Abstract
Genes encoding wheat prolamins belong to complicated multi-gene families in the wheat genome. To understand the structural complexity of storage protein loci, we sequenced and analyzed orthologous regions containing both gliadin and LMW-glutenin genes from the A and B genomes of a tetraploid wheat species, Triticum turgidum ssp. durum. Despite their physical proximity to one another, the gliadin genes and LMW-glutenin genes are organized quite differently. The gliadin genes are found to be more clustered than the LMW-glutenin genes which are separated from each other by much larger distances. The separation of the LMW-glutenin genes is the result of both the insertion of large blocks of repetitive DNA owing to the rapid amplification of retrotransposons and the presence of genetic loci interspersed between them. Sequence comparisons of the orthologous regions reveal that gene movement could be one of the major factors contributing to the violation of microcolinearity between the homoeologous A and B genomes in wheat. The rapid sequence rearrangements and differential insertion of repetitive DNA has caused the gene islands to be not conserved in compared regions. In addition, we demonstrated that the i-type LMW-glutenin originated from a deletion of 33-bps in the 5′ coding region of the m-type gene. Our results show that multiple rounds of segmental duplication of prolamin genes have driven the amplification of the ω-gliadin genes in the region; such segmental duplication could greatly increase the repetitive DNA content in the genome depending on the amount of repetitive DNA present in the original duplicate region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prolamins, rich in proline and glutamine, are major grass seed storage proteins present in the endosperm of the grain (Shewry and Tatham 1990). Wheat prolamins can be divided into two major groups, glutenins and gliadins, according to their polymerization properties. Polymeric glutenins, consisting of the high molecular weight (HMW) glutenin and low molecular weight (LMW) glutenin, are held together by intermolecular disulphide bonds and form gluten polymers, whereas gliadins, consisting of the α-, γ- and ω-gliadins, are mainly monomeric proteins. The gliadins associate with glutenin polymers in the presence of water through non-covalent interactions, with the resulting gluten dough possessing the visco-elastic characteristics important for many of wheat’s end-use products. Therefore, the bread-making quality of wheat flour is largely determined by the complex relationships of different prolamin components present in the endosperm of the grain (Shewry and Halford 2002).
Because of the importance of prolamins in determining wheat quality, research has been directed towards characterizing and understanding the genomic organization of prolamin genes. The HMW-glutenins are mapped to the Glu-1 loci on the long arm of the homoeologous group 1 chromosomes, while the LMW-glutenins are mapped to the Glu-3 loci on the short arm of the same chromosomes (Payne et al. 1982; Cassidy et al. 1998). Tightly linked to the Glu-3 are the Gli-1 and Gli-3 loci that encode the γ- and ω-gliadins, respectively (Garcia-Olmedo et al. 1982; Singh and Shepherd 1988; Dubcovsky et al. 1997; Metakovsky et al. 1997). The α-gliadin genes are located at the Gli-2 loci of the short arm of the homoeologous group 6 chromosomes (Anderson et al. 1984). One of the challenges in characterizing wheat prolamin genes is the fact that these genes are members of multi-gene families present in each of the three homoeologous A, B, and D genomes of hexaploid wheat. The HMW-glutenin belongs to the smallest prolamin gene family and contains only two copies, named x-type and y-type, in each homoeologous genome (Payne et al. 1982). The α-gliadins are the most abundant, with the estimated number up to 150 different copies (Anderson et al. 1997). The LMW-glutenin gene family contains 30–40 members (Sabelli and Shewry 1991; Cassidy et al. 1998), while the two remaining families, the γ-gliadins and the ω-gliadins, contain 16–39 and 15–18 copies (Sabelli and Shewry 1991), respectively. The fact that such a large number of prolamin genes are located in just few major chromosomal regions suggests that prolamin genes are physically closely linked or clustered. However, little is known about the physical spacing between tightly linked prolamin loci.
Recent genomics studies have provided new tools to elucidate the complex prolamin gene families. A large number of EST collections derived from developing seed cDNA libraries have allowed identification of new classes of prolamin genes and novel seed storage proteins (Anderson et al. 2001; Nagy et al. 2005). The employment of large insert BAC libraries can permit us to study the complex structural organization of prolamin gene families (Gu et al. 2004a; Johal et al. 2004; Ozdemir and Cloutier 2005). Characterization of wheat BAC clones using a α-gliadin gene probe revealed that positive BAC clones carry anywhere from one to five copies of the α-gliadin genes per BAC (Gu et al. 2004a). In contrast, when the BAC clones were used to characterize the LMW-glutenins, no single BAC clone was found to carry more than one copy of the LMW-glutenin gene, suggesting that the seven copies of LMW-glutenin genes in Ae. tauschii, the D genome donor of hexaploid wheat, are likely to be separated by more than a BAC insert’s length (Johal et al. 2004).
Comparative sequence analysis of large orthologous genomic regions from closely related genomes provides an important view of sequence changes that have occurred in recent evolutionary history. To further understand genomic complexity of prolamin genes, comparative analysis on the orthologous regions spanning the two paralogous HMW-glutenin genes from the A, B and D genomes of wheat indicated that distances separating the two genes are variable from 50 to 180 kb in three genomes (Gu et al. 2004b, 2006). In addition, although gene colinearity is generally retained, the intergenic regions consisting of large blocks of nested retroelements are not colinear, suggesting that the insertion of the repetitive DNA occurred after the differentiation of these three genomes between 2.5 and 4.5 million years ago (MYA) (Huang et al. 2002; Gu et al. 2004b; Kong et al. 2004). A detailed sequence comparison of the Glu-3 regions of closely related A genome of a tetraploid wheat (T. turgidum ssp. durum) and Am genome of T. monococcum also revealed a dynamic sequence change at LMW-glutenin region (Wicker et al. 2003). To date, a detail sequence analysis on Gli-1 and Gli-3 locus regions has not been reported, and little is known about the genomic organization of the LMW-glutenin locus region in other homoeologous wheat genomes and its physical relationship to the Gli-1 and Gli-3 locus regions.
In this study, we sequenced two large-insert BAC clones containing both gliadin and LMW-glutenin genes from the A and B genomes of T. durum wheat. Our results revealed that the gliadin and LMW-glutenin genes are clustered immediately adjacent to one another. In addition, non-prolamin genes are present in the same region and are interspersed between them. We were able to identify a series of sequence duplication events that resulted in an increased copy number of prolamin genes. Furthermore, comparison of the orthologous prolamin gene-containing regions from the A and B genome of T. durum allowed us to further examine dynamic sequence evolution in the two homoeologous wheat genomes.
Materials and methods
BAC selection and sequencing
A half million clone BAC library of tetraploid durum wheat, T. turgidum ssp. durum (2n = 4x = 28, AABB), cultivar “Langdon” was screened with a probe mixture containing the coding sequences of both LMW-glutenin and γ-gliadin according to the method described by Kong et al. (2004). A total of 148 positive BAC clones were retrieved from the screening. Positive BAC clones were fingerprinted to assemble BAC contigs as described previously (Gu et al. 2004a). Positive BAC clones were also hybridized with LMW-glutenin and γ-gliadin probes separately to search for clones carrying both probe sequences. BAC clones 790O10 and 1144H03 showed hybridization with both probes and are overlapping clones associated in the same contig with BAC107G22 previously characterized by Wicker et al. (2003). BAC790O10 has a larger insert size (∼158 kb) as compared to the insert of BAC1144H03 (∼116 kb) and low-pass shotgun sequencing of BAC790O10 identified the presence of both LMW-glutenin and gliadin gene sequences from the A genome. To search for BAC clones containing LMW-glutenin gene from the B genome, a set of primers reported for specific detection of the B genome LMW-glutenin gene was used to screen all positive clones (Gale et al. 2003). Using the primer set, BAC419P13 provided a PCR product derived from the LMW-glutenin in the B genome (data not shown). Further characterization of this BAC clone by Southern hybridization and shotgun sequencing indicated that it represents a region orthologous with BAC790O10 and BAC107G22. Therefore, BAC790O10 and BAC419P13 were selected for sequence completion.
The sequencing of the two Triticum turgidum BAC clones was carried out as described previously (Kong et al. 2004). In brief, shotgun sequencing libraries for selected BAC clones were first constructed with randomly sheared BAC DNA isolated with a Large Construct Kit (Qiagen). The sheared DNA was blunt ended by incubation with a mung bean exonuclease (BioLab) and dephosphorylated using a shrimp alkaline phosphatase (USB). Single “A” tails were added by incubating with Taq polymerase in the presence of dNTPs. The resulting DNA was fractionated in agarose gel and a fraction of DNA with size of 3–5 kb was selected for purification using Gel Extraction Kit (Qiagen). The DNA was ligated into pCR4TOPO vectors and transformed into DH10B electroMAX cells (Invitrogen). Inserts of plasmid DNA were sequenced from both directions with T7 and T3 primers using BigDye terminator chemistry (Applied Biosystems) on an ABI3730xl capillary sequencer.
Sequence analysis
For sequence assembly, a target of ∼10-fold coverage was chosen. The sequence data generated from each BAC clone was used to assemble continuous contigs using the Lasergene SeqMan module (DNAStar) (www.DNAStar.com). In this module, a high stringency parameter for base calling and quality assessment was selected to generate the most accurate consensus sequence reads possible. Phrap assembly engine (http://www.phrap.org) was also used for contig assembly. In some cases, assemblies with two different programs helped resolve gap regions and the order of contig. For gap filling, a primer-walking procedure was employed to sequence clones whose sequences are located at the ends of different contigs, and the dGTP BigDye terminator chemistry (Applied Biosystems) was used in the sequencing reaction to resolve the problem of potential secondary structures caused by high G/C content in the gap regions. In the regions where low sequence coverage was observed, primers were designed to amplify the regions from BAC DNA to improve the sequence coverage by sequencing the PCR products. The accuracy of the sequence assembly was also verified by comparing digestion pattern of BAC DNA with HindIII, EcoRI, and NotI with the predicted restriction pattern of the computer-assembled sequence. The contiguous sequences from BAC790O10 and BAC419P13 were deposited in the NCBI database with Accession Nos. of EF426564 and EF426565, respectively.
For annotation, the contiguous sequence of BAC insert was searched against the Triticeae Repeat Sequence Database (TREP) at the GrainGenes web site at http://wheat.pw.usda.gov/ITMI/Repeats/ to identify known repetitive DNA. The long terminal repeat sequences of LTR retrotransposable elements were delineated using Dotter analysis and BLAST search. For the sequence unmasked with repetitive DNA, a homology search was performed against NCBI non-redundant and dbEST databases using BLASTN, BLASTX, and TBLASTX algorithms. FGENESH (http://www.softberry.com/nucleo.html) and GENESCAN (http://genemark.mit.edu/GENESCAN.htm) were used for gene prediction. Sequences identified as candidate genes by the gene-finding programs were further investigated to determine if they are actually gene sequence by a homology search of predicted exons against protein database using BLASTX. They were considered likely genes only if a match with an expect value of <e−10 was found in the database.
Southern hybridization
For Southern hybridization of BAC clones, the HindIII-digested BAC DNA was size fragmented in 1% agarose gel and then blotted onto Hybond N+ membranes (Amersham, Piscataway, NJ). Probes were labeled with 32P isotope using the DECAprimeII DNA labeling Kit (Ambion, Austin, Texas). After hybridization for 16 h in a solution containing 0.5 M sodium phosphate (pH 7.2), 7% SDS, and 1% bovine serum albumin (BSA), the blot was washed three times in 0.2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) plus 0.1% SDS. The images were detected by phosphorimaging.
PCR-based mapping of BAC clones to specific chromosomes
To map BAC clones to specific wheat chromosomes, PCR primers were designed from the junction regions between two repetitive DNA elements. Because the insertion of a specific repetitive DNA element into another element is unique, the primers targeting the junction region will be specific for the region present in a BAC clone, and such primers can be used to map sequenced BAC clones onto specific chromosomes using chromosome deletion/substitution lines available for wheat (Devos et al. 2005). To map BAC790O10 and BAC419P13, two primers sets, 790O10F-TTAATTGCAAGGAACTACTACAC and 790O10R-AAATTATAGATACGTTGGAGACAT, and 419P13F-CAACGTAGACAAGGGTAAAAATGG and 419P13R-ACGGTAGGTGTTGCGGTTAGTA, were designed, respectively, using the strategy described above. These two primer sets were used to amplify products from DNA templates extracted from T. durum cultivar “Langdon” and Langdon substitution lines, LDN1D(1A) and LDN1D(1B). BAC DNA templates were used as controls in the PCR.
For PCR amplification of specific m-type and i-type LMW-glutenin genes, LMW-glutenin sequences from the sequenced BACs and retrieved from GenBank (AB062862, AB062861, AY585350, AY585355, AY585349, AB062877, and AB062878) representing both types of genes were used for Clustal W analyses. Primers were designed based on the sequence around the deletion region. Forward primer LMW-i-F-GCCGTTGCGCAAATTTCACAGC is specific for the i-type and LMW-m-F-CAAGTGCCATTGCACAAATGGAG for the m-type LMW-glutenin genes. The reverse primer LMW-R1-GGAGGAATACCTTGCATGGGT is derived from a conversed region and can be used for both types of LMW-glutenin.
Results
Identification and characterization of wheat BACs carrying both gliadin and LMW-glutenin genes
To discern the structural organization of the genomic regions containing prolamin genes, a half million clone BAC library of tetraploid T. durum wheat was screened with a probe mixture containing both LMW-glutenin and γ-gliadin coding sequences (Cenci et al. 2003). Among 148 positive clones, two BAC clones (BAC790O10 and BAC1144H03) hybridized to both the LMW-glutenin and γ-gliadin probes. Positive BACs were also fingerprinted for contig assembly according to the method described previously for characterization of BAC clones containing α-gliadin genes (Gu et al. 2004a). BAC790O10 and BAC1144H03 are overlapping clones associated in a contig that consists of 13 BAC clones (Fig. 1A). Southern hybridization results indicated that in this contig, six BACs (Fig. 1B, Lanes 1–6) hybridized to the γ-gliadin probes, while five BACs (Fig. 1B, Lanes 9–13) strongly hybridized to LMW-glutenin probe only. Based on Southern hybridizations and further sequence analysis (see below), it is likely that the contig contains at least five γ-gliadin and two LMW-glutenin genes (Fig. 1A and B). The contig includes BAC107G22, which has been previously characterized by Wicker et al. (2003). BAC107G22 from the A genome of T. durum wheat contains a LMW-glutenin gene and an allelic region of the Pm3 locus that confers race-specific resistance in hexaploid wheat to the powdery mildew fungal pathogen (Blumeria graminis f. sp. tritici) (Wicker et al. 2003; Srichumpa et al. 2005; Yahiaoui et al. 2006). Therefore, the identified contig represents a region that contains at least three genetic loci, Gli-1, Glu-3, and Pm3 from the A genome of T. durum (Fig. 1A). BAC790O10 was selected for sequencing because it contains two γ-gliadin genes and an additional LMW-glutenin gene (Fig. 1A). This BAC will also identify junction sequences between same types of prolamin genes and between different types of prolamin genes (γ-gliadin and LMW-glutenin) for understanding the genomic organization of regions containing multiple prolamin genes.
Characterization of BAC clones containing LMW-glutenin and gliadin genes. (A) A BAC contig spans three genetic loci. BAC clones positive for γ-gliadin and/or LMW-glutenin probes were fingerprinted for contig assembly according to the method described previously (Gu et al. 2004a). BAC790O10 and BAC1144H03 were hybridized with both the probes and associated in the same contig, Contig5. BAC790O10 also overlaps with BAC107G22 containing LMW-glutenin and putative RGL-like resistance genes (Wicker et al. 2003). Both BACs sequenced previously and in this study were colored with gray in the contig. The LMW-glutenin, γ-gliadin, and putative RGL-like resistance genes are labeled with different shapes as indicated, and their relative positions and numbers of copy in the BACs are estimated according to the Southern hybridization (see B) and BAC sequencing results. (B) Southern hybridization of BAC clones in the contig. BAC DNA was digested with HindIII restriction enzyme. The restriction fragments were separated by agarose gel electrophoresis. Duplicate gels were blotted onto Hybond N+ membranes and hybridized with either the γ-gliadin (left panel) or LMW-glutenin (right panel) probes. BAC DNA in each lane is as follow: Lane 1, 865A04; lane 2, 930M22; lane 3, 1185B11; lane 4, 1140K04; lane 5, 548O20; lane 6, 1108C02; lane 7, 1144H03; lane 8, 790O10; lane 9, 107G22; lane 10, 403L21; lane 11, 1164N12; lane 12, 216M03; lane 13, 032E20. Positions for DNA size standard markers are provided. Positive bands for each probe are indicated with solid triangles. The positive γ-gliadin band in lanes 6, 7, and 8 consisted of two 1.82-kb fragments, each containing one γ-gliadin gene based on the BAC sequence analysis. (C) Mapping sequenced BAC clones onto specific chromosomes. Primers designed from repetitive DNA junction regions present in BAC790O10 (left panel) and BAC419P13 (right panel) were used in PCRs using DNA templates extracted from T. durum cultivar “Langdon” (lanes 1 and 5), two Langdon substitution lines, LDN1D(1A) (lanes 2 and 6), and LDN1D(1B) (lanes 3 and 7). BAC790O10 (lane 4) and BAC419P13 (lane 8) were used as positive controls. DNA size standard (lane M) was used to determine the size of amplified PCR products
In a search for LMW-glutenin genes from the B genome, BAC419P13 and BAC317N07 were identified using a set of primers specific for the B genome LMW-glutenin genes (Gale et al. 2003). These are also the only positive BAC clones that overlapped each other. Further characterization indicated that BAC419P13 has a larger insert that hybridized not only with the LMW-glutenin and ω-gliadin probes, but also the Pm3-related sequence probe (data not shown), suggesting that BAC419P13 represent an orthologous Gli-1/3-Glu-3-Pm3 region from the B genome of T. durum.
To confirm that BAC790O10 and BAC419P13 are derived from the A and B genomes, respectively, we employed a PCR method developed by Devos et al. (2005) to map BAC clones using specific primers designed from repeat boundary/junction regions. The primer set derived from BAC790O10 amplified a product from Langdon and from a substitution line, LDN1D(1B), with the 1A chromosome substituted by the homoeologous chromosome from the D genome (Joppa and Williams 1988), but did not produce a PCR product in the substitution line, LDN1D(1A) (Fig. 1C, left panel). Meanwhile, the other primer set, derived from BAC419P13, produced a PCR product from Langdon and LDN1D(1A), but failed to amplify a product in LDN1D(1B) line (Fig. 1C, right panel). These results verified the chromosomal locations of BAC790O10 and BAC419P13.
Sequence organization of BAC clones
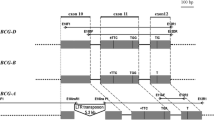
BAC790O10 and BAC419P13 were completely sequenced and annotated using a combination of various BLAST searches and bioinformatic analyses. The A genome BAC clone, BAC790O10, is 158 kb in size and contains 13 genes (Table 1), which are mainly located in two gene islands. The first gene island contains 8 genes (Gene1–Gene8) within a 30-kb region, with a gene density of one gene per 3.8 kb, the highest gene density yet reported in wheat. The second gene island contains four putative genes (Gene9–Gene12) in a 26-kb region with a gene density of one gene per 6.5 kb. The two gene islands are separated from each other by a ∼19-kb repetitive DNA region (Fig. 2). The 3′ end of the BAC insert consists mainly of repetitive DNA and overlaps BAC107G22 by a region of 35 kb (Fig. 2). The available sequence from BAC107G22 allowed us to extend the repetitive region and resolve the complex structure of the nested repetitive DNA elements (Supplement 1). This large block of repetitive DNA region (∼100 kb) separates the two LMW genes (Gene12 and Gene14) residing on different BACs. A pseudogene fragment (Gene13) of 111 bp that has 70% homology with the protein sequence encoded by the last exon of a Glabra2-like gene is located inside a gypsy retrotransposon, Fatimah-p (Supplement 1). This pseudogene sequence was previously identified in BAC107G22 and suggested to have been acquired by the gypsy retrotransposon (Wicker et al. 2003).
Structural organization in the orthologous prolamin gene-containing regions from the A and B genomes of T. turgidum. BAC107G22 that overlaps with BAC790O10 was sequenced and annotated by Wicker et al. (2003). Different types of genes represented by different color boxes are labeled with numbers and their relative positions are indicated along the contiguous sequences of the BAC clones from T. turgidum. The arrows below the gene numbers indicate the transcriptional orientation for each corresponding gene. The predicted identity of each gene is showed in Table 1. Boxes filled with dark gray represent regions containing repetitive DNA. The box filled with line pattern represents a region with few nested repetitive elements as described by Wicker et al. (2003). Orthologous genes from the A and B genomes are connected with a dash line. The RFLP marker SFR159 was indicated with a crossed box
BAC419P13 from the B genome is about 140 kb in length. A total of 12 genes were identified in the 140-kb region with an average gene density of one gene per 11.7 kb (Fig. 2). However, it is worthy to note that several of the prolamin genes are actually pseudogenes (Gene20, Gene22, Gene24, and Gene27) (see below). The gene density based on intact genes would be much lower in this region. The total amount of repetitive DNA in BAC419P13 is about 70 kb, accounting for approximate 51% of the sequenced region. The repetitive DNA content in this region is considerably lower than the average 80% repetitive DNA content reported in most other sequenced wheat BACs (SanMiguel et al. 2002; Wicker et al. 2003; Gu et al. 2004b; Isidore et al. 2005). This may be attributed to the observation that large blocks of nested retrotransposons (>40 kb) were not identified; instead, regions with one or two genes separated by relatively small blocks of repetitive DNA (∼20 kb) were often present (Fig. 2).
Comparison of orthologous Gli-1/3-Glu-3-Pm3 regions between the A and B genomes of T. durum
Combining the sequences of BAC790O10 and BAC107G22 through their overlapping sequences yields a region of approximately 265 kb in length (Fig. 2). BAC107G22 contains a single LMW-glutenin gene, TdGluA3-1(Gene14), which is followed by a marker, SFR159. The presence of unique SFR159 immediately downstream of a LMW-glutenin gene was used by Wicker et al. (2003) to verify the orthologous relationship of BAC107G22 from the A genome of T. durum wheat with a region from the Am genome of T. monococcum. We found that in the B genome region of BAC419P13, a segment following the LMW-glutenin (Gene30) has an 85% sequence identity over a 0.8-kb region with the SFR159 from the A genome (Fig. 2), further supporting that the two LMW-glutenin locus regions from the A and B genomes are orthologous. Colinear genes between the two genomes also include a GPI-anchored protein gene (Gene8 and Gene28) and an RGL-related Pm3 resistance gene analog (Gene19 and Gene31). The TdLRR-1 pseudogene (Gene15) in the A genome was identified in the B genome (Gene29); its position with respect to the LMW-glutenin gene (Gene30) was inverted (Fig. 2). Taken together, the data suggests that the B genome region from BAC419P13 is orthologous to the A genome region represented by BAC790O10 and BAC107G22.
Despite some conservation of the above genes, violation of gene colinearity is present in the orthologous regions from the A and B genomes. In addition to the duplicate copies of the LMW-glutenin genes in the A genome and the inversion of the TdLRR-1 pseudogene, Gene16, Gene17, and Gene18 are missing in the B genome BAC419P13. Gene9–Gene11 are also present only in the A genome. The acquisition of the Glabra2-like gene fragment (Gene13) by the gypsy retrotransposon occurred only in the A genome. Furthermore, large number of repetitive DNA elements have differentially inserted into intergenic regions, which significantly expanded the distance between certain genes in the A genome. Because of this, and along with considerable violation of gene colinearity, the gene island containing Gene28–31 in the B genome appear to correspond to several regions in the A genome (Fig. 2), suggesting that gene islands might not be conserved among the homoeologous wheat genomes.
More strikingly, we found that upstream of the GPI-anchored protein gene (Gene8 and Gene28), there are eight ω-gliadin genes (Gene20–27) (see discussion) in the B genome, while in the A genome multiple γ-gliadin and γ-gliadin-like genes are mainly present. Although the γ-gliadins encoded by Gli-1 and the ω-gliadins encoded by Gli-3 are evolutionarily more closely related to each other than either are to other prolamin genes (Sabelli and Shewry 1991; Hsia and Anderson 2001), it is more likely that considerable sequence changes have occurred after the split of the two homoeologous wheat genomes and caused rearrangements of prolamin genes in these genomic regions.
Prolamin and non-prolamin genes in the sequenced regions
Our data indicated several prolamin genes can be physically associated within a BAC insert size. BAC790O10 from the A genome contains six prolamin gene sequences. The first two (Gene1 and Gene2) are both γ-gliadin genes and are 7.7-kb apart and share 98% identity. In addition to the two typical γ-gliadin genes, sequence analyses revealed the presence of gliadin-related genes in the sequenced region. A BLASTx search identified Gene3, which is a gliadin-related gene since the E value against γ-gliadins is only 6.0 × 10−13. We named Gene3 a gliadin-like gene. A strong match of Gene3 to a γ-gliadin related EST, CD919876, was identified with an E value of 1.5 × 10−96 (Table 1). However, it is likely that Gene3 is a pseudogene or gene fragment since a correct translational initiation site was not identified and mutations have caused translational disruption of the coding region. Similarly, Gene5 is another inactive gliadin-like gene. Gene6 encodes a gliadin product that lacks a prominent repeat domain. Such genes related to wheat prolamin genes have already been identified and termed low molecular weight gliadins (LMW-gliadin) (Anderson et al. 2001). Given the presence of the two gliadin-like genes and one LMW-gliadin gene in this sequenced region, we might expect much more gliadin-related gene sequences in the wheat genome.
In BAC419P13 of the B genome, there are a total of 9 prolamin genes and gene fragments. Eight of them (Gene20–Gene27) belong to the ω-gliadin gene family (Table 1). Gene21 and Gene23 are likely to be intact genes since they are both translatable into two proteins with 444 and 439 amino acids, respectively. They share a 98% sequence identity with each other at the nucleotide level. The other six ω-gliadin genes are likely inactive caused by different types of sequence changes. Although Gene26 has a full-length ω-gliadin gene sequence and shares a ∼96% nucleotide identity with Gene21 and Gene23, it contains multiple in-frame premature stop codons in the coding region. Gene25, which contains both the start codon at the 5′ end and a stop codon at the 3′ end, is only 390-bp in length, missing a large portion of the ω-gliadin repeat in the coding region. Four ω-gliadin genes (Gene20, Gene22, Gene24, and Gene27) are clearly fragments since they only contain a ∼100-bp segment of the coding sequence from the 3′ end of the gene. Their similar structures suggest that these gene fragments are paralogous and that a deletion to an ancestor ω-gliadin gene occurred before the duplication of these gene fragments.
In contrast to gliadins, only one LMW-glutenin gene was found in each sequenced BAC. Although in the region of the A genome, there is second LMW-glutenin gene (Gene14), it is located in different BAC and more than 100 kb apart from the first LMW-glutenin gene (Gene12) (Fig. 2). It appears that LMW-glutenin genes are not clustered like those of gliadin genes.
Previously, it was not clear if the prolamin gene regions are devoid of non-prolamin genes. We identified several non-prolamin genes in the sequenced genomic regions (Fig. 2 and Table 1). These include genes encoding a cyclophilin-like protein (Gene4), a UDP-glycosyltransferase (Gene9), GPI-anchored proteins (Gene8 and Gene28), and putative genes (Gene7, Gene10 and Gene11) encoding unknown proteins (Table 1). These non-prolamin genes are interspersed between the two different types of prolamin genes (Fig. 2). For example, between γ-gliadin and LMW-glutenin genes in the A genome, there are six non-prolamin genes (Gene4, 7, 8, 9, 10, 11). Two non-prolamin genes are present between ω-gliadin and LMW-glutenin genes in the B genome (Gene28 and 29). The insertion of non-prolamin genes can also occur between the same types of prolamin genes. For instance, in the gliadin region of the A genome, a full-length cyclophilin gene (Gene4) resides between two gliadin-like genes (between Gene3 and Gene5).
A deletion in the m-type LMW-glutenin resulted in the i-type gene in the A genome
The two LMW-glutenin genes (Gene14 and Gene30) are clearly orthologous due to the presence of SFR159 marker in the same position following the two genes. The two genes share more than 90% nucleotide identity at the 5′ and 3′ coding regions. The middle regions are less conserved (∼80% identity) likely due to the frequent indel events in the repetitive domain region of the LMW-glutenin genes (data not shown). In the A genome, the first LMW-glutenin gene (Gene12) shows 94% nucleotide identity with its paralogous gene (Gene14). However, it does not have the SFR159 marker following the 3′ end (Fig. 2) and its coding region contains three premature stop codons. Because of a higher sequence identify, it is likely that the duplication of the paralogous genes (Gene12 and Gene14) occurred after the separation of the orthologous genes (Gene14 and Gene30) in A and B genomes.
LMW-glutenins can be grouped into three different types (m, s, and i) based on the first amino acid residue of the mature protein—methionine, serine, or isoleucine, respectively (D’Ovidio and Masci 2004). It is proposed that the s-type LMW-glutenin with the N-terminal sequence of SHIPGL probably originates through differential gene processing on the m-type LMW-glutenin due to the presence of an asparagine residue (N) at the N-terminal region (MENSHIPGL) (D’Ovidio and Masci 2004). The evolution of i-type LMW-glutenin genes is not clear. Analysis of the translated product of Gene30 shows that the N-terminal sequence after the signal peptide is _“MENSHIPGL”_ (Fig. 3), indicating that it belongs to the m-type LMW-glutenin, while both Gene12 and Gene14 are the i-type LMW-glutenin since their amino acid sequences after the signal peptide are ISQQQQ (Fig. 3). The result suggests that the orthologous genes from the A and B genomes encode different types of LMW-glutenins. This allowed us to have a more direct comparison to understand the evolutionary relationship between the two LMW-glutenin genes. When the 5′ end sequences of the LMW-glutenin genes from the A and B genomes were aligned (Fig. 3), a sequence of 33 bps flanking by a short direct repeat (AT) in the B genome LMW-glutenin was absent in the A genome immediately following the signal peptide sequence. This sequence change caused a codon change from a M to I and a removal of 11 amino acids in the LMW-glutenin gene from the A genome (Fig. 3).
Alignment of m-type and i-type LMW-glutenin sequences. The sequences from the 5′ coding regions of the LMW-glutenin genes from the A genome (Gene14) and A genomes (Gene30) were aligned with Clustral W program. The dash lines represent gap regions between two sequences. The translated peptide sequences are indicated either above or below the corresponding coding sequence. The signal peptide sequence and the start of mature peptide are labeled
The result from Fig. 3 suggests that the i-type LMW-glutenin gene in the A genome resulted from a deletion event in the m-type gene. If multiple LMW-glutenin genes existed prior to this indel event, we might expect that the m-type LWM-glutenin gene(s) are present in the A genome and the indel only occurred to a specific Glu-3 locus region. To test this hypothesis, forward primers specific for the m-type and i-type LMW-glutenin genes were designed, and the reverse primer is derived from a conserved region of LMW-glutenin genes from the A, B and D genomes of wheat (see Materials and methods). The primer set specific for the i-type LMW-glutenin gene only produced a product(s) from genomic DNA containing the A genome of wheat (Fig. 4). It failed to amplify products from either the S or D genomes of different diploid wheats. In addition, using Chinese Spring and its group 1 nulli-tetrasomic lines, a PCR product was amplified with Chinese Spring, N1BT1A (Fig. 4, lane 7) and N1DT1A (Fig 4, lane 8), but no product was detected with N1AT1B (Fig. 4, lane 6). Taken together, it is likely that the deletion event occurred in the diploid A genome before wheat polyploidization.
PCR amplification of specific m-type and i-type LMW-glutenin genes in different wheat genomes. Primer sets specific for m-type and i-type LMW-glutenin genes were used in PCRs with genomic DNA extracted from different wheats. Lane 1, diploid T. urartu (AA genome); lane 2, diploid Ae. speltoides (SS genome); lane 3, diploid Ae. tauschii (DD genome); lane 4, tetraploid T. turgidum durum wheat (AABB genome); lane 5, Hexaploid T. aestivum Chinese Spring wheat; lane 6, CSN1AT1B (Chinese Spring nullisomic 1A, tetrasomic 1B); lane 7, CSN1BT1A); lane 8, CSN1DT1A. Lane M, 1-kb Plus DNA Ladder (Invitrogen)
When the primer set specific for the m-type LMW-glutenin gene was used, we detected PCR products from all the wheat genomic DNA examined in this experiment, indicating that the m-type LMW-glutenin genes are present in all three wheat genomes. This also suggests that the sequence change was a deletion event since it is unlikely that the same insertion would occur to the three wheat genomes. Multiple PCR bands from some genomic DNA were anticipated; even single band in the gel could be derived from PCR products of different LMW-glutenin genes, since primers were designed from the conserved regions for LMW-glutenin genes. The fact that both the primer sets amplify products from the diploid A genome suggests that both types of LMW-glutenin genes are present in the A genome (Fig. 4). The deletion event resulting in the conversion of m-type to i-type genes might only happened to one of the m-type LMW-glutenin genes in the diploid A genome. This result provides an explanation for the previous finding that the i-type LMW-glutenins in hexaploid wheats are A genome specific (Zhang et al. 2004; Ikeda et al. 2006).
Gene duplication
Prolamin genes exist in multiple copies, suggesting gene duplication was an active process during their evolution. In BAC 790O10, the two γ-gliadin genes (Gene1 and Gene2) shared ∼98% sequence identity. Analysis of the flanking sequences revealed that a 3.5-kb segment containing the γ-gliadin gene and a repetitive region consisting of two fragments of LTR retrotransposons, Erika and Sabrina were also a part of the duplication event. Detailed analysis of the two paralogous regions identified a 235-bp segment that repeated three times in the duplicated region. Such structure suggests that a cross-over between D1 and D3 through homologous recombination resulted in the duplication of the complete ∼7.5 kb region, and the D2 region in the middle is likely to be a derivative of this event (Fig. 5A).
Model for the evolution of prolamin gene-containing regions by duplication. (A) Duplication of a gliadin-containing region through unequal cross-over recombination. D1, D2, and D3 are repeat regions sharing 95% sequence identify. γ-gliadin genes are indicated by black boxes, with transcription orientation indicated by an arrow. The partial Erika and Sabrina retroelements are labeled in the color boxes. (B) Multiple duplication events in the ω-gliadin gene region. Both ω-gliadin genes and gene fragments are indicated with black boxes with different length and their gene numbers corresponding to the designation in Fig. 2 are provided. Putative transcription orientation of each gene or gene fragment is indicated by an arrow. Transposable elements are labeled with different color boxes. The number in the circle represents the each duplication event, and the origin of duplication and duplicated products are bracketed
The multiple ω-gliadin genes in the B genome also appear to have been derived from gene duplication events. Given that four of the ω-gliadin gene fragments (Gene20, Gene22, Gene24, and Gene27) all contain only the last 100 bps from the 3′ end of the gene, and considering the patterns of duplication and insertion of repetitive DNA elements, we propose a model that involves at least 4 rounds of duplications, resulting in 8 ω-gliadin genes in the sequenced BAC region (Fig. 5B). The first duplication occurred in an ancestor locus containing two ω-gliadin genes, one an intact copy and the other a 100-bp gene fragment. The first duplication resulted in four ω-gliadin genes; two fragments and two that are intact. The second duplication occurred after the deletion of one of the intact genes and insertion of a WIS retroelement between the two ω-gliadin genes (Gene20 and Gene 21). After the second duplication, a CACTA element inserted in front of the WIS element into one of the duplicated regions. Hence, the region involved in the third duplication contained two ω-gliadin genes separated by a 15-kb repetitive DNA region. The fourth duplication involved a region containing only one ω-gliadin gene. After this duplication, a deletion occurred, leading to another fragmented version of the original intact ω-gliadin gene, Gene25 (see previous discussion). During the evolution of this ω-gliadin gene region, three retroelements (Jela, Fatima, and WIS) have inserted upstream of Gene27. Taken together, frequent sequence duplication, along with insertion of retroelements, might have reshaped this region. Basically, in addition of the increase of the ω-gliadin genes, the size of the segment has also been expanded from ∼10 kb to a much larger ∼110 kb.
Discussion
In this study, we performed a detailed sequence comparison of the orthologous regions harboring four important genetic loci, Gli-1, Gli-3, Glu-3, and Pm3, from the A and B genomes of T. durum wheat. This analysis not only reveals the dynamics of sequence rearrangements in the orthologous regions from two homoeologous wheat genomes, but also provides insight into the molecular mechanisms underlying the evolution of the prolamin genes. Furthermore, this study reveals the extent of the physical linkage between the gliadin genes and LMW-glutenin genes, and the unexpected presence of non-prolamin genes within different types of prolamin gene loci.
Clustering of prolamin genes in wheat genomes
Despite the genetic evidence prior to this study that indicated Gli-1, Gli-3, and Glu-3 loci were tightly linked, the genomic organization and physical spacing between any two prolamin loci had not been well characterized at the sequence level. In this study, sequencing large-insert BAC clones revealed that in the B genome, there are eight ω-gliadin genes/gene fragments in a ∼100 kb region. Such a tightly linked organization for ω-gliadin genes has not been previously reported. Gene clustering also holds true for the γ-gliadin genes (Fig. 1A). We showed that the clustering of the ω-gliadin genes is the consequence of multiple rounds of segmental duplications (Fig. 5B).
Gene duplication can occur at different levels. While whole genome duplication can result in duplicate copies of all the genes in the genome, smaller scale duplications only involve individual genes or genomic segments (Lawton-Rauh 2003; Moore and Purugganan 2005). Multiple segmental duplications that result in a high density of duplicate genes has been described; however, it is usually difficult to delineate the segmental region involved in each duplications (Yuan et al. 2002; Zhang et al. 2005). Therefore, the approach to understanding the mechanism of gene family evolution is to construct phylogenies for the gene families based on sequence identities (Cannon et al. 2004). In our study, evidence based on the four ω-gliadin gene fragments derived from the same deletion event and the repetitive DNA insertion patterns helped identify four rounds of segmental duplication events in this region, resulting in eight copies of ω-gliadin sequences. Multiple duplications suggest that the genomic region containing these ω-gliadin sequences might be a hot spot for segmental duplication. Interestingly, the Pm3 resistant locus carrying multiple copies of the Pm3 genes resides in the nearby region (Yahiaoui et al. 2006). However, large gene families such as MADS-box and NBS-LRR resistance gene clusters are often present at multiple genomic locations on different chromosomes, suggesting that single-gene and whole-genome duplications both contribute to the diversity of plant species (Baumgarten et al. 2003; Moore and Purugganan 2005; Rijpkema et al. 2007). Wheat prolamin gene families are primarily localized in three genomic regions, two on chromosome group1 and one on chromosome group 6. It is likely that whole-genome duplication did not significantly contribute to the increase of prolamin genes, whereas, segmental duplication is the major force in the evolution of prolamin gene families in the wheat genome.
In contrast to the clustering of the gliadin genes, LMW-glutenin genes are often separated from each other by much larger distances. The two paralogous A genome LMW-glutenin genes (Gene12 and Gene14) are approximately 100 kb apart (Fig. 2 and Supplement 1). The study on the Glu-3 region from the Am genome of T. monococcum showed that the three paralogous LMW-glutenin genes resided on three different BAC clones and the two LMW-glutenin genes, TmGlu-A3-2 and TmGlu-A3-3, are separated by 150 kb (Wicker et al. 2003). Previous studies on LMW-glutenin genes from the D genome of Ae. tauschii also indicated that all seven of the LMW-glutenin genes in the genome are located on seven different BACs (Johal et al. 2004), again implying a large distance between any two LMW-glutenin genes. It has been shown that the rapid amplification of retrotransposable elements in the wheat genomes can greatly increase the size of intergenic regions (SanMiguel et al. 2002; Wicker et al. 2003; Gu et al. 2006). In this study, we found that the intergenic region between two paralogous LMW-glutenin genes (Gene12 and Gene14) was significantly expanded by the insertion of large blocks of nested repetitive DNA since the gene duplication. It is possible that rapid amplification of repetitive DNA might have also increased the intergenic regions between some gliadin genes. We realized that both sequenced BACs carry only portions of the gliadin gene families, and our Southern hybridization experiment revealed that almost half of the gliadin BACs (approximately 34 BAC clones) contained single gliadin gene (data not shown). Previous analyses on the α-gliadin family indicated that 70% of end sequences of the α-gliadin positive BACs are repetitive DNA (Gu et al. 2004b).
Structural organization of prolamin genes in wheat genome
Wheat prolamin genes are unique to Triticeae species, suggesting their recent evolutionary history. Model species such as rice will not provide insights about the creation and evolution of species-specific genes. We also could not identify orthologous regions in the rice genome based on sequence comparison. Direct sequencing of the wheat BACs provided an important view of structural organization of the genomic regions containing multiple prolamin genes. One interesting finding in this study is the identification of several LMW-gliadin gene and γ-gliadin-like genes in the sequenced A genome region. While the LMW-gliadin genes are known to be expressed in the endosperm and are usually lacking a prominent repeat domain (Anderson et al. 2001), the γ-gliadin-like genes have not been characterized. Our sequence data now revealed that at least some are mapped near the Gli-1 locus. The evolutionary relationship among the LMW-gliadins, γ-gliadin-like genes, and typical γ-gliadin genes, such as Gene1 and Gene2, is not clear. However, it appears that they are similar enough to the gliadins to be included as member of the gliadin family of genes (Anderson et al. 2001). Despite the fact that the γ-gliadin-like genes identified here are likely pseudogenes or inactive, it is possible that these types of genes are active in other genomic regions or different Triticeae genomes. Furthermore, the discovery of new types of prolamin related sequences in the prolamin regions suggests that prolamin superfamily might be more complex than originally thought (Shewry and Tatham 1990).
Another notable finding is that prolamin genes are interspersed with non-prolamin genes (Fig. 2). This structural organization provides further assistance for explaining the previous observation that fingerprinting and subsequent assembly of prolamin BAC clones did not result in large contigs spanning the entire locus region; instead multiple contigs and singletons were produced, likely owing to the large distance separating the prolamin genes (Gu et al. 2004a; Ozdemir and Cloutier 2005). Similar result was obtained in this study when both the gliadin and LMW-glutenin positive BAC clones were fingerprinted and assembled together (data not shown). The mosaic organization of prolamin and non-prolamin genes also makes it difficult to determine the size of the genomic regions spanning the prolamin gene loci within the wheat genomes. For example, we only analyzed two orthologous prolamin regions from the A and the B genomes. Other prolamin genes closely linked to the studied regions are likely present in each wheat genomes. It would be interesting to examine if the genomic organization of the other prolamin regions are similar to the analyzed BACs, and what are the physical distances between different types of prolamin genes in other regions?
Rapid sequence changes in homoeologous wheat genomes
Our results provided further support to the notion that the intergenic regions between wheat homoeologous genome are not conserved (Gu et al. 2004b; Chantret et al. 2005). Transposable elements comprise of over 75% of the wheat genome with a size of 17 Gb (Li et al. 2004; Paux et al. 2006) and the majority of retrotransposable elements inserted into their current positions within the last ∼2 million years (SanMiguel et al. 1998, 2002; Ma et al. 2004; Brunner et al. 2005). In addition, transposable elements have the tendency to insert into similar elements. Therefore, large and complex structures with multiple tiers of nested insertions can be formed in the intergenic regions within a short revolutionary history, resulting in differential expansion in local genomic regions (Gu et al. 2004b). In this study, we found that the two LMW-glutenin genes in the A genome are separated by more than 100 kb of repetitive DNA, while this local expansion is not present in the orthologous region from the B genome (Fig. 2 and Supplement 1).
While retroelement amplification is the primary means by which repetitive DNA causes large-scale genome expansion, segmental duplication also contributes to the increase of repetitive DNA content in the genome. Two of the four segmental duplications in the B genome involved regions containing repetitive DNA (Fig. 3B). The repetitive DNA contributed by the segmental duplication accounts for 30% of the total repetitive DNA in BAC419P13. The segmental duplication in the A genome BAC790O10 also involved a region containing a gliadin gene and 3.5 kb of repetitive DNA (Fig. 3A). Wicker et al. (2003) described a segmental duplication of a 54-kb region of mostly repetitive DNA which is present in the Am genome of T. monococcum. The frequent segmental duplication events must contribute to the rapid increase of repetitive DNA in the wheat genome evolution.
Detailed comparative analyses on the orthologous regions between wheat homoeologous genomes have been conducted only in the HMW-glutenin and the Hardness locus regions (Gu et al. 2004b; Chantret et al. 2005). It appears that gene colinearity is maintained in the HMW-glutenin region, while loss of two genes caused by a deletion event occurred in the Hardness region. However, in the orthologous regions studied here violations of gene colinearity seem more frequent. In addition to the possible rearrangement of Gli-1 and Gli-3 in the genomes, gene duplication (Gene12 and Gene14) and inversion (between Gene29 and Gene30) also caused disruption of microcolinearity in the compared region. Moreover, seven genes appear to be specific for the A genome of T. durum wheat since they are not present in the B genome of T. durum (Fig. 2) or Am genome of T. monococcum (Wicker et al. 2003). The significant violation of gene colinearity in the region could be explained by the notion that different genomic regions are subjected to different rate of sequence arrangements. In wheat, it has been demonstrated that the rate of evolution in genomic regions is correlated to the recombination rate along the chromosomes (Akhunov et al. 2003). The distal regions with higher recombination rates tend to have more sequence arrangements. However, recent studies indicate that almost 20% of predicted genes have moved to different locations from their original genomic locations in maize (Bruggmann et al. 2006). One of the well-characterized mechanisms for gene movement is mediated by Mutator-like transposons such as PACK-MULEs in rice (Jiang et al. 2004) and Helitrons in maize (Lai et al. 2005; Morgante et al. 2005; Morgante 2006). Nevertheless, because of the rapid sequence rearrangements, differential amplification/deletion of transposable elements, and their active roles in shuffling genomic sequences, we might expect that many gene islands, like those in the compared regions (Fig. 2), are not conserved between the homoeologous wheat genomes.
The homoeologous wheat genomes, which diverged from a common ancestor only 3–4 million years ago (Huang et al. 2002), are not as conserved as previously thought owing largely to the differential insertion of transposable elements (Gu et al. 2004b; Chantret et al. 2005). The result from the present study further supports the rapid sequence changes after the split of wheat genomes. The knowledge on the great sequence divergence could have practical usefulness in designing effective strategies to tackle the wheat’s large and complex genome. Because of the limited sequence conservation between homoeologous wheat genomes, it might be possible to conduct global fingerprinting of polyploid wheat genome for constructing separating physical maps for individual subgenomes, although such strategy needs experimental validation by detailed analyses on multiple orthologous regions from the wheat genomes.
References
Akhunov ED, Goodyear AW, Geng S, Qi LL, Echalier B, Gill BS, Miftahudin, Gustafson JP, Lazo G, Chao S et al (2003) The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res 13:753–763
Anderson OD, Litts JC, Gautier MF, Greene FC (1984) Nucleic acid sequence and chromosome assignment of a wheat storage protein gene. Nucleic Acids Res 12:8129–8144
Anderson OD, Litts JC, Greene FC (1997) The α-gliadin gene family. I. Characterization of ten new wheat α-gliadin genomic clones, evidence for limited sequence conservation of flanking DNA, and Southern analysis of the gene family. Theor Appl Genet 95:50–58
Anderson OD, Hsia CC, Adalsteins AE, Lew EJ-L, Kasarda DD (2001) Identification of several new class of low-molecular-weight wheat gliadin-related proteins and genes. Theor Appl Genet 103:307–315
Baumgarten A, Cannon S, Spangler R, May G (2003) Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165:309–319
Bruggmann R, Bharti AK, Gundlach H, Lai J, Young S, Pontaroli AC, Wei F, Haberer G, Fuks G, Du C et al (2006) Uneven chromosome contraction and expansion in the maize genome. Genome Res 16:1241–1251
Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17:343–360
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10–31
Cassidy BG, Dvorak J, Anderson OD (1998) The wheat low-molecular-weight glutenin genes: characterization of six new genes and progress in understanding gene family structure. Theor Appl Genet 96:743–750
Cenci A, Chantret N, Kong X, Gu Y, Anderson OD, Fahima T, Distelfeld A, Dubcovsky J (2003) Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp. durum). Theor Appl Genet 107:931–939
Chantret N, Salse J, Sabot F, Rahman S, Bellec A, Laubin B, Dubois I, Dossat C, Sourdille P, Joudrier P et al (2005) Molecular basis of evolutionary events that shaped the Hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17:1033–1045
D’Ovidio R, Masci S (2004) The low-molecular-weight glutenin subuntis of wheat gluten. J Cereal Sci 39:321–339
Devos KM, Ma J, Pontaroli AC, Pratt LH, Bennetzen JL (2005) Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc Natl Acad Sci USA 102:19243–19248
Dubcovsky J, Echaide M, Giancola S, Rousset M, Luo M, Joppa LR, Dvorak J (1997) Seed-storage-protein loci in RFLP maps of diploid, tetraploid, and hexaploid wheat. Theor Appl Genet 95:1169–1180
Gale KR, Ma W, Zhang W, Johal J, Butow BJ (2003) Simple DNA markers for genes influencing wheat quality. In: Proceedings of the tenth international wheat genetics symposium, vol 1, pp 435–438
Garcia-Olmedo F, Carbonero P, Jone BL (1982) Chromosomal locations of genes that control wheat endsperm proteins. Adv Cereal Sci Technol 5:1–47
Gu YQ, Crossman C, Kong X, Luo M, You FM, Coleman-Derr D, Dubcovsky J, Anderson OD (2004a) Genomic organization of the complex alpha-gliadin gene loci in wheat. Theor Appl Genet 109:648–657
Gu YQ, Coleman-Derr D, Kong X, Anderson OD (2004b) Rapid genome evolution revealed by comparative sequence analysis of orthologous regions from four triticeae genomes. Plant Physiol 135:459–470
Gu YQ, Salse J, Coleman-Derr D, Dupin A, Crossman C, Lazo GR, Huo N, Belcram H, Ravel C, Charmet G, Charles M, Anderson OD, Chalhoub B (2006) Types and rates of sequence evolution at the high-molecular-weight glutenin locus in hexaploid wheat and its ancestral genomes. Genetics 174:1493–1504
Hsia CC, Anderson OD (2001) Isolation and characterization of wheat omega-gliadin genes. Theor Appl Genet 103:37–44
Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA 99:8133–8138
Ikeda TM, Araki E, Fujita Y, Yano H (2006) Characterization of low-molecular-weight glutenin subunit genes and their protein products in common wheats. Theor Appl Genet 112:327–334
Isidore E, Scherrer B, Chalhoub B, Feuillet C, Keller B (2005) Ancient haplotypes resulting from extensive molecular rearrangements in the wheat A genome have been maintained in species of three different ploidy levels. Genome Res 15:526–536
Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431:569–573
Johal J, Gianibelli MC, Rahman S, Morell MK, Gale KR (2004) Characterization of low-molecular-weight glutenin genes in Aegilops tauschii. Theor Appl Genet 109:1028–1040
Joppa LR, Williams ND (1988) Landgon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30:222–228
Kong XY, Gu YQ, You FM, Dubcovsky J, Anderson OD (2004) Dynamics of the evolution of orthologous and paralogous portions of a complex locus region in two genomes of allopolyploid wheat. Plant Mol Biol 54:55–69
Lai J, Li Y, Messing J, Dooner HK (2005) Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA 102:9068–9073
Lawton-Rauh A (2003) Evolutionary dynamics of duplicated genes in plants. Mol Phylogenet Evol 29:396–409
Li W, Zhang P, Fellers JP, Friebe B, Gill BS (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40:500–511
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Metakovsky EV, Branlard G, Chernakov VM, Upelniek VP, Redaelli R, Pogna NE (1997) Recombination mapping of some chromosome 1A-, 1B-, 1D-, and 6B-controlled gliadins and low-molecular-weight glutenin subunits in common wheat. Theor Appl Genet 94:788–795
Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8:122–128
Morgante M (2006) Plant genome organization and diversity: the year of the junk! Curr Opin Biotechnol 17:168–173
Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A (2005) Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet 37:997–1002
Nagy IJ, Takacs I, Juhasz A, Tamas L, Bedo Z (2005) Identification of a new class of recombinant prolamin genes in wheat. Genome 48:840–847
Ozdemir N, Cloutier S (2005) Expression analysis and physical mapping of low-molecular-weight glutenin loci in hexaploid wheat (Triticum aestivum L.). Genome 48:401–410
Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48:463–474
Payne PI, Holt LM, Worland AG, Law CN (1982) Structural and genetical studies on the high-molecular-weight subunits of wheat glutenin. Part 3: telocentric mapping of the subunit genes on the long arms of the homoeologous group 1 chromosomes. Theor Appl Genet 63:129–138
Rijpkema AS, Gerats T, Vandenbussche M (2007) Evolutionary complexity of MADS complexes. Curr Opin Plant Biol 10:32–38
Sabelli P, Shewry PR (1991) Characterization and organization of gene families at the Gli-1 loci of bread and durum wheat by restriction fragment analysis. Theor Appl Genet 83:209–216
SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20:43–45
SanMiguel PJ, Ramakrishna W, Bennetzen JL, Busso CS, Dubcovsky J (2002) Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5Am. Funct Integr Genomics 2:70–80
Shewry PR, Halford NG (2002) Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot 53:947–958
Shewry PR, Tatham AS (1990) The prolamin storage proteins of cereal seeds: structure and evolution. Biochem J 267:1–12
Singh NK, Shepherd KW (1988) Linkage mapping of genes controlling endosperm storage proteins in wheat. 1. Genes on the short arm of group 1 chromosomes. Theor Appl Genet 75:628–641
Srichumpa P, Brunner S, Keller B, Yahiaoui N (2005) Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 139:885–895
Wicker T, Yahiaoui N, Guyot R, Schlagenhauf E, Liu ZD, Dubcovsky J, Keller B (2003) Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15:1186–1197
Yahiaoui N, Brunner S, Keller B (2006) Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J 47:85–98
Yuan Q, Hill J, Hsiao J, Moffat K, Ouyang S, Cheng Z, Jiang J, Buell CR (2002) Genome sequencing of a 239-kb region of rice chromosome 10L reveals a high frequency of gene duplication and a large chloroplast DNA insertion. Mol Genet Genomics 6:713–720
Zhang W, Gianibelli MC, Rampling LR, Gale KR (2004) Characterisation and marker development for low molecular weight glutenin genes from Glu-A3 alleles of bread wheat (Triticum aestivum L.). Theor Appl Genet 108:1409–1419
Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng X-W, He Z-H, Lemaux PG (2005) Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol 139:1107–1124
Acknowledgements
We thank Mingcheng Luo for the assistance in BAC fingerprinting and Gerald Lazo for bioinformatics support. This work was partially supported by grants from National Basic Research Program of China (2002CB111301) and National Natural Science Foundation of China (30571158). Work at WRRC is supported by US, Department of Agriculture-Agriculture Research Service Grant CRIS 5325022100-011.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Shuangcheng Gao and Yong Qiang Gu contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2007_9208_MOESM1_ESM.pdf
Nested insertion of retroelements and subsequent deletion in the intergenic region between two LMW-glutenin genes. Two LMW-glutenin genes are represented by black boxes. The start position of each LMW-glutenin gene in the contiguous 265-kb region is provided. Retroelements were annotated based on BLAST searches against the TREP database (http://wheat.pw.usda.gov/ITMI/Repeats/). A repetitive DNA with “s” stands for a solo retroelement and “p” for a partial retroelement. Deletion(s) has occurred in the region. The putative deleted portion from each retroelement is indicated with a blue region. The exact sequencelength deleted is difficult to determine. However, It can be expected that without the deletion event, the two LMW-glutenin genes could be separated by a much larger distance. Insertion of Glabra2-like gene into a Fatimah retroelement is indicated. (PDF 19 kb)
Rights and permissions
About this article
Cite this article
Gao, S., Gu, Y.Q., Wu, J. et al. Rapid evolution and complex structural organization in genomic regions harboring multiple prolamin genes in the polyploid wheat genome. Plant Mol Biol 65, 189–203 (2007). https://doi.org/10.1007/s11103-007-9208-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9208-1