, many of the MADS genes, when overexpressed under 35S control, result in dramatic leaf phenotypes that are unrelated to normal gene function (Krizek and Meyerowitz 1996; Goodrich et al. 1997).
Although a tie to cytokinin signaling has been demonstrated in DRNL overexpression lines, several lines of suggestive evidence hint that DRNL may function in auxin signaling. First, DRNL contains three consensus auxin response elements (ARE): two are in the promoter at −1.1 kb and −3.0 kb, and the third is within the coding sequence (Fig. 2B). AREs are bound by a family of transcription factors called ARFs. Activity of the ARFs is controlled by a second set of proteins, the AUX/IAA proteins, which directly bind to the ARFs and block their ability to bind to DNA. In response to auxin, the AUX/IAA proteins are degraded by an SCFTIR1-mediated mechanism (Gray et al. 2001; Dharmasiri et al. 2005a,b; Kepinski and Leyser 2005), allowing the ARFs to bind to AREs and mediate transcriptional activation or repression of auxin target genes. Recent genetic analysis of the members of the ARF gene family indicates that several ARFs function in stamen development. For example, both ARF1 and ARF2 function in stamen filament elongation; in arf2-8 and arf1-5 arf2-8 double mutants, stamen filaments do not fully elongate (Ellis et al. 2005). Similarly, arf6-2 arf8-3 double mutants exhibit reduced petal and stamen length (Nagpal et al. 2005). Using pDRNL::GUS, we were unable to detect changes in the DRNL expression pattern in arf1-5 arf2-8 and arf6-2 arf8-3 double mutants. It is possible that pDRNL::GUS expression is altered in other arf mutants that we have not yet tested. It is also possible, due to the large size of the ARF family, that the ARFs function redundantly, and changes in DRNL expression will not be observed until DRNL expression is analyzed in a higher-order arf mutant background.
A second reason that we postulate that auxin plays a role in DRNL function comes from similarity of the DRNL spatial expression pattern to the locations of highest concentrations of auxins, as determined by indole-3-acetic acid (IAA) antibody staining and expression of readouts of the auxin response such as DR5::GUS (Ulmasov et al. 1997; Aloni et al. 2003). In the embryo, the highest concentrations of DRNL are at the tip of the developing cotyledons. This region of the embryo has been demonstrated to have high concentrations of active auxins. Similarly, in the seedling, high levels of auxin are observed in developing leaf primordia. DRNL is expressed at the distal tip of developing leaves. As leaves enlarge, DRNL expression is detected in the hydathodes, during the time when expression at the distal tip of the leaf is decreasing. Again, the regions where DRNL is expressed mirror the regions of the seedling that have been demonstrated to possess high levels of active auxins (Aloni et al. 2003).
In the flower, the correlation between DRNL expression and auxin is less striking. DR5::GUS is expressed at low levels in flowers prior to stage 7, but IAA antibody staining suggests that inactive auxin conjugates are present throughout young flowers (Aloni et al. 2006). At stage 8, high levels of DR5::GUS activity are present in the anthers and stamen filaments. At present, we do not know if the floral developmental defects in drnl-2 are auxin-mediated effects. Future work will help elucidate this issue.
Additional evidence that DRNL may be involved in the auxin response comes from a microarray study where gene expression in leaves of 35S::BOLITA/DRNL was compared to wild type (Marsch-Martinez et al. 2006). Expression of 25 auxin-related genes was altered more than two fold, and most of these genes were downregulated in response to DRNL overexpression. Included in this list are the three IAA genes: IAA17/AXR3, IAA3/SHY2, and IAA7/AXR2.
Despite suggestive evidence DRNL is involved in the auxin response, direct evidence that DRNL is auxin responsive is lacking. We and others have been unable to detect direct transcriptional changes in DRNL transcript levels or expression pattern in response to auxin treatment (Marsch-Martinez et al. 2006) (T. Jack and A. Nag, unpublished). In addition, our failure to observe an obvious change in pDRNL::GUS expression pattern in arf1-5 arf2-8 and arf6-2 arf8-3 mutants (Supplementary Figure 1) does not support a connection between auxin and DRNL.
However, a connection to auxin has been demonstrated for the related AP2 gene DRN. Specifically, DRN has been shown to act upstream of the auxin transport and response during embryo development (Chandler et al. 2007). The relationship between DRNL and auxin in the embryo has yet to be rigorously examined.
In summary, in this study we characterized a loss-of-function allele of the DRNL gene. Previous studies on this gene focused on the overexpression phenotypes, which may not reflect the actual function of this gene. Here we demonstrate that DRNL is critical for proper stamen development, but, at present, the mechanism of DRNL function is unknown. This study is the first to clearly demonstrate an important role in flower development for a single AP2 domain protein.
Abbreviations
- IAA:
-
Indole-3-acetic acid
- EMS:
-
Ethylmethanesulfonate
- kb:
-
Kilobase
- ARE:
-
Auxin response elements
- ARF:
-
Auxin response factor
- SAM:
-
Shoot apical meristem
- X-gluc:
-
5-bromo-4chloro-3indolyl-beta-O-glucuronic acid
- SEM:
-
Scanning electron microscope
References
Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119:109–120
Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of auxin in regulating Arabidopsis flower development. Planta 223:315–328
Aloni R, Schwalm K, Langhans M, Ullrich CI (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216:841–853
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Banno H, Ikeda Y, Niu QW, Chua NH (2001) Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13:2609–2618
Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126:2387–2396
Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1:37–52
Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112:1–20
Campisi L, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T (1999) Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J 17:699–707
Chandler JW, Cole M, Flier A, Grewe B, Werr W (2007) The AP2 transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134:1653–1662
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101:15243–15248
Dellaporta SL, Wood VP, Hicks JB (1983) A plant DNA mini-preparation: version II. Plant Mol Biol Report 4:19–21
Dharmasiri N, Dharmasiri S, Estelle M (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445
Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9:109–119
Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8:155–168
Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132:4563–4574
Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386:44–51
Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8:1548–1560
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414:271–276
Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7:465–471
Hill JP, Lord EM (1989) Floral development in Arabidopsis thaliana: a comparison of the wild-type and the homeotic pistillata mutant. Can J Bot 67:2922–2936
Hill TA, Day CD, Zondlo SC, Thackeray A, Irish VF (1998) Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125:1711–1721
Ikeda Y, Banno H, Niu QW, Howell SH, Chua NH (2006) The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol 47:1443–1456
Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and post-transcriptional regulation determine floral organ identity. Cell 76:703–716
Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Phys 129:440–550
Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6:1211–1225
Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435:446–451
Kim S, Soltis PS, Wall K, Soltis DE (2006) Phylogeny and domain evolution in the APETALA2-like gene family. Mol Biol Evol 23:107–120
Kirch T, Simon R, Grunewald M, Werr W (2003) The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell 15:694–705
Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8:137–153
Knopf R, Trebitsh T (2006) The female-specific CS-ACS1G gene of cucumber. A case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched-chain amino acid transaminase gene. Plant Cell Physiol 47:1217–1228
Komaki MK, Okada K, Nishino E, Shimura Y (1988) Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development 104:195–203
Koornneef M, van Elden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ (1983) Linkage map of Arabidopsis thaliana. J Hered 74:265–272
Krizek BA, Fletcher JC (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6:688–698
Krizek BA, Meyerowitz EM (1996) The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122:11–22
Krizek BA, Prost V, Macias A (2000) AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12:1357–1366
Lebel-Hardenack S, Grant S (1997) Genetics of sex determination in flowering plants. Trends Plant Sci 2:130–136
Liu Y, Zhao T-J, Liu J-M, Liu W-Q, Liu Q, Yan Y-B, Zhou H-M (2006) The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett 580:1303–1308
Liu Z, Meyerowitz EM (1995) LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121:975–991
Long JA, Barton MK (1998) The development of apical embryonic pattern in Arabidopsis. Development 125:3027–3035
Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Phys 123:795–805
Magnani E, Sjolander K, Hake S (2004) From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell 16:2265–2277
Marsch-Martinez N, Greco R, Becker JD, Dixit S, Bergervoet JH, Karaba A, de Folter S, Pereira A (2006) BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol Biol 62:825–843
Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, Ecker JR, Reed JW (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132:4107–4118
Pruitt RE, Bowman JL, Grossniklaus U (2003) Plant genetics: a decade of integration. Nat Genet (Suppl) 33:294–304
Riechmann JL, Meyerowitz EM (1998) The AP2/EBEBP family of plant transcription factors. Biol Chem 379:633–646
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309
Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönning W-E, Saedler H, Sommer H (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11:251–263
Simon MA, Bowtell DDL, Dodson GS, Laverty TR, Rubin GM (1991) Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tryosine kinase. Cell 67:701–716
Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2:755–767
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971
van der Graaff E, Dulk-Ras AD, Hooykaas PJ, Keller B (2000) Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127:4971–4980
Ward JM, Smith AM, Shah PK, Galanti SE, Yi H, Demianski AJ, van der Graaff E, Keller B, Neff MM (2006) A new role for the Arabidopsis AP2 transcription factor, LEAFY PETIOLE, in gibberellin-induced germination is revealed by the misexpression of a homologous gene, SOB2/DRN-LIKE. Plant Cell 18:29–39
Yang Y, Xiang H, Jack T (2003) pistillata-5, an Arabidopsis B class mutant with strong defects in petal but not in stamen development. Plant J 33:177–188
Yi Y, Jack T (1998) An intragenic suppressor of the Arabidopsis floral organ identity mutant apetala3-1 functions by suppressing defects in splicing. Plant Cell 10:1465–1477
Zachgo S, de Andrade Silva E, Motte P, Tröbner W, Saedler H, Schwarz-Sommer Z (1995) Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121:2861–2875
Zluvova J, Nicolas M, Berger A, Negrutiu I, Moneger F (2006) Premature arrest of the male flower meristem precedes sexual dimorphism in the dioecious plant Sliene latifolia. Proc Natl Acad Sci USA 103:18854–18859
Acknowledgements
We thank John Chandler and Wolfgang Werr for sharing the drnl-1 allele prior to publication, and for productive discussions. We also thank Jason Reed for the arf mutant lines, Hong Ma for his opinion of the anther expression pattern in DRNL, Chuck Daghlian for his help with SEM, Eileen Piwarzyk and Stacey King for comments on the manuscript, and the Arabidopsis Biological Resource Center for seed stocks. This work was supported by a grant from the US National Science Foundation (IBN-0516736).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nag, A., Yang, Y. & Jack, T. DORNRÖSCHEN-LIKE, an AP2 gene, is necessary for stamen emergence in Arabidopsis. Plant Mol Biol 65, 219–232 (2007). https://doi.org/10.1007/s11103-007-9210-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9210-7